Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants
Abstract
1. Introduction
2. Transcription of Drought-Responsive Genes
2.1. Drought-Responsive Genes
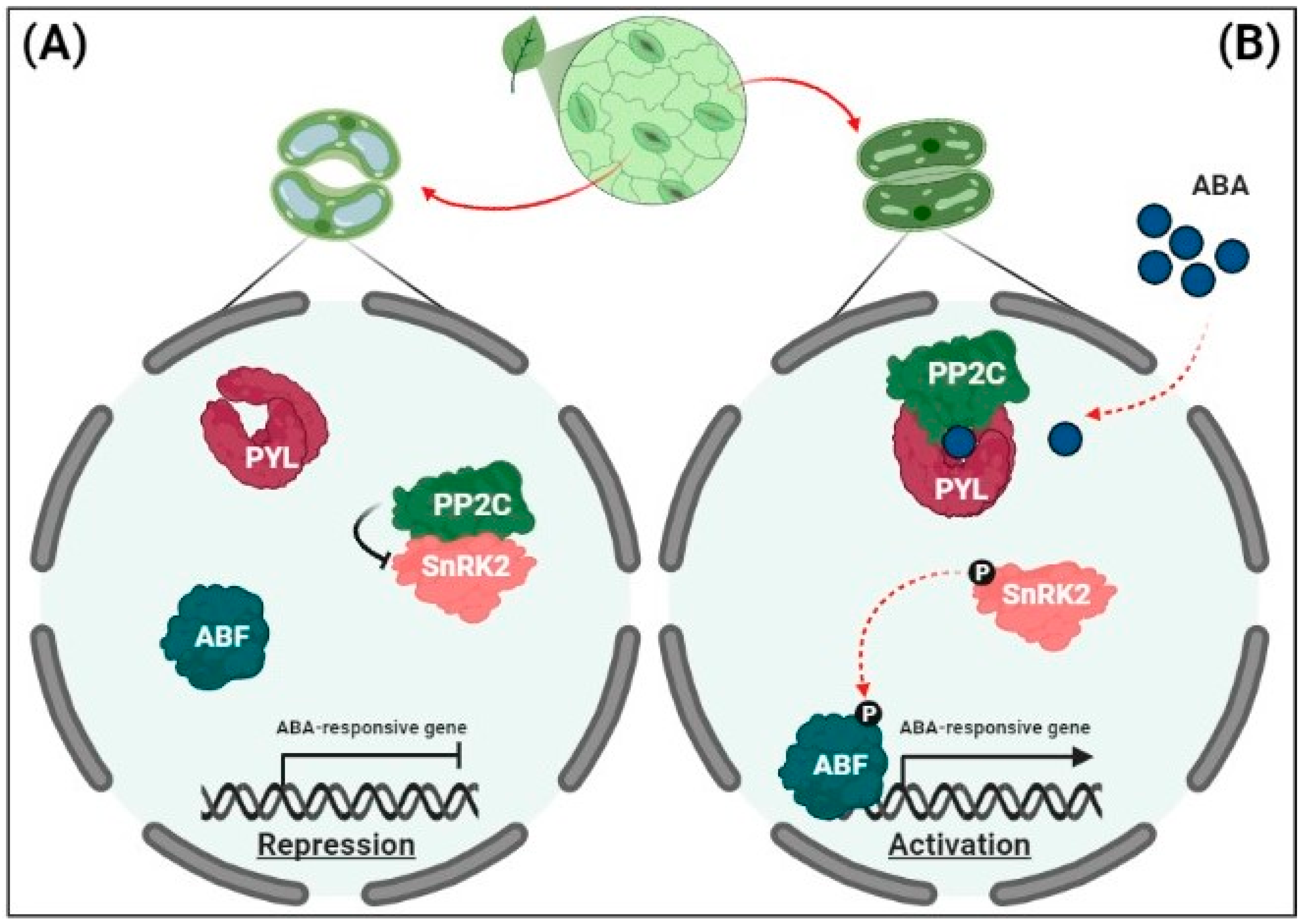
2.2. Regulation of ABA Signaling
3. Chromatin Modifications for Drought-Responsive Gene Transcription
3.1. Drought Stress Tolerance and DNA Methylation
3.2. Chromatin Remodeling Implicated in Drought-Responsive Gene Expression
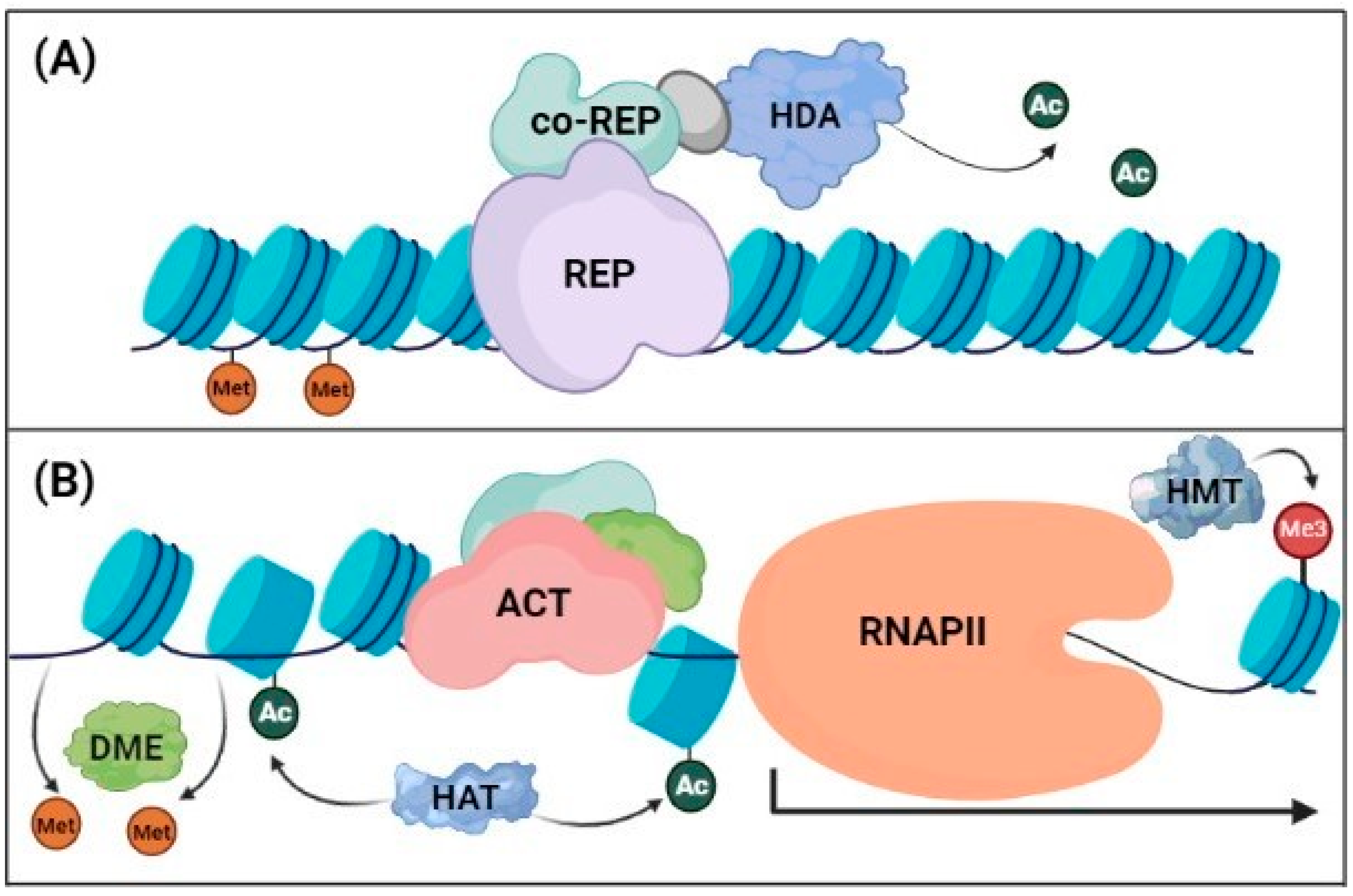
3.2.1. Histone Modifications
3.2.2. Chromatin Remodeling Complexes
3.3. Non-Coding RNAs
3.4. Chromatin Loop Formation
4. Transcriptional Memory of Drought Tolerance
4.1. Drought Stress Memory
4.2. Drought Stress Memory Genes
4.3. Mechanism of Transcriptional Stress Memory
4.3.1. Epigenetic Marks for Stress Memory
4.3.2. Memory Transmission by Mitosis
5. Transgenerational Inheritance of Memory
5.1. Transgenerational Transmission of Drought Tolerance
5.2. DNA Methylation for Transgenerational Inheritance
5.3. Overcoming Meiosis
6. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 1996, 7, 161–167. [Google Scholar] [CrossRef]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genom. 2014, 14, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic acid-induced stomatal closure: An important component of plant defense against abiotic and biotic stress. Front. Plant Sci. 2021, 12, 615114. [Google Scholar] [CrossRef]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef]
- Hoth, S.; Morgante, M.; Sanchez, J.-P.; Hanafey, M.K.; Tingey, S.V.; Chua, N.H. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 2002, 115, 4891–4900. [Google Scholar] [CrossRef]
- Cairns, B.R. The logic of chromatin architecture and remodeling at promoters. Nature 2009, 461, 193–198. [Google Scholar] [CrossRef]
- Yamamuro, C.; Zhu, J.-K.; Yang, Z. Epigenetic modifications and plant hormone action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef]
- Kim, J.-H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, F.; Zhou, B. The characters of non-coding RNAs and their biological roles in plant development and abiotic stress response. Int. J. Mol. Sci. 2022, 23, 4124. [Google Scholar] [CrossRef] [PubMed]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Godwin, J.; Farrona, S. Plant epigenetic stress memory induced by drought: A physiological and molecular perspective. Methods Mol. Biol. 2020, 2093, 243–259. [Google Scholar] [PubMed]
- Choudhary, M.; Singh, A.; Rakshit, S. Coping with low moisture stress: Remembering and responding. Physiol. Plant. 2021, 172, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef]
- Marthandan, V.; Geetha, R.; Kumutha, K.; Renganathan, V.G.; Karthikeyan, A.; Ramalingam, J. Seed priming: A feasible strategy to enhance drought tolerance in crop plants. Int. J. Mol. Sci. 2020, 21, 8258. [Google Scholar] [CrossRef]
- Luo, M.; Liu, X.; Singh, P.; Cui, Y.; Zimmerli, L.; Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 129–136. [Google Scholar] [CrossRef]
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 8, 114. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Kumar, P.; Verma, V.; Sharma, R.; Bhargava, B.; Irfan, M. Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol. Biochem. 2022, 179, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Naumova, N.; Imakaev, M.; Fudenberg, G.; Zhan, Y.; Lajoie, B.R.; Mirny, L.A.; Dekker, J. Organization of the mitotic chromosome. Science 2013, 342, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Higgins, J.M. Histone modifications and mitosis: Countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013, 23, 175–184. [Google Scholar] [CrossRef]
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049. [Google Scholar] [CrossRef]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef]
- Springer, N.M. Epigenetics and crop improvement. Trends Genet. 2013, 29, 241–247. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef]
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092. [Google Scholar] [CrossRef]
- Yao, T.; Zhang, J.; Xie, M.; Yuan, G.; Tschaplinski, T.J.; Muchero, W.; Chen, J.-G. Transcriptional regulation of drought response in Arabidopsis and woody plants. Front. Plant Sci. 2021, 11, 572137. [Google Scholar] [CrossRef] [PubMed]
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Majee, M.; Datta, A. Chromatin-based transcriptional reprogramming in plants under abiotic stresses. Plants 2022, 11, 1449. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of epigenetics for improvement of drought and other stress resistance in crops: A review. Plants 2021, 10, 1226. [Google Scholar] [CrossRef]
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ABA-responsive gene expression in response to drought stress: Cellular regulation and long-distance signaling. Adv. Bot. Res. 2019, 92, 83–113. [Google Scholar]
- Cheong, J.-J. Modulation of abscisic acid signaling for stomatal operation under salt stress conditions. Adv. Bot. Res. 2022, 103, 89–121. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Cutler, A.; Krochko, J. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Merilo, E.; Jalakas, P.; Laanemets, K.; Mohammadi, O.; Hõrak, H.; Kollist, H.; Brosché, M. Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol. Plant 2015, 8, 1321–1333. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA transport and plant water stress responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Marion-Poll, A. Abscisic acid metabolism and transport. Adv. Bot. Res. 2019, 92, 1–49. [Google Scholar]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Nguyen, N.H.; Cheong, J.-J. Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. Int. J. Mol. Sci. 2020, 21, 9517. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signaling. Essays Biochem. 2015, 58, 29–48. [Google Scholar]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Furner, I.J.; Matzke, M. Methylation and demethylation of the Arabidopsis genome. Curr. Opin. Plant Biol. 2011, 14, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In response to abiotic stress, DNA methylation confers epigenetic changes in plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef]
- Li, X.; Zhu, J.; Hu, F.; Ge, S.; Ye, M.; Xiang, H.; Zhang, G.; Zheng, X.; Zhang, H.; Zhang, S.; et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genom. 2012, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhou, S.; Gong, X.; Song, Y.; Van Nocker, S.; Ma, F.; Guan, Q. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol. J. 2018, 16, 672–687. [Google Scholar] [CrossRef]
- Van Dooren, T.J.M.; Silveira, A.B.; Gilbault, E.; Jiménez-Gómez, J.M.; Martin, A.; Bach, L.; Tisné, S.; Quadrana, L.; Loudet, O.; Colot, V. Mild drought in the vegetative stage induces phenotypic, gene expression and DNA methylation plasticity in Arabidopsis but no transgenerational effects. J. Exp. Bot. 2020, 71, 3588–3602. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Satish, L.; Kalendar, R.; Narayanan, M.; Kandasamy, S.; Sharma, A.; Emamverdian, A.; Wei, Q.; Zhou, M. The dynamism of transposon methylation for plant development and stress adaptation. Int. J. Mol. Sci. 2021, 22, 11387. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-S.; Pan, Y.-J.; Zhao, X.-Q.; Dwivedi, D.; Zhu, L.-H.; Ali, J.; Fu, B.-Y.; Li, Z.-K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef]
- Wang, W.; Qin, Q.; Sun, F.; Wang, Y.; Xu, D.; Li, Z.; Fu, B. Genome-wide differences in DNA methylation changes in two contrasting rice genotypes in response to drought conditions. Front. Plant Sci. 2016, 7, 1675. [Google Scholar] [CrossRef]
- Gayacharan; Joel, A.J. Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2013, 19, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Li, M.; Lou, Q.; Xia, H.; Wang, P.; Li, T.; Liu, H.; Luo, L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 2013, 8, e80253. [Google Scholar] [CrossRef]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 2016, 5, e13546. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Ricardi, M.M.; Iusem, N.D. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 2013, 8, 864–872. [Google Scholar] [CrossRef]
- Liang, D.; Zhang, Z.; Wu, H.; Huang, C.; Shuai, P.; Ye, C.-Y.; Tang, S.; Wang, Y.; Yang, L.; Wang, J.; et al. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014, 15 (Suppl. S1), S9. [Google Scholar] [CrossRef] [PubMed]
- Chwialkowska, K.; Nowakowska, U.; Mroziewicz, A.; Szarejko, I.; Kwasniewski, M. Water-deficiency conditions differently modulate the methylome of roots and leaves in barley (Hordeum vulgare L.). J. Exp. Bot. 2016, 67, 1109–1121. [Google Scholar] [CrossRef]
- Lu, X.; Wang, X.; Chen, X.; Shu, N.; Wang, J.; Wang, D.; Wang, S.; Fan, W.; Guo, L.; Guo, X.; et al. Single-base resolution methylomes of upland cotton (Gossypium hirsutum L.) reveal epigenome modifications in response to drought stress. BMC Genom. 2017, 18, 297. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef]
- Kim, J.-S.; Lim, J.Y.; Shin, H.; Kim, B.-G.; Yoo, S.-D.; Kim, W.T.; Huh, J.H. ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 2019, 179, 1810–1821. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 2016, 28, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-T.; Luo, M.; Wang, Y.Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, Y.-Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Cheong, J.-J. AtMYB44 interacts with TOPLESS-RELATED corepressors to suppress protein phosphatase 2C gene transcription. Biochem. Biophys. Res. Commun. 2018, 499, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Khan, I.U.; Ali, A.; Khan, H.A.; Baek, D.; Park, J.; Lim, C.J.; Zareen, S.; Jan, M.; Lee, S.Y.; Pardo, J.M.; et al. PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in Arabidopsis. Front. Plant Sci. 2020, 11, 623. [Google Scholar] [CrossRef]
- Struhl, K.; Segal, E. Determinants of nucleosome positioning. Nat. Struc. Mol. Biol. 2013, 20, 267–273. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, W.; Jiang, J. Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiol. 2015, 168, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Li, H.; Wei, Q.; Zhao, X.; Wang, C.; Zhu, Q.; Yi, X.; Xu, W.; Liu, X.S.; Jin, W.; et al. Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica. Mol. Plant 2013, 6, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Jung, C.; Cheong, J.-J. Chromatin remodeling for the transcription of type 2C protein phosphatase genes in response to salt stress. Plant Physiol. Biochem. 2019, 141, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, C.; Peng, J.; Li, C.; Wan, F.; Zhang, S.; Zhou, Y.; Yan, Y.; Qi, L.; Sun, K.; et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef] [PubMed]
- To, T.K.; Kim, J.M. Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 2014, 4, 58. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ueda, M.; Seki, M. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, X.; Gao, Y.; Zhou, C.; Chiang, V.L.; Li, W. Histone acetylation changes in plant response to drought stress. Genes 2021, 12, 1409. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Ishida, J.; Morosawa, T.; Kawashima, M.; Matsui, A.; Toyoda, T.; Kimura, H.; Shinozaki, K.; Seki, M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Avramova, Z.; Fromm, M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 2011, 66, 735–744. [Google Scholar] [CrossRef]
- Widiez, T.; Symeonidi, A.; Luo, C.; Lam, E.; Lawton, M.; Rensing, S.A. The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J. 2014, 79, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.-S.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ichihashi, Y.; Suzuki, T.; Shibata, A.; Shirasu, K.; Yamaguchi, N.; Ito, T. Abscisic acid-dependent histone demethylation during postgermination growth arrest in Arabidopsis. Plant Cell Environ. 2019, 42, 2198–2214. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Shang, J.-Y.; He, X.-J. Chromatin-remodeling complexes: Conserved and plant-specific subunits in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 499–515. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Thouly, C.; Masson, M.L.; Lai, X.; Carles, C.C.; Vachon, G. Unwinding BRAHMA functions in plants. Genes 2020, 11, 90. [Google Scholar] [CrossRef]
- Farrona, S.; Hurtado, L.; Reyes, J.C. A nucleosome interaction module is required for normal function of Arabidopsis thaliana BRAHMA. J. Mol. Biol. 2007, 373, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Archacki, R.; Yatusevich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A.; et al. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucl. Acids Res. 2017, 45, 3116–3129. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gu, L.; Gao, L.; Chen, C.; Wei, C.-Q.; Qiu, Q.; Chien, C.-W.; Wang, S.; Jiang, L.; Ai, L.-F.; et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Han, S.-K.; Kim, H.J.; Wu, M.-F.; Steiner, E.; Birnbaum, K.D.; Hong, J.C.; Eshed, Y.; Wagner, D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 2013, 24, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-K.; Sang, Y.; Rodrigues, A.; BIOL425 F2010; Wu, M.-F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–4906. [Google Scholar] [CrossRef]
- Peirats-Llobet, M.; Han, S.-K.; Gonzalez-Guzman, M.; Jeong, C.W.; Rodriguez, L.; Belda-Palazon, B.; Wagner, D.; Rodriguez, P.L. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant 2016, 9, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, D.; Tian, G.; Sun, L.; Yang, M.; Yin, X.; Xiao, J.; Sheng, Y.; Zhu, D.; He, H.; et al. Chromatin remodeling complexes regulate genome architecture in Arabidopsis. Plant Cell 2022, 34, 2638–2651. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Gelaw, T.A.; Sanan-Mishra, N. Non-coding RNAs in response to drought stress. Int. J. Mol. Sci. 2021, 22, 12519. [Google Scholar] [CrossRef]
- Nosaka, M.; Itoh, J.I.; Nagato, Y.; Ono, A.; Ishiwata, A.; Sato, Y. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 2012, 8, e1002953. [Google Scholar] [CrossRef] [PubMed]
- Cho, J. Transposon-derived non-coding RNAs and their function in plants. Front. Plant Sci. 2018, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Kumar, S.; Sanan-Mishra, N. Osa-miR820 regulatory node primes rice plants to tolerate salt stress in an agronomically advantageous manner. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, Z.; Hong, H.; Chen, Z.; Wu, C.; Li, X.; Xiao, J.; Wang, S. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2016, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Reyes, J.L.; Chua, N.-H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007, 49, 592–606. [Google Scholar] [CrossRef]
- Li, W.-X.; Oono, Y.; Zhu, J.; He, X.-J.; Wu, J.-M.; Iida, K.; Lu, X.-Y.; Cui, X.; Jin, H.; Zhu, J.-K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, X.; Zhou, Y.; Xie, J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol. Lett. 2016, 38, 711–721. [Google Scholar] [CrossRef]
- Chung, P.J.; Jung, H.; Jeong, D.-H.; Ha, S.-H.; Choi, Y.D.; Kim, J.-K. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom. 2016, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, Y.-C.; Wang, C.-Y.; Luo, Y.-C.; Huang, Q.-J.; Chen, S.-Y.; Zhou, H.; Qu, L.-H.; Chen, Y.-Q. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 2009, 583, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants 2020, 9, 1377. [Google Scholar] [CrossRef]
- Yu, Y.; Ni, Z.; Wang, Y.; Wan, H.; Hu, Z.; Jiang, Q.; Sun, X.; Zhang, H. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019, 285, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef]
- Zubko, E.; Meyer, P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J. 2007, 52, 1131–1139. [Google Scholar] [CrossRef]
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.; Poirier, Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 2013, 25, 4166–4182. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Urquiaga, M.C.O.; Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. From trash to luxury: The potential role of plant lncRNA in DNA methylation during abiotic stress. Front. Plant Sci. 2021, 11, 603246. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Dean, A. In the loop: Long range chromatin interactions and gene regulation. Brief Funct. Genom. 2011, 10, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, B.A.; De Laat, W. Getting the genome in shape: The formation of loops, domains and compartments. Genome Biol. 2015, 16, 154. [Google Scholar] [CrossRef]
- Mirkovitch, J.; Mirault, M.E.; Laemmli, U.K. Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 1984, 39, 223–232. [Google Scholar] [CrossRef]
- Rodriguez-Granados, N.Y.; Ramirez-Prado, J.S.; Veluchamy, A.; Latrasse, D.; Raynaud, C.; Crespi, M.; Ariel, F.; Benhamed, M. Put your 3D glasses on: Plant chromatin is on show. J. Exp. Bot. 2016, 67, 3205–3221. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Mapendano, C.K.; Jensen, T.H. An ending is a new beginning: Transcription termination supports re-initiation. Cell Cycle 2011, 10, 863–865. [Google Scholar] [CrossRef][Green Version]
- Tan-Wong, S.M.; Zaugg, J.B.; Camblong, J.; Xu, Z.; Zhang, D.W.; Mischo, H.E.; Ansari, A.Z.; Luscombe, N.M.; Steinmetz, L.M.; Proudfoot, N.J. Gene loops enhance transcriptional directionality. Science 2012, 338, 671–675. [Google Scholar] [CrossRef]
- Gagliardi, D.; Manavella, P.A. Short-range regulatory chromatin loops in plants. New Phytol. 2020, 228, 466–471. [Google Scholar] [CrossRef]
- Tan-Wong, S.M.; Wijayatilake, H.D.; Proudfoot, N.J. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009, 23, 2610–2624. [Google Scholar] [CrossRef] [PubMed]
- Crevillén, P.; Sonmez, C.; Wu, Z.; Dean, C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013, 32, 140–148. [Google Scholar] [CrossRef]
- Jégu, T.; Latrasse, D.; Delarue, M.; Hirt, H.; Domenichini, S.; Ariel, F.; Crespi, M.; Bergounioux, C.; Raynaud, C.; Benhamed, M. The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2014, 26, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Misteli, T. Long-range chromatin interactions. Cold Spring Harb. Perspect. Biol. 2015, 7, a019356. [Google Scholar] [CrossRef]
- Deng, W.; Blobel, G.A. Do chromatin loops provide epigenetic gene expression states? Curr. Opin. Genet. Dev. 2010, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.; Jung, I. Long-range chromatin interactions in pathogenic gene expression control. Transcription 2020, 11, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Cao, X.; Liu, Y.; Li, J.; Li, Y.; Li, D.; Zhang, K.; Gao, C.; Dong, A.; Liu, X. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 2018, 94, 1083–1097. [Google Scholar] [CrossRef]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2022, 27, 699–716. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Prasad, S.S.; Mitra, J.; Siddiqui, N.; Sahoo, L.; Kobayashi, Y.; Koyama, H. How do plants remember drought? Planta 2022, 256, 7. [Google Scholar] [CrossRef]
- Walter, J.; Nagy, L.; Hein, R.; Rascher, U.; Beierkuhnlein, C.; Willner, E.; Jentsch, A. Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 2011, 71, 34–40. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Vignjevic, M.; Jiang, D.; Jacobsen, S.; Wollenweber, B. Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 2014, 65, 6441–6456. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32. [Google Scholar] [CrossRef]
- Abdallah, M.B.; Methenni, K.; Nouairi, I.; Zarrouk, M.; Youssef, N.B. Drought priming improves subsequent more severe drought in a drought-sensitive cultivar of olive cv. Chétoui. Sci. Hortic. 2017, 221, 43–52. [Google Scholar] [CrossRef]
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A.; Shahid, M. Terminal drought and seed priming improves drought tolerance in wheat. Physiol. Mol. Biol. Plants 2018, 24, 845–856. [Google Scholar] [CrossRef]
- Goh, C.-H.; Nam, H.G.; Park, Y.S. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003, 36, 240–255. [Google Scholar] [CrossRef]
- Virlouvet, L.; Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 2015, 205, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Chae, S.; Oh, N.-I.; Nguyen, N.H.; Cheong, J.-J. Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front. Genet. 2020, 11, 576086. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Brzezinka, K.; Altmann, S.; Czesnick, H.; Nicolas, P.; Gorka, M.; Benke, E.; Kabelitz, T.; Jähne, F.; Graf, A.; Kappel, C.; et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 2016, 5, e17061. [Google Scholar] [CrossRef]
- Kou, S.Y.; Gu, Q.Y.; Duan, L.; Liu, G.J.; Yuan, P.R.; Li, H.H.; Wu, Z.G.; Liu, W.H.; Huang, P.; Liu, L. Genome-wide bisulphite sequencing uncovered the contribution of DNA methylation to rice short-term drought memory formation. J. Plant Growth Regul. 2021, 1–15. [Google Scholar] [CrossRef]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833. [Google Scholar] [CrossRef]
- Baile, F.; Gómez-Zambrano, Á.; Calonje, M. Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 2022, 3, 100267. [Google Scholar] [CrossRef] [PubMed]
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shen, W.-H. Chromatin modulation and gene regulation in plants: Insight about PRC1 function. Biochem. Soc. Trans. 2018, 46, 957–966. [Google Scholar] [CrossRef]
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb repressive complex 2 in eukaryotes-An evolutionary perspective. Epigenomes 2022, 6, 3. [Google Scholar] [CrossRef]
- Mozgova, I.; Hennig, L. The Polycomb group protein regulatory network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewski, T.; Xiao, J.; Yang, Y.; Wagner, D. PRC2 activity, recruitment, and silencing: A comparative perspective. Trends Plant Sci. 2021, 26, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.; Farrona, S. The importance of networking: Plant Polycomb Repressive Complex 2 and its interactors. Epigenomes 2022, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Mylne, J.S.; Barrett, L.; Tessadori, F.; Mesnage, S.; Johnson, L.; Bernatavichute, Y.V.; Jacobsen, S.E.; Fransz, P.; Dean, C. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. USA 2006, 103, 5012–5017. [Google Scholar] [CrossRef]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-binding factor basic pentacysteine6 recruits the polycomb-repressive complex1 component like heterochromatin protein1 to GAGA DNA motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ding, Y.; Fromm, M.; Avramova, Z. Different gene-specific mechanisms determine the ‘revised-response’ memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res. 2014, 42, 5556–5566. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Fromm, M.; Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant 2014, 7, 502–513. [Google Scholar] [CrossRef]
- Kim, J.-M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, aad9780. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.-M.; Croce, L.D.; Cavalli, G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Fletcher, J.C. State of the art: TrxG factor regulation of post-embryonic plant development. Front. Plant Sci. 2017, 8, 1925. [Google Scholar] [CrossRef] [PubMed]
- Grini, P.E.; Thorstensen, T.; Alm, V.; Vizcay-Barrena, G.; Windju, S.S.; Jørstad, T.S.; Wilson, Z.A.; Aalen, R.B. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 2009, 4, e7817. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, Y.; Law, J.A.; Zhang, X. SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 18557–18562. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.; Henderson, J.T.; Svedin, E.; Fiers, M.; McCarthy, K.; Smith, A.; Guo, C.; Bishop, B.; Zhang, H.; Riksen, T.; et al. Cross-talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 2016, 203, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Luo, J.-H.; Cui, Z.-H.; Xue, M.; Wang, L.; Zhang, X.-Y.; Pawlowski, W.P.; He, Y. ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 2017, 174, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ndamukong, I.; Xu, Z.; Lapko, H.; Fromm, M.; Avramova, Z. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012, 8, e1003111. [Google Scholar] [CrossRef] [PubMed]
- Fromm, M.; Avramova, Z. ATX1/AtCOMPASS and the H3K4me3 marks: How do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 2014, 21, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 instructive for transcription activation? Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Horn, P.J.; Peterson, C.L. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007, 21, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Bellec, M.; Radulescu, O.; Lagha, M. Remembering the past: Mitotic bookmarking in a developing embryo. Curr. Opin. Sys. Biol. 2018, 11, 41–49. [Google Scholar] [CrossRef]
- Petruk, S.; Sedkov, Y.; Johnston, D.M.; Hodgson, J.W.; Black, K.L.; Kovermann, S.K.; Beck, S.; Canaani, E.; Brock, H.W.; Mazo, A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 2012, 150, 922–933. [Google Scholar] [CrossRef]
- Black, K.L.; Petruk, S.; Fenstermaker, T.K.; Hodgson, J.W.; Caplan, J.L.; Brock, H.W.; Mazo, A. Chromatin proteins and RNA are associated with DNA during all phases of mitosis. Cell Discov. 2016, 2, 16038. [Google Scholar] [CrossRef]
- Costa, S.; Dean, C. Storing memories: The distinct phases of Polycomb-mediated silencing of Arabidopsis FLC. Biochem. Soc. Trans. 2019, 47, 1187–1196. [Google Scholar] [CrossRef]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kovalchuk, I. Genome instability and epigenetic modification--heritable responses to environmental stress? Curr. Opin. Plant Biol. 2011, 14, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Verkest, A.; Byzova, M.; Martens, C.; Willems, P.; Verwulgen, T.; Slabbinck, B.; Rombaut, D.; Van de Velde, J.; Vandepoele, K.; Standaert, E.; et al. Selection for improved energy use efficiency and drought tolerance in canola results in distinct transcriptome and epigenome changes. Plant Physiol. 2015, 168, 1338–1350. [Google Scholar] [CrossRef]
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A. Seed priming and transgenerational drought memory improves tolerance against salt stress in bread wheat. Plant Physiol. Biochem. 2017, 118, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Raju, S.K.K.; Shao, M.R.; Sanchez, R.; Xu, Y.Z.; Sandhu, A.; Graef, G.; Mackenzie, S. An epigenetic breeding system in soybean for increased yield and stability. Plant Biotechnol. J. 2018, 16, 1836–1847. [Google Scholar] [CrossRef]
- Campos, E.I.; Stafford, J.M.; Reinberg, D. Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol. 2014, 24, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Garvis, S.; Palladino, F. Histone methylation and memory of environmental stress. Cells 2019, 8, 339. [Google Scholar] [CrossRef]
- Šrut, M. Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere 2021, 282, 131026. [Google Scholar] [CrossRef]
- Zenk, F.; Loeser, E.; Schiavo, R.; Kilpert, F.; Bogdanović, O.; Iovino, N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 2017, 357, 212–216. [Google Scholar] [CrossRef]
- Molla-Herman, A.; Matias, N.R.; Huynh, J.R. Chromatin modifications regulate germ cell development and transgenerational information relay. Curr. Opin. Insect Sci. 2014, 1, 10–18. [Google Scholar] [CrossRef]
- Weiser, N.E.; Kim, J.K. Multigenerational regulation of the Caenorhabditis elegans chromatin landscape by germline small RNAs. Annu. Rev. Genet. 2019, 53, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, O.; Reinders, J.; Caikovski, M.; Smathajitt, C.; Paszkowski, J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 2007, 130, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145–1148. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Morgado, L.; Preite, V.; Oplaat, C.; Anava, S.; de Carvalho, J.F.; Rechavi, O.; Johannes, F.; Verhoeven, K.J.F. Small RNAs reflect grandparental environments in apomictic dandelion. Mol. Biol. Evol. 2017, 34, 2035–2040. [Google Scholar] [CrossRef]
- Kuhlmann, M.; Finke, A.; Mascher, M.; Mette, M.F. DNA methylation maintenance consolidates RNA-directed DNA methylation and transcriptional gene silencing over generations in Arabidopsis thaliana. Plant J. 2014, 80, 269–281. [Google Scholar] [CrossRef]
- Ganguly, D.R.; Crisp, P.A.; Eichten, S.R.; Pogson, B.J. The Arabidopsis DNA methylome is stable under transgenerational drought stress. Plant Physiol. 2017, 175, 1893–1912. [Google Scholar] [CrossRef]
- Braun, R.E. Packaging paternal chromosomes with protamine. Nat. Genet. 2001, 28, 10–12. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335–E8343. [Google Scholar] [CrossRef] [PubMed]
- Calarco, J.P.; Borges, F.; Donoghue, M.T.; Van Ex, F.; Jullien, P.E.; Lopes, T.; Gardner, R.; Berger, F.; Feijó, J.A.; Becker, J.D.; et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 2012, 151, 194–205. [Google Scholar] [CrossRef]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Berger, F. Chromatin remodelling during male gametophyte development. Plant J. 2015, 83, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendía, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Paro, R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.H.; Vu, N.T.; Cheong, J.-J. Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants. Int. J. Mol. Sci. 2022, 23, 12918. https://doi.org/10.3390/ijms232112918
Nguyen NH, Vu NT, Cheong J-J. Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants. International Journal of Molecular Sciences. 2022; 23(21):12918. https://doi.org/10.3390/ijms232112918
Chicago/Turabian StyleNguyen, Nguyen Hoai, Nam Tuan Vu, and Jong-Joo Cheong. 2022. "Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants" International Journal of Molecular Sciences 23, no. 21: 12918. https://doi.org/10.3390/ijms232112918
APA StyleNguyen, N. H., Vu, N. T., & Cheong, J.-J. (2022). Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants. International Journal of Molecular Sciences, 23(21), 12918. https://doi.org/10.3390/ijms232112918





