BaZFP1, a C2H2 Subfamily Gene in Desiccation-Tolerant Moss Bryum argenteum, Positively Regulates Growth and Development in Arabidopsis and Mosses
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis and Classification of BaZFP Proteins
2.2. Identification and Physicochemical Properties of BaZFPs
2.3. Conserved Motif Composition Analysis of C2H2 Proteins
2.4. Bioinformatics Analyses of the BaZFP1 Gene and Its Encoded Protein
2.5. Transactivation Activity and Subcellular Localization Analysis of BaZFP1
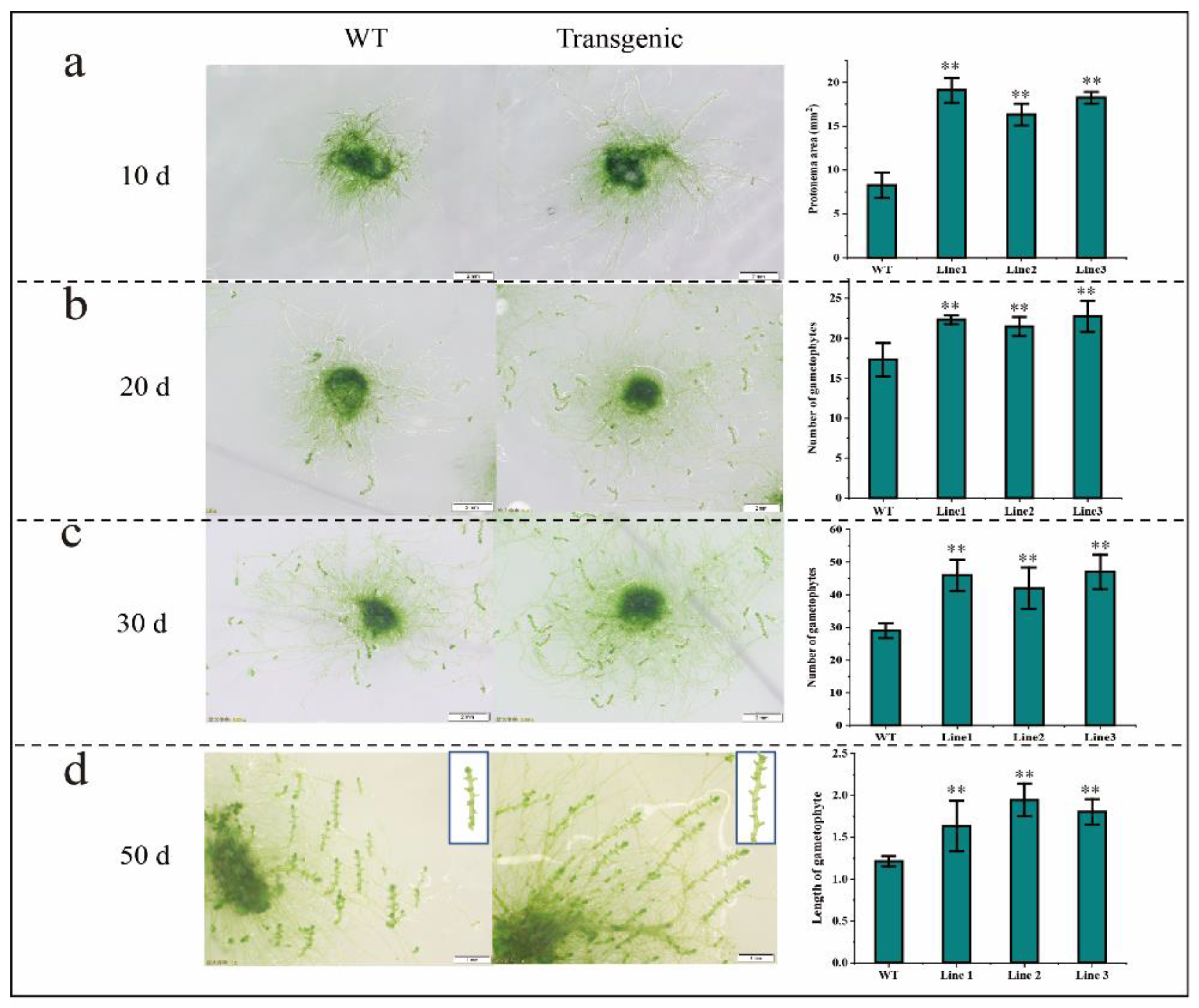
2.6. BaZFP1 Overexpression Promotes Growth and Gametophyte Differentiation in B. argenteum and P. patens
2.7. BaZFP1 Overexpression Regulates Arabidopsis Growth and Development
2.8. Quantitative Analysis of Growth- or Development-Related Gene Expression Profiling
3. Discussion
3.1. Conserved Motif Composition and Classification of BaZFPs
3.2. Overexpression of BaZFP1 Regulates the Growth and Development of B. argenteum and P. patens
3.3. Heterologous Expression of BaZFP1 Regulates the Growth and Reproduction Process in Arabidopsis
4. Methods and Materials
4.1. DNA/Protein Sequence and Phylogenetic Analyses
4.2. Vector Construction and Plant Transformation
4.3. Transactivation Activity in Yeast
4.4. Subcellular Localization
4.5. Plant Materials and Growth Conditions
4.6. QRT-PCR Analysis to Analyze the Expression Pattern of Growth- or Development-Related Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, W.; Zhu, W.; Yang, L.; Liang, W.; Li, H.; Chen, M.; Luo, Z.; Huang, G.; Duan, L.; Dreni, L.; et al. Small reproductive organs, a superman-like transcription factor, regulates stamen and pistil growth in rice. New Phytol. 2021, 233, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-R.; Wang, X.-C.; Li, X.-M.; Yang, C.-D. Transcription factors in higher plant. Hereditas 2004, 26, 403–408. [Google Scholar]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Takatsuji, H. Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 1999, 39, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Böhm, S.; Frishman, D.; Mewes, H.-W. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997, 25, 2464–2469. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.R.; Schäfer, U.; Jäckle, H.; Böhm, S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. Embo Rep. 2002, 3, 1158–1162. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wang, J.; Sun, Q.; Li, W.; Yu, Y.; Zhao, M.; Meng, Z. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct. Integr. Genom. 2017, 17, 653–666. [Google Scholar] [CrossRef]
- Jiao, Z.; Wang, L.; Du, H.; Wang, Y.; Wang, W.; Liu, J.; Huang, J.; Huang, W.; Ge, L. Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula. BMC Plant Biol. 2020, 20, 1–17. [Google Scholar] [CrossRef]
- Iuchi, S. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 2001, 58, 625–635. [Google Scholar] [CrossRef]
- Pabo, C.O.; Peisach, E.; Grant, R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001, 70, 313–340. [Google Scholar] [CrossRef]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-Wide Analysis of C2H2 Zinc-Finger Family Transcription Factors and Their Responses to Abiotic Stresses in Poplar (Populus trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsuji, H.; Mori, M.; Benfey, P.; Ren, L.; Chua, N. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 1992, 11, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Joseph, M.P.; Papdi, C.; Kozma-Bognár, L.; Nagy, I.; López-Carbonell, M.; Rigó, G.; Koncz, C.; Szabados, L. The Arabidopsis zinc finger protein3 Interferes with Abscisic Acid and Light Signaling in Seed Germination and Plant Development. Plant Physiol. 2014, 165, 1203–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chrispeels, H.E.; Oettinger, H.; Janvier, N.; Tague, B.W. AtZFP1, encoding Arabidopsis thaliana C2H2 zinc-finger protein 1, is expressed downstream of photomorphogenic activation. Plant Mol. Biol. 2000, 42, 279–290. [Google Scholar] [CrossRef]
- Sakai, H.; Medrano, L.J.; Meyerowitz, E.M. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 1995, 378, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Wagner, D.R. The Arabidopsis SERRATE Gene Encodes a Zinc-Finger Protein Required for Normal Shoot Development. Plant Cell 2001, 13, 1263. [Google Scholar] [CrossRef] [Green Version]
- Dinkins, R.; Pflipsen, C.; Thompson, A.; Collins, G.B. Ectopic Expression of an Arabidopsis Single Zinc Finger Gene in Tobacco Results in Dwarf Plants. Plant Cell Physiol. 2002, 43, 743–750. [Google Scholar] [CrossRef] [Green Version]
- Lyu, T.; Cao, J. Cys2/His2 Zinc-Finger Proteins in Transcriptional Regulation of Flower Development. Int. J. Mol. Sci. 2018, 19, 2589. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, M.; Miyazaki, S.; Kawaide, H. Hormonal Diterpenoids Distinct to Gibberellins Regulate Protonema Differentiation in the Moss Physcomitrium patens. Plant Cell Physiol. 2020, 61, 1861–1868. [Google Scholar] [CrossRef]
- Rensing, S.A.; Goffinet, B.; Meyberg, R.; Wu, S.Z.; Bezanilla, M. The Moss Physcomitrium (Physcomitrella) patens: A Model Organism for Non-Seed Plants. Plant Cell 2020, 32, 1361–1376. [Google Scholar] [CrossRef] [PubMed]
- Reski, R. Development, Genetics and Molecular Biology of Mosses. Bot. Acta 1998, 111, 1–15. [Google Scholar] [CrossRef]
- Cove, D. The moss Physcomitrella patens. Annu. Rev. Genet. 2005, 39, 339–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.S.; Gao, B.; Zhang, D.Y.; Liang, Y.Q.; Liu, X.J.; Zhao, J.Y.; Zhang, J.H.; Wood, A.J. Identification, Classification, and Functional Analysis of AP2/ERF Family Genes in the Desert Moss Bryum argenteum. Int. J. Mol. Sci. 2018, 19, 3637. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, J.; Wang, L.; Wang, X.; Gu, Z. The spatial distribution patterns of biological soil crusts in the Gurbantunggut Desert, Northern Xinjiang, China. J. Arid Environ. 2007, 68, 599–610. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.; Yang, R.; Gao, B.; Yao, J.; Oliver, M.J.; Zhang, D. BaDBL1, a unique DREB gene from desiccation tolerant moss Bryum argenteum, confers osmotic and salt stress tolerances in transgenic Arabidopsis. Plant Sci. 2021, 313, 111047. [Google Scholar] [CrossRef]
- Gao, B.; Li, X.; Zhang, D.; Liang, Y.; Yang, H.; Chen, M.; Zhang, Y.; Zhang, J.; Wood, A.J. Desiccation tolerance in bryophytes: The dehydration and rehydration transcriptomes in the desiccation-tolerant bryophyte Bryum argenteum. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Acereto-Escoffié, P.; Chi-Manzanero, B.; Echeverría-Echeverría, S.; Grijalva, R.; Kay, A.J.; González-Estrada, T.; Castaño, E.; Rodríguez-Zapata, L. Agrobacterium-mediated transformation of Musa acuminata cv. “Grand Nain” scalps by vacuum infiltration. Sci. Hortic. 2005, 105, 359–371. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Hu, R.; Hua, C.; Ali, I.; Zhang, A.; Liu, B.; Wu, M.; Huang, L.; Gan, Y. AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem. Biophys. Res. Commun. 2017, 483, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, J.; Zhang, H. Rice ZFP15 Gene Encoding for a Novel C2H2-type Zinc Finger Protein Lacking DLN box, is Regulated by Spike Development but not by Abiotic Stresses. Mol. Biol. Rep. 2005, 32, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Huang, P.; Zhang, L.; Shi, Y.; Sun, D.; Yan, Y.; Liu, X.; Dong, B.; Chen, G.; Snyder, J.H.; et al. Characterization of 47 Cys(2)-His(2) zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 2016, 211, 1035–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, K.; Sakamoto, A.; Kobayashi, A.; Rybka, Z.; Kanno, Y.; Nakagawa, H.; Nishino, T.; Takatsuji, H. Cys(2)/His(2) zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 1998, 26, 608–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourcilleau, D.; Lenne, C.; Armenise, C.; Moulia, B.; Julien, J.-L.; Bronner, G.; Leblanc-Fournier, N. Phylogenetic Study of Plant Q-type C2H2 Zinc Finger Proteins and Expression Analysis of Poplar Genes in Response to Osmotic, Cold and Mechanical Stresses. DNA Res. 2011, 18, 77–92. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Liang, W.; Gu, P.; Huang, Z. Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol. Biochem. 2016, 106, 129–140. [Google Scholar] [CrossRef]
- Shi, X.; Wu, Y.; Dai, T.; Gu, Y.; Wang, L.; Qin, X.; Xu, Y.; Chen, F. JcZFP8, a C2H2 zinc finger protein gene from Jatropha curcas, influences plant development in transgenic tobacco. Electron. J. Biotechnol. 2018, 34, 76–82. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Aoyama, T.; Hiwatashi, Y.; Shigyo, M.; Kofuji, R.; Kubo, M.; Ito, M.; Hasebe, M. AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development 2012, 139, 3120–3129. [Google Scholar] [CrossRef] [Green Version]
- Brejšková, L.; Hála, M.; Rawat, A.; Soukupová, H.; Cvrčková, F.; Charlot, F.; Nogué, F.; Haluška, S.; Žárský, V. SEC6 exocyst subunit contributes to multiple steps of growth and development of Physcomitrella (Physcomitrium patens). Plant J. 2021, 106, 831–843. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional Control of Gene Expression by MicroRNAs. Cell 2010, 140, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.A.; Fattash, I.; Ma, Z.; Cho, S.H.; Beike, A.K.; Reski, R.; Axtell, M.J.; Frank, W. DICER-LIKE3 Activity in Physcomitrella patens DICER-LIKE4 Mutants Causes Severe Developmental Dysfunction and Sterility. Mol. Plant 2012, 5, 1281–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially Redundant Functions of Arabidopsis DICER-like Enzymes and a Role for DCL4 in Producing trans-Acting siRNAs. Curr. Biol. 2005, 15, 1494–1500. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Guo, H. On hormonal regulation of the dynamic apical hook development. New Phytol. 2018, 222, 1230–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Zhang, D.; Qiu, Y.; Xiao, Z.; Ji, Y.; Li, W.; Xia, Y.; Wang, Y.; Guo, H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J. Integr. Plant Biol. 2021, 64, 5–22. [Google Scholar] [CrossRef]
- Zhang, X.; Ji, Y.; Xue, C.; Ma, H.; Xi, Y.; Huang, P.; Wang, H.; An, F.; Li, B.; Wang, Y.; et al. Integrated Regulation of Apical Hook Development by Transcriptional Coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 2018, 30, 1971–1988. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R.; Chen, H.; Deng, X.W.; Wei, N. SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis. Plant Cell 2020, 32, 3792–3811. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Dill, A.; Sun, T.-P. Synergistic Derepression of Gibberellin Signaling by Removing RGA and GAI Function in Arabidopsis thaliana. Genetics 2001, 159, 777–785. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Verkest, A.; Gonzalez, N.; Heyndrickx, K.S.; Eeckhout, D.; Han, S.-K.; Jégu, T.; Archacki, R.; Van Leene, J.; Andriankaja, M.; et al. ANGUSTIFOLIA3 Binds to SWI/SNF Chromatin Remodeling Complexes to Regulate Transcription during Arabidopsis Leaf Development. Plant Cell 2014, 26, 210–229. [Google Scholar] [CrossRef] [Green Version]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Nole-Wilson, S.; Tranby, T.L.; Krizek, B.A. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 2005, 57, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.J.; Yu, Y.; Liu, X.; Zhang, X.S.; Su, Y.H. The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell 2021, 33, 1907–1926. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation Tagging of the Floral Inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [Green Version]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Lister, C.; Dean, C. Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 2021, 599, 657–661. [Google Scholar] [CrossRef]
- Schultz, E.A.; Haughn, G.W. LEAFY, a Homeotic Gene That Regulates Inflorescence Development in Arabidopsis. Plant Cell 1991, 3, 771–781. [Google Scholar] [CrossRef] [Green Version]
- Siriwardana, N.S.; Lamb, R.S. A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana. Plant J. 2012, 71, 736–749. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.-F.; Sun, Y.; Zhu, R.-L. In vitro micropropagation of Bryum argenteum Hedw. Cryptogam. Bryol. 2010, 31, 233–239. [Google Scholar]
- Gao, B.; Zhang, D.; Li, X.; Yang, H.; Zhang, Y.; Wood, A.J. De novo transcriptome characterization and gene expression profiling of the desiccation tolerant moss Bryum argenteum following rehydration. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ashton, N.W.; Cove, D.J. Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss, physcomitrella-patens. Mol. Gen. Genet. 1977, 154, 87–95. [Google Scholar] [CrossRef]
- Linsmaier, E.M.; Skoog, F. Organic Growth Factor Requirements of Tobacco Tissue Cultures. Physiol. Plant. 1965, 18, 100–127. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, P.; Liu, X.; Li, X.; Zhang, D. BaZFP1, a C2H2 Subfamily Gene in Desiccation-Tolerant Moss Bryum argenteum, Positively Regulates Growth and Development in Arabidopsis and Mosses. Int. J. Mol. Sci. 2022, 23, 12894. https://doi.org/10.3390/ijms232112894
Zhou P, Liu X, Li X, Zhang D. BaZFP1, a C2H2 Subfamily Gene in Desiccation-Tolerant Moss Bryum argenteum, Positively Regulates Growth and Development in Arabidopsis and Mosses. International Journal of Molecular Sciences. 2022; 23(21):12894. https://doi.org/10.3390/ijms232112894
Chicago/Turabian StyleZhou, Ping, Xiujin Liu, Xiaoshuang Li, and Daoyuan Zhang. 2022. "BaZFP1, a C2H2 Subfamily Gene in Desiccation-Tolerant Moss Bryum argenteum, Positively Regulates Growth and Development in Arabidopsis and Mosses" International Journal of Molecular Sciences 23, no. 21: 12894. https://doi.org/10.3390/ijms232112894
APA StyleZhou, P., Liu, X., Li, X., & Zhang, D. (2022). BaZFP1, a C2H2 Subfamily Gene in Desiccation-Tolerant Moss Bryum argenteum, Positively Regulates Growth and Development in Arabidopsis and Mosses. International Journal of Molecular Sciences, 23(21), 12894. https://doi.org/10.3390/ijms232112894






