Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome
Abstract
1. Introduction
2. Results and Discussion
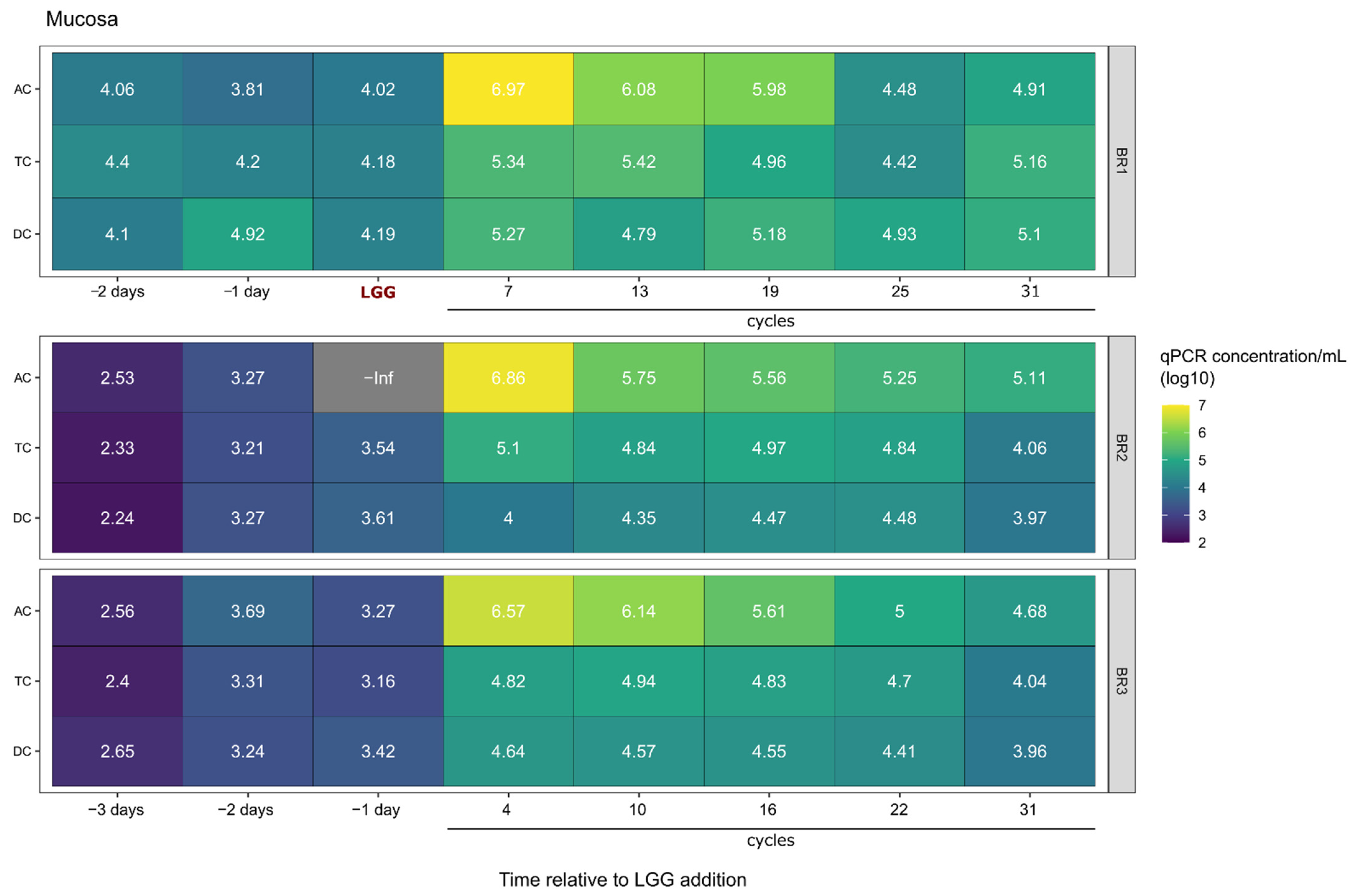
2.1. Survival and Growth of LGG in a Multi-Compartment In Vitro Model of the Colon
2.2. LGG Persistence and Growth Versus Transient Presence over Time
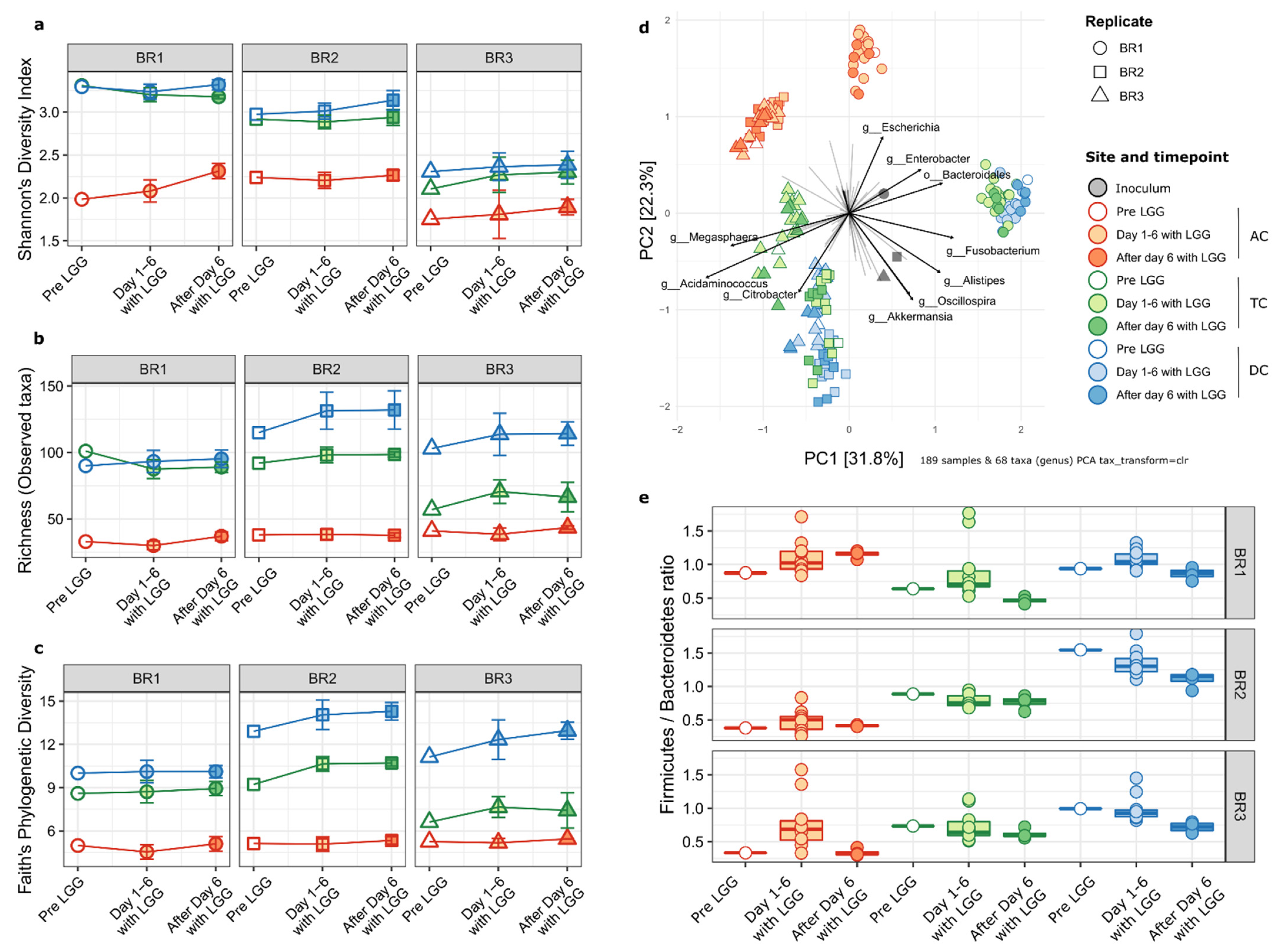
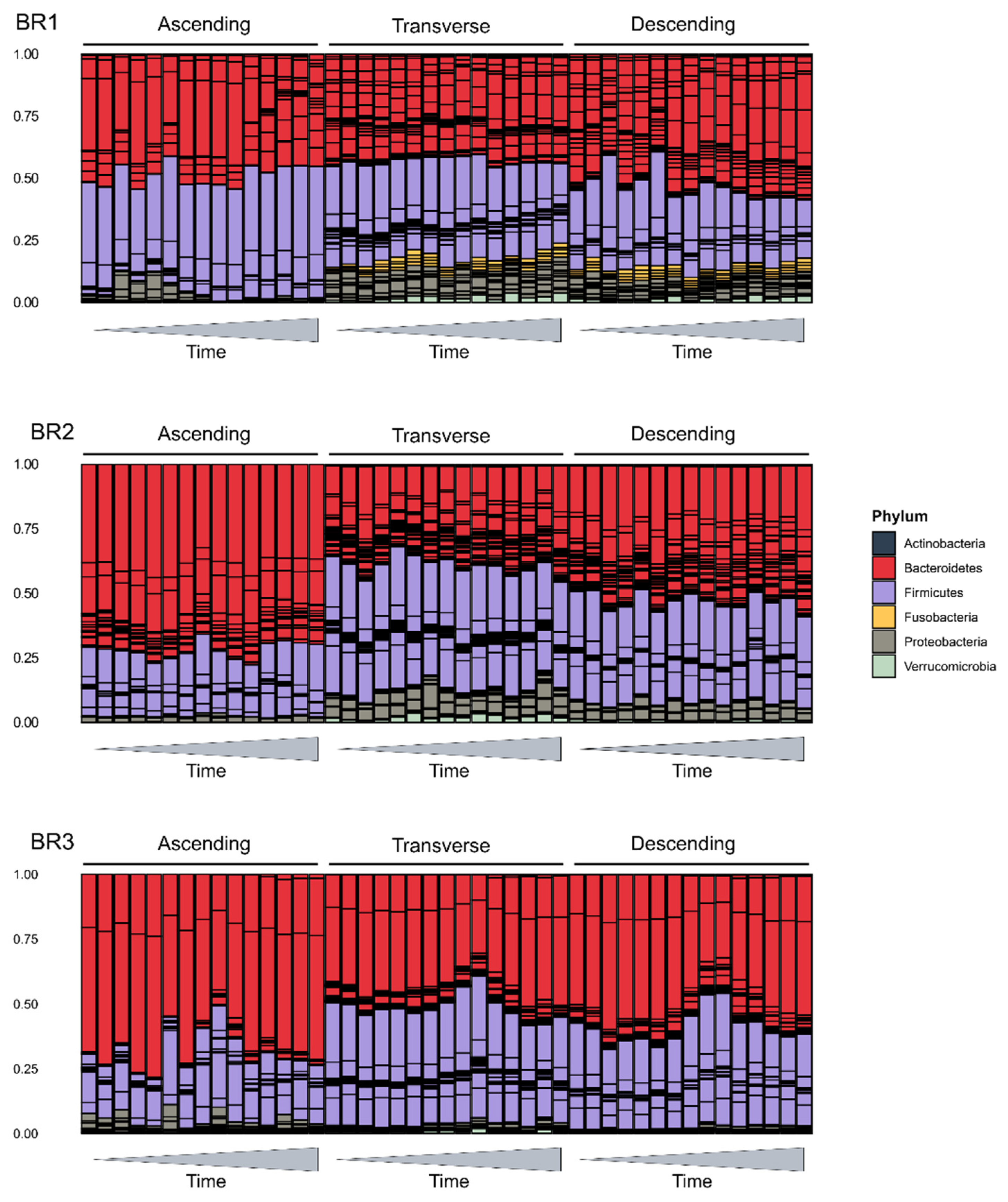
2.3. Impact on the Host Microbial Community
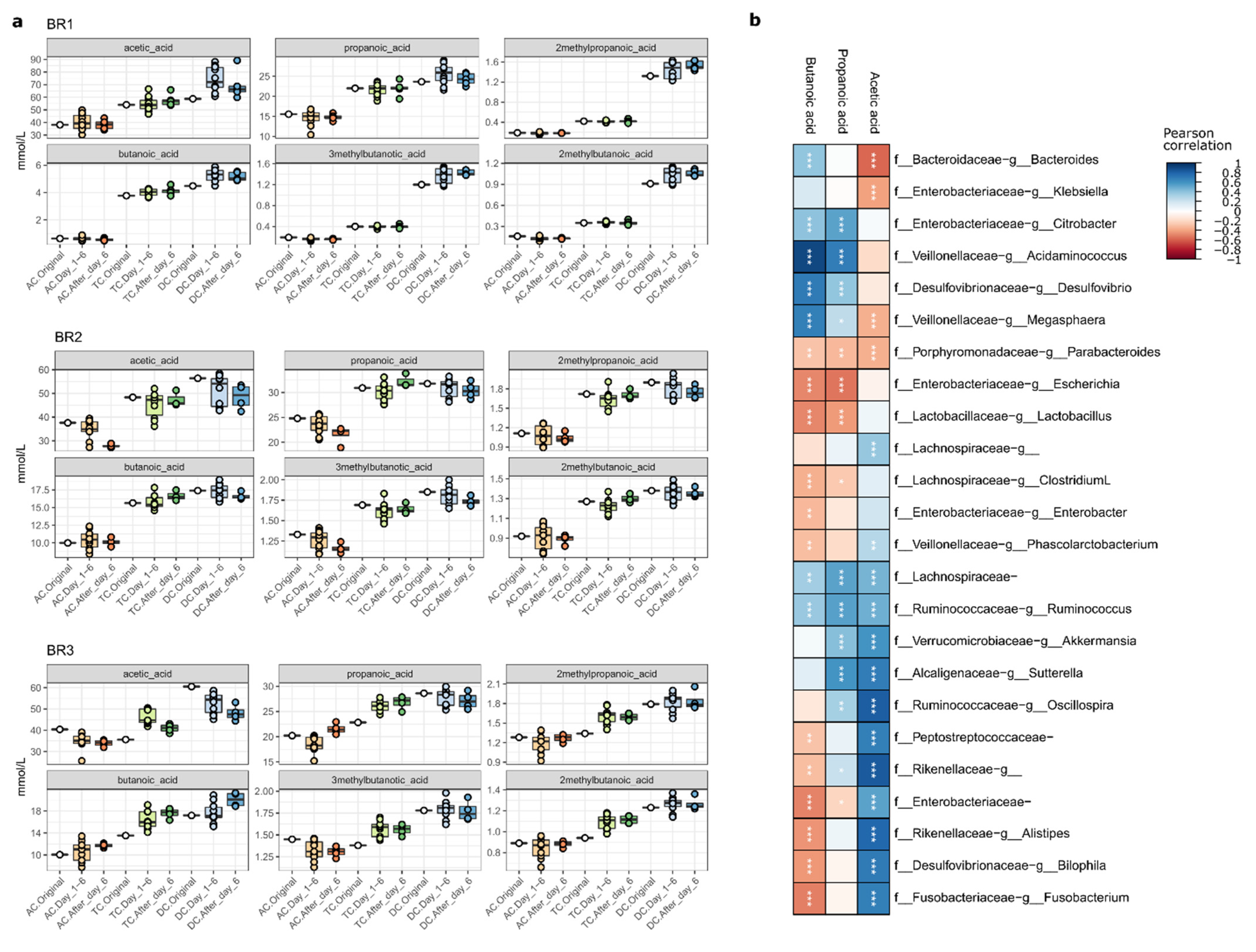
2.4. Impact of LGG Addition on SCFA Synthesis
3. Materials and Methods
3.1. Materials
3.2. Establishment of a Stable Gut Microbial Community in SHIME®
3.3. Culturing LGG for Inoculation
3.4. Nucleotide Analysis
3.4.1. DNA Extraction
3.4.2. Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR) Detection of LGG
3.4.3. DNA Sequencing and Bioinformatics Analysis
3.5. Short Chain Fatty Acids Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cameron, D.; Hock, Q.S.; Kadim, M.; Mohan, N.; Ryoo, E.; Sandhu, B.; Yamashiro, Y.; Jie, C.; Hoekstra, H.; Guarino, A. Probiotics for gastrointestinal disorders: Proposed recommendations for children of the Asia-Pacific region. World J. Gastroenterol. 2017, 23, 7952–7964. [Google Scholar] [CrossRef] [PubMed]
- Pace, F.; Pace, M.; Quartarone, G. Probiotics in digestive diseases: Focus on Lactobacillus GG. Minerva Gastroenterol. E Dietol. 2015, 61, 273–292. [Google Scholar]
- Chen, L.; Li, H.; Li, J.; Chen, Y.; Yang, Y. Lactobacillus rhamnosus GG treatment improves intestinal permeability and modulates microbiota dysbiosis in an experimental model of sepsis. Int. J. Mol. Med. 2019, 43, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-B.; Lee, H.-C.; Hu, J.-J.; Hou, S.-Y.; Liu, H.-L.; Fang, H.-W. Dose-dependent effect of Lactobacillus rhamnosus on quantitative reduction of faecal rotavirus shedding in children. J. Trop. Pediatr. 2009, 55, 297–301. [Google Scholar] [CrossRef]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef]
- Chibbar, R.; Dieleman, L.A. The gut microbiota in celiac disease and probiotics. Nutrients 2019, 11, 2375. [Google Scholar] [CrossRef]
- Girbovan, A.; Sur, G.; Samasca, G.; Lupan, I. Dysbiosis a risk factor for celiac disease. Med. Microbiol. Immunol. 2017, 206, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Verstraelen, H.; Osowska, S.; Doerffel, Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology 2008, 135, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, Y.; Polk, D.B.; Tomasula, P.M.; Yan, F.; Liu, L. Preserving viability of Lactobacillus rhamnosus GG in vitro and in vivo by a new encapsulation system. J. Control. Release 2016, 230, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M., Jr.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Liu, L.; Dempsey, P.J.; Tsai, Y.H.; Raines, E.W.; Wilson, C.L.; Cao, H.; Cao, Z.; Liu, L.; Polk, D.B. A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J. Biol. Chem. 2013, 288, 30742–30751. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Rapozo, D.C.; Bernardazzi, C.; de Souza, H.S. Diet and microbiota in inflammatory bowel disease: The gut in disharmony. World J. Gastroenterol. 2017, 23, 2124–2140. [Google Scholar] [CrossRef]
- Rigsbee, L.; Agans, R.; Shankar, V.; Kenche, H.; Khamis, H.J.; Michail, S.; Paliy, O. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am. J. Gastroenterol. 2012, 107, 1740–1751. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Qian, Z.; Yin, J.; Xu, W.; Zhou, X. The role of intestinal microbiota in cardiovascular disease. J. Cell. Mol. Med. 2019, 23, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef]
- Everard, A.; Cani, P.D. Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 73–83. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Obesity, diabetes, and gut microbiota: The hygiene hypothesis expanded? Diabetes Care 2010, 33, 2277–2284. [Google Scholar] [CrossRef]
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J. Diabetes Res. 2018, 2018, 3462092. [Google Scholar] [CrossRef]
- Sudo, N. Role of gut microbiota in brain function and stress-related pathology. Biosci. Microbiota Food Health 2019, 38, 75–80. [Google Scholar] [CrossRef]
- Tremlett, H.; Bauer, K.C.; Appel-Cresswell, S.; Finlay, B.B.; Waubant, E. The gut microbiome in human neurological disease: A review. Ann. Neurol. 2017, 81, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, probiotics and neurodegenerative diseases: Deciphering the gut brain axis. Cell. Mol. Life Sci. CMLS 2017, 74, 3769–3787. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Galland, L. The gut microbiome and the brain. J. Med. Food 2014, 17, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.C.; Rehman, A.; Yu, S.; Andino, N.M. Brain fogginess, gas and bloating: A link between SIBO, probiotics and metabolic acidosis. Clin. Transl. Gastroenterol. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Schnadower, D.; Tarr, P.I.; Casper, T.C.; Gorelick, M.H.; Dean, J.M.; O’Connell, K.J.; Mahajan, P.; Levine, A.C.; Bhatt, S.R.; Roskind, C.G.; et al. Lactobacillus rhamnosus GG versus Placebo for Acute Gastroenteritis in Children. N. Engl. J. Med. 2018, 379, 2002–2014. [Google Scholar] [CrossRef]
- Kuisma, J.; Mentula, S.; Jarvinen, H.; Kahri, A.; Saxelin, M.; Farkkila, M. Effect of Lactobacillus rhamnosus GG on ileal pouch inflammation and microbial flora. Aliment. Pharmacol. Ther. 2003, 17, 509–515. [Google Scholar] [CrossRef]
- Doron, S.; Snydman, D.R. Risk and safety of probiotics. Clin. Infect. Dis. 2015, 60 (Suppl. 2), S129–S134. [Google Scholar] [CrossRef]
- Preidis, G.A.; Versalovic, J.J.G. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 2009, 136, 2015–2031. [Google Scholar] [CrossRef]
- Walker, M.E.; Simpson, J.B.; Redinbo, M.R. A structural metagenomics pipeline for examining the gut microbiome. Curr. Opin. Struct. Biol. 2022, 75, 102416. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Firrman, J.; Tanes, C.; Bittinger, K.; Thomas-Gahring, A.; Wu, G.D.; Van den Abbeele, P.; Tomasula, P.M. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS ONE 2018, 13, e0197692. [Google Scholar] [CrossRef] [PubMed]
- Patrignani, F.; Parolin, C.; D’Alessandro, M.; Siroli, L.; Vitali, B.; Lanciotti, R. Evaluation of the fate of Lactobacillus crispatus BC4, carried in Squacquerone cheese, throughout the simulator of the human intestinal microbial ecosystem (SHIME). Food Res. Int. 2020, 137, 109580. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.J.A.; et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Vázquez, L.J.; Fernández, F.J. Optimal control of a bioreactor. Appl. Math. Comput. 2010, 216, 2559–2575. [Google Scholar] [CrossRef]
- Von Ossowski, I.; Reunanen, J.; Satokari, R.; Vesterlund, S.; Kankainen, M.; Huhtinen, H.; Tynkkynen, S.; Salminen, S.; de Vos, W.M.; Palva, A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 2010, 76, 2049–2057. [Google Scholar] [CrossRef]
- Walter, J. Ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl. Environ. Microbiol. 2008, 74, 4985–4996. [Google Scholar] [CrossRef]
- Nishiyama, K.; Sugiyama, M.; Mukai, T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms 2016, 4, 34. [Google Scholar] [CrossRef]
- Hardin, G. The Competitive Exclusion Principle: An idea that took a century to be born has implications in ecology, economics, and genetics. Science 1960, 131, 1292–1297. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Ma, X. Dietary Amino Acids and the Gut-Microbiome-Immune Axis: Physiological Metabolism and Therapeutic Prospects. Compr. Rev. Food Sci. Food Saf. 2019, 18, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.I.; Imholz, N.C.; Verhoeven, T.L.; Balzarini, J.; Van Damme, E.J.; Schols, D.; Vanderleyden, J.; Lebeer, S. Lectin-like molecules of Lactobacillus rhamnosus GG inhibit pathogenic Escherichia coli and Salmonella biofilm formation. PLoS ONE 2016, 11, e0161337. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, Y.-H.; Yang, J.-C.; Yang, G.-Y.; Zhou, D.; Wang, J.-F. A selected Lactobacillus rhamnosus strain promotes EGFR-independent Akt activation in an enterotoxigenic Escherichia coli K88-infected IPEC-J2 cell model. PLoS ONE 2015, 10, e0125717. [Google Scholar] [CrossRef]
- Firrman, J.; Liu, L.; Tanes, C.; Friedman, E.S.; Bittinger, K.; Daniel, S.; van den Abbeele, P.; Evans, B. Metabolic analysis of regionally distinct gut microbial communities using an in vitro platform. J. Agric. Food Chem. 2019, 68, 13056–13067. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Molenaar, D.; van IJcken, W.; Venema, K.; Kort, R. Genome instability in Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2013, 79, 2233–2239. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahalak, K.K.; Firrman, J.; Bobokalonov, J.; Narrowe, A.B.; Bittinger, K.; Daniel, S.; Tanes, C.; Mattei, L.M.; Zeng, W.-B.; Soares, J.W.; et al. Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome. Int. J. Mol. Sci. 2022, 23, 12973. https://doi.org/10.3390/ijms232112973
Mahalak KK, Firrman J, Bobokalonov J, Narrowe AB, Bittinger K, Daniel S, Tanes C, Mattei LM, Zeng W-B, Soares JW, et al. Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome. International Journal of Molecular Sciences. 2022; 23(21):12973. https://doi.org/10.3390/ijms232112973
Chicago/Turabian StyleMahalak, Karley K., Jenni Firrman, Jamshed Bobokalonov, Adrienne B. Narrowe, Kyle Bittinger, Scott Daniel, Ceylan Tanes, Lisa M. Mattei, Wei-Bin Zeng, Jason W. Soares, and et al. 2022. "Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome" International Journal of Molecular Sciences 23, no. 21: 12973. https://doi.org/10.3390/ijms232112973
APA StyleMahalak, K. K., Firrman, J., Bobokalonov, J., Narrowe, A. B., Bittinger, K., Daniel, S., Tanes, C., Mattei, L. M., Zeng, W.-B., Soares, J. W., Kobori, M., Lemons, J. M. S., Tomasula, P. M., & Liu, L. (2022). Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome. International Journal of Molecular Sciences, 23(21), 12973. https://doi.org/10.3390/ijms232112973





