Fatty Acid Biosynthesis Pathways Are Downregulated during Stigma Development and Are Critical during Self-Incompatible Responses in Ornamental Kale
Abstract
:1. Introduction
2. Results
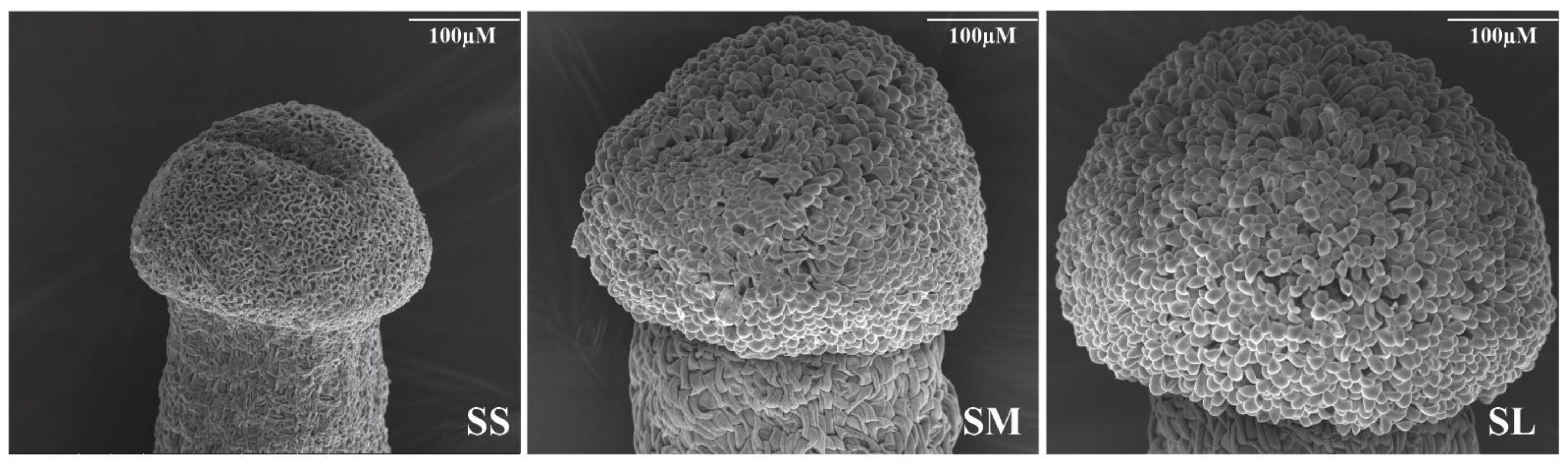
2.1. Phenotypic Observation of Different Stages of Development and after Pollination in Stigma
2.2. Full-Length Transcriptome Analysis Based on RNA-seq and SMRT-seq
2.3. Identification of Fusion Transcript, AS, APA, SSR, ORFs, and lncRNA
2.4. Identification and Functional Analysis of TFs and DEGs
2.5. Cluster Analysis and qRT-PCR Validation of Genes Related in the Flavonoid Biosynthesis Pathway
2.6. Functional Analysis of Related Genes in the Fatty Acid Biosynthesis Pathway
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Total RNA Extraction
4.3. PacBio SMRT-Seq and Illumina RNA-seq Library Preparation and Sequencing
4.4. PacBio SMRT-seq and Illumina RNA-seq Data Processing
4.5. Prediction of Fusion Transcript, AS, APA, SSR, ORFs, and lncRNAs
4.6. Identification and Functional Annotation of DEGs
4.7. Construction of WGCNA
4.8. qRT-PCR Analysis
4.9. Pollination Experiment
4.10. FASN Inhibitor Treatment and Pollination Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| SI | Self-Incompatibility |
| CP | Compatible Pollination |
| BUSCO | Benchmarking Universal Single-Copy Orthologs |
| FLNC | Full-Length Non-Chimeric |
| CCS | Circular Consensus Sequencing |
| lncRNAs | Long Non-Coding RNAs |
| AS | Alternative Splicing |
| APA | Alternative Polyadenylation |
| SSR | Simple Sequence Repeat |
| ICE | Iterative Clustering for Error Correction |
| NR | NCBI Non-Redundant Protein Sequences |
| HQ | High Quality |
| LQ | Low Quality |
| ORF | Open Reading Frame |
| SMRT | Single-Molecule, Real-Time |
| FPKM | Fragment Per Kilobase of Transcript Per Million Fragments mapped |
| DEG | Differentially Expressed Gene |
| TF | Transcription Factor |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | Real-Time Quantitative PCR |
| FASN | Fatty Acid Synthase |
References
- Abhinandan, K.; Sankaranarayanan, S.; Macgregor, S.; Goring, D.R.; Samuel, M.A. Cell-cell signaling during the Brassicaceae self-incompatibility response. Trends Plant Sci. 2021, 27, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, Q.; Hu, G.; Qin, Y. Comparative transcriptional survey between self-incompatibility and self-compatibility in Citrus reticulata Blanco. Gene 2017, 609, 52–61. [Google Scholar] [CrossRef]
- Wang, L.; Peng, H.; Ge, T.; Liu, T.; Hou, X.; Li, Y. Identification of differentially accumulating pistil proteins associated with self-incompatibility of non-heading C hinese cabbage. Plant Biol. 2014, 16, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H.; Elleman, C. Structural changes in the pollen grain of Brassica oleracea during dehydration in the anther and development on the stigma as revealed by anhydrous fixation techniques. Micron Microsc. Acta 1985, 16, 255–270. [Google Scholar] [CrossRef]
- Matsuda, T.; Matsushima, M.; Nabemoto, M.; Osaka, M.; Sakazono, S.; Masuko-Suzuki, H.; Takahashi, H.; Nakazono, M.; Iwano, M.; Takayama, S. Transcriptional characteristics and differences in Arabidopsis stigmatic papilla cells pre-and post-pollination. Plant Cell Physiol. 2015, 56, 663–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jany, E.; Nelles, H.; Goring, D.R. The molecular and cellular regulation of Brassicaceae self-incompatibility and self-pollen rejection. Int. Rev. Cell Mol. Biol. 2019, 343, 1–35. [Google Scholar]
- Johnson, M.A.; Harper, J.F.; Palanivelu, R. A fruitful journey: Pollen tube navigation from germination to fertilization. Annu. Rev. Plant Biol. 2019, 70, 809–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Shen, L.; Xiao, Y.; Vyshedsky, D.; Peng, C.; Sun, X.; Liu, Z.; Cheng, L.; Zhang, H.; Han, Z. Pollen PCP-B peptides unlock a stigma peptide-eceptor kinase gating mechanism for pollination. Science 2021, 372, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen acceptance or rejection: A tale of two pathways. Trends Plant Sci. 2016, 21, 1058–1067. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Thorsness, M.K.; Rundle, S.J.; Goldberg, M.L.; Nasrallah, J.B.; Nasrallah, M.E. Ablation of papillar cell function in Brassica flowers results in the loss of stigma receptivity to pollination. Plant Cell 1993, 5, 263–275. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, H. Dry stigmas, water and self-incompatibility in Brassica. Sex. Plant Reprod. 1995, 8, 1–10. [Google Scholar] [CrossRef]
- Swanson, R.; Clark, T.; Preuss, D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 2005, 18, 163–171. [Google Scholar] [CrossRef]
- Broz, A.K.; Bedinger, P.A. Pollen-Pistil Interactions as Reproductive Barriers. Annu. Rev. Plant Biol. 2021, 72, 615–639. [Google Scholar] [CrossRef]
- Bateman, A. Self-incompatibility systems in angiosperms. Heredity 1952, 6, 285–310. [Google Scholar] [CrossRef] [Green Version]
- Takayama, S.; Shiba, H.; Iwano, M.; Shimosato, H.; Che, F.S.; Kai, N.; Watanabe, M.; Suzuki, G.; Hinata, K.; Isogai, A. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 2000, 97, 1920–1925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasaki, T.; Hatakeyama, K.; Suzuki, G.; Watanabe, M.; Isogai, A.; Hinata, K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 2000, 403, 913–916. [Google Scholar] [CrossRef]
- Takayama, S.; Shimosato, H.; Shiba, H.; Funato, M.; Che, F.S.; Watanabe, M.; Iwano, M.; Isogai, A. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature 2001, 413, 534–538. [Google Scholar] [CrossRef]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005, 56, 467–489. [Google Scholar] [CrossRef] [Green Version]
- Murase, K.; Shiba, H.; Iwano, M.; Che, F.S.; Watanabe, M.; Isogai, A.; Takayama, S. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 2004, 303, 1516–1519. [Google Scholar] [CrossRef] [PubMed]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakita, M.; Shimosato, H.; Murase, K.; Isogai, A.; Takayama, S. Direct interaction between S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol. 2007, 24, 185–190. [Google Scholar] [CrossRef]
- Samuel, M.A.; Chong, Y.T.; Haasen, K.E.; Aldea-Brydges, M.G.; Stone, S.L.; Goring, D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 2009, 21, 2655–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15185. [Google Scholar] [CrossRef] [PubMed]
- Scandola, S.; Samuel, M.A. A flower-specific phospholipase D is a stigmatic compatibility factor targeted by the self-incompatibility response in Brassica napus. Curr. Biol. 2019, 29, 506–512. [Google Scholar] [CrossRef] [Green Version]
- Lan, X.; Yang, J.; Cao, M.; Wang, Y.; Kawabata, S.; Li, Y. Isolation and characterization of a J domain protein that interacts with ARC1 from ornamental kale (Brassica oleracea var. acephala). Plant Cell Rep. 2015, 34, 817–829. [Google Scholar] [CrossRef]
- Iwano, M.; Ito, K.; Fujii, S.; Kakita, M.; Asano-Shimosato, H.; Igarashi, M.; Kaothien-Nakayama, P.; Entani, T.; Kanatani, A.; Takehisa, M. Calcium signalling mediates self-incompatibility response in the Brassicaceae. Nat. Plants 2015, 1, 15128. [Google Scholar] [CrossRef] [Green Version]
- Silva, N.; Goring, D. Mechanisms of self-incompatibility in flowering plants. Cell. Mol. Life Sci. 2001, 58, 1988–2007. [Google Scholar] [CrossRef]
- Haffani, Y.; Gaude, T.; Cock, J.; Goring, D. Antisense suppression of thioredoxinhmRNA in Brassica napus cv. Plant Mol. Biol. 2004, 55, 619–630. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, Y.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Dai, C.; Ma, C. The Brassica napus GATA transcription factor BnA5.ZML1 is a stigma compatibility factor. J. Integr. Plant Biol. 2020, 62, 1112–1131. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, J.; Su, S.; Wei, X.; Yang, L.; Zhao, H.; Yu, J.; Wang, J.; Hui, J.; Hao, S. FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol. 2021, 31, 3004–3016. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Tang, W.; Jamshed, M.; Northey, J.; Patel, D.; Smith, D.; Siu, K.M.; Muench, D.G.; Wang, Z.Y.; Goring, D.R. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role for microtubule dynamics during pollen responses. Mol. Cell. Proteom. 2011, 10, M111.011338. [Google Scholar] [CrossRef] [PubMed]
- Nazemof, N.; Couroux, P.; Xing, T.; Robert, L.S. Proteomic analysis of the mature Brassica stigma reveals proteins with diverse roles in vegetative and reproductive development. Plant Sci. 2016, 250, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Yang, J.; Abhinandan, K.; Nie, Y.; Li, X.; Li, Y.; Samuel, M.A. Flavonoids and ROS play opposing roles in mediating pollination in ornamental kale (Brassica oleracea var. acephala). Mol. Plant 2017, 10, 1361–1364. [Google Scholar] [CrossRef] [Green Version]
- Sankaranarayanan, S.; Jamshed, M.; Deb, S.; Chatfield-Reed, K.; Kwon, E.J.G.; Chua, G.; Samuel, M.A. Deciphering the stigmatic transcriptional landscape of compatible and self-incompatible pollinations in Brassica napus reveals a rapid stigma senescence response following compatible pollination. Mol. Plant 2013, 6, 1988–1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osaka, M.; Matsuda, T.; Sakazono, S.; Masuko-Suzuki, H.; Maeda, S.; Sewaki, M.; Sone, M.; Takahashi, H.; Nakazono, M.; Iwano, M. Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol. 2013, 54, 1894–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Gao, C.; Yue, Y.; Liu, Z.; Ma, C.; Zhou, G.; Yang, Y.; Duan, Z.; Li, B.; Wen, J. Time-course transcriptome analysis of compatible and incompatible pollen-stigma interactions in Brassica napus L. Front. Plant Sci. 2017, 8, 682–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y.; Cao, L.; Zhang, Q.; Amee, M.; Chen, K.; Chen, L. SMRT and Illumina RNA sequencing reveal novel insights into the heat stress response and crosstalk with leaf senescence in tall fescue. BMC Plant Biol. 2020, 20, 366. [Google Scholar] [CrossRef]
- An, D.; Cao, H.X.; Li, C.; Humbeck, K.; Wang, W. Isoform sequencing and state-of-art applications for unravelling complexity of plant transcriptomes. Genes 2018, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016, 7, 12060. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, E.P.; Wilkins, C.; Demidchik, V.; Davies, J.M.; Glover, B.J. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 2010, 61, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Van Blokland, R.; Quattrocchio, F.; Van Tunen, A.J.; Mol, J.N. Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. Plant Cell 1990, 2, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Kehrel, B.; Wiermann, R. Immunochemical localization of phenylalanine ammonia-lyase and chalcone synthase in anthers. Planta 1985, 163, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.E.; Quattrocchio, F.; Mol, J.N. The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 1994, 16, 123–132. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Nishihara, M.; Mishiba, K.; Yamamura, S. Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci. 2005, 168, 1309–1318. [Google Scholar] [CrossRef]
- Piffanelli, P.; Ross, J.H.; Murphy, D.J. Intra-and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J. 1997, 11, 549–562. [Google Scholar] [CrossRef]
- Evans, D.E.; Sang, J.P.; Cominos, X.; Rothnie, N.E.; Knox, R.B. A study of phospholipids and galactolipids in pollen of two lines of Brassica napus L.(rapeseed) with different ratios of linoleic to linolenic acid. Plant Physiol. 1990, 93, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, R.-l.; Feng, J.; Zhang, B.-X.; Huang, Y.; Cheng, J.; Zhang, C.X. Transcriptome and gene expression analysis of an oleaginous diatom under different salinity conditions. BioEnergy Res. 2014, 7, 192–205. [Google Scholar] [CrossRef]
- Yu, L.; Tan, X.; Jiang, B.; Sun, X.; Gu, S.; Han, T.; Hou, W. A peroxisomal long-chain acyl-CoA synthetase from Glycine max involved in lipid degradation. PLoS ONE 2014, 9, e100144. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Pérez, A.J.; Venegas-Calerón, M.; Vaistij, F.E.; Salas, J.J.; Larson, T.R.; Garcés, R.; Graham, I.A.; Martínez-Force, E. Reduced expression of FatA thioesterases in Arabidopsis affects the oil content and fatty acid composition of the seeds. Planta 2012, 235, 629–639. [Google Scholar] [CrossRef]
- Liu, Y.; Han, J.; Li, Z.; Jiang, Z.; Luo, L.; Zhang, Y.; Chen, M.; Yang, Y.; Liu, Z. Heterologous Expression of Jatropha curcas Fatty Acyl-ACP Thioesterase A (JcFATA) and B (JcFATB) Affects Fatty Acid Accumulation and Promotes Plant Growth and Development in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 4209. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Kimura, F.; Hirakata, K.; Tsuda, K.; Takasugi, T.; Eiguchi, M.; Nakagawa, K.; Kurata, N. Fatty acid elongase is required for shoot development in rice. Plant J. 2011, 66, 680–688. [Google Scholar] [CrossRef]
- Xiao, R.; Zou, Y.; Guo, X.; Li, H.; Lu, H. Fatty acid desaturases (FADs) modulate multiple lipid metabolism pathways to improve plant resistance. Mol. Biol. Rep. 2022, 49, 9997–10011. [Google Scholar] [CrossRef] [PubMed]
- Omura, S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol. Rev. 1976, 40, 681–697. [Google Scholar] [CrossRef] [PubMed]
- D’agnony, G.; Rosenfeld, I.S.; Awaya, J.; Ōmura, S.; Vagelos, P.R. Inhibition of fatty acid synthesis by the antibiotic cerulenin: Specific inactivation of β-ketoacyl-acyl carrier protein synthetase. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1973, 326, 155–166. [Google Scholar] [CrossRef]
- Vance, D.; Goldberg, I.; Mitsuhashi, O.; Bloch, K.; Ōmura, S.; Nomura, S. Inhibition of fatty acid synthetases by the antibiotic cerulenin. Biochem. Biophys. Res. Commun. 1972, 48, 649–656. [Google Scholar] [CrossRef]







| DEG Set | DEG Number | Up-Regulated | Down-Regulated |
|---|---|---|---|
| SS vs. SM | 4178 | 2166 | 2012 |
| SS vs. SL | 6349 | 3063 | 3286 |
| SM vs. SL | 5185 | 2870 | 2315 |
| Total: 15,712 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, H.; Li, H.; Abhinandan, K.; Xun, B.; Yao, K.; Shi, J.; Zhao, R.; Li, M.; Wu, Y.; Lan, X. Fatty Acid Biosynthesis Pathways Are Downregulated during Stigma Development and Are Critical during Self-Incompatible Responses in Ornamental Kale. Int. J. Mol. Sci. 2022, 23, 13102. https://doi.org/10.3390/ijms232113102
Qin H, Li H, Abhinandan K, Xun B, Yao K, Shi J, Zhao R, Li M, Wu Y, Lan X. Fatty Acid Biosynthesis Pathways Are Downregulated during Stigma Development and Are Critical during Self-Incompatible Responses in Ornamental Kale. International Journal of Molecular Sciences. 2022; 23(21):13102. https://doi.org/10.3390/ijms232113102
Chicago/Turabian StyleQin, Hongtao, Hang Li, Kumar Abhinandan, Baoru Xun, Kun Yao, Jiayuan Shi, Ruoxi Zhao, Mugeng Li, Ying Wu, and Xingguo Lan. 2022. "Fatty Acid Biosynthesis Pathways Are Downregulated during Stigma Development and Are Critical during Self-Incompatible Responses in Ornamental Kale" International Journal of Molecular Sciences 23, no. 21: 13102. https://doi.org/10.3390/ijms232113102






