From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction
Abstract
1. Introduction
2. A Brief Review of the 2D Co-Culture Model in the Research of Neural Cell–Cell Interaction
3. Application of 3D Co-Culture Models in the Study of Neural Cells Interactions
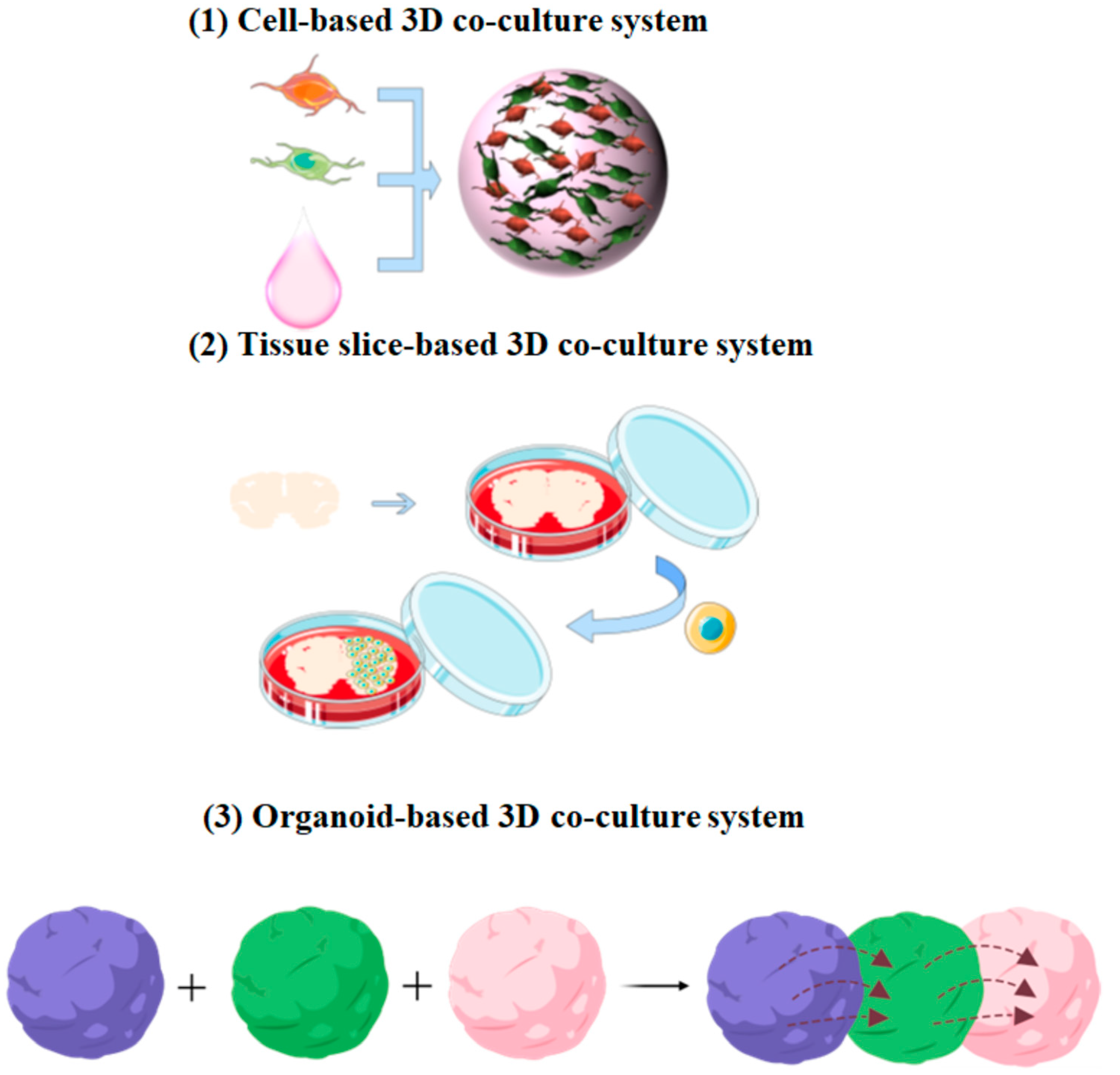
3.1. Cell-Based 3D Co-Culture System
3.2. Tissue Slice-Based 3D Co-Culture System
| Co-Culture System | Objective | Main Results | Investigation | Refs |
|---|---|---|---|---|
| Purkinje progenitors and human fetal cerebellar slices. | To direct human iPSCs differentiate toward Purkinje neurons. | Fetal cerebellar slices promote differentiation and maturation of Purkinje neurons. | The degree of neuronal differentiation and electrophysiology analysis. | [41] |
| DPCs and adult mouse hippocampal slices. | To investigate whether human DPCs can promote neuroregeneration. | DPCs stimulated the growth of neuronal cells (especially neurons) in the edges of the hippocampal slices. | Dendrite length and cell viability. | [42] |
| NPCs and auditory brainstem slice. | To evaluate the potential of using DPSC as a therapeutic procedure for hearing disability patients. | Co-culture with auditory brainstem slices promotes DPSCs differentiation into neurons. | Neuronal differentiation and intracellular calcium oscillation. | [43] |
| ESNPs and hippocampal slices. | To explore the mechanism of ESNPs migration. | Chemokines secreted by vascular-associated astrocytes direct ESNPs migration. | Cell number and cell morphology. | [44] |
| MSCs and rat organotypic hippocampal slice. | To determine the neuroprotective potential of MSCs. | MSCs reduced cell death in hippocampal slices. | Cell number and function of cell secretion. | [45] |
| OPCs and cerebellar slice. | To investigate OPCs differentiation and myelin formation. | OPCs could efficiently differentiate into oligodendrocytes and form compact myelin in the cerebellar slice. | Myelin thickness and the area of myelinated axons. | [48] |
| GSCs and whole adult brain coronal slice. | To investigate distinct responses of engrafted GSCs to diverse microenvironments in the brain tissue. | Patient-derived GSCs have distinct responses to region-specific adult brain microenvironments. | Cell proliferation, differentiation, and migration. | [47] |
| Mouse spinal cord slices and aortic fragments. | To analyze the mechanisms of interaction between vascular and neural structures. | Nerve tissue has a significant positive effect on aortic sprouting. | Cell ratio and axon growth. | [51] |
3.3. Organoid-Based 3D Co-Culture System
3.3.1. Neuromuscular Co-Culture System
3.3.2. Assembloids: Multi-Organism Co-Culture System
| Seed Cells | System Composition | Objective | Main Results | Advantages | Refs |
|---|---|---|---|---|---|
| hPSC-derived axial stem cells | NMO and NMJ. | To build NMOs and model NMDs. | A functional NMJ was generated in the constructed NMO, and a functional spinal cord network was formed. | Simultaneous differentiation of the mesoderm and ectoderm was achieved. | [56] |
| iPSCs | Motor neuron spheroids and 3D muscle fiber bundles. | To investigate the pathogenesis of ALS. | Formation of NMJ that can control muscle contraction. | Developed a 3D human motor unit model in a microfluidic device. | [57] |
| Human muscle progenitors and hPSCs | Motor neuron endplates and muscle fibers. | To model and evaluate adult human NMJ development or disease in culture. | Human muscle progenitors mixed with motor neurons self-organize to form functional NMJ connections. | Functional connectivity is confirmed with calcium imaging and electrophysiological recordings. | [58] |
| PSCs | 3D spheroids resembling either the dorsal or ventral forebrain. | To recapitulate the saltatory migration of interneurons. | After migration, interneurons functionally integrate with glutamatergic neurons to form a microphysiological system. | The intricate connections between different CNS and local circuits were perfectly reproduced in vitro. | [59] |
| Human ESCs | hThOs and hCOs. | To Understand human thalamic development and model circuit organizations in the brain. | The fusion of the organoid forms a reciprocal projection. | Fused disparate regionally specified human brain organoids. | [60] |
| Human ESCs | Cerebral organoids, mouse spinal cord, and muscle. | To investigate whether brain organoids can produce functional neuronal output. | Cerebral organoids exhibit active neuronal networks and can innervate the mouse spinal cord. | Air-liquid interface culture of cerebral organoids leads to improved survival and maturation. | [62] |
4. Microfluidic Platform-Based Neural-Glial Cell Co-Culture System
5. Using Co-Culture System to Establish Microbial Infected Neural Disease Model
5.1. Using Brain Organoids Co-Culture System to Study Zika Virus-Impaired CNS
5.2. Virus-Brain Organoids Co-Culture System Provides Initial Insights into the Potential Neurotoxic Effects of SARS-CoV-2
6. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Rahman, N.I.A.; Shimizu, A.; Ogita, H. Cell-to-cell contact-mediated regulation of tumor behavior in the tumor microenvironment. Cancer Sci. 2021, 112, 4005–4012. [Google Scholar] [CrossRef] [PubMed]
- Goshi, N.; Morgan, R.K.; Lein, P.J.; Seker, E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J. Neuroinflamm. 2020, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Arora, D.; Gupta, P.; Jaglan, S.; Roullier, C.; Grovel, O.; Bertrand, S. Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnol. Adv. 2020, 40, 107521. [Google Scholar] [CrossRef] [PubMed]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, F.H.; Sharma, P.; Gibbons, A.; Goggans, T.; Erzurum, S.C.; Haque, S.J. Human airway epithelial cell determinants of survival and functional phenotype for primary human mast cells. Proc. Natl. Acad. Sci. USA 2005, 102, 14380–14385. [Google Scholar] [CrossRef]
- MacDonald, K.P.; Rowe, V.; Clouston, A.D.; Welply, J.K.; Kuns, R.D.; Ferrara, J.L.; Thomas, R.; Hill, G.R. Cytokine expanded myeloid precursors function as regulatory antigen-presenting cells and promote tolerance through IL-10-producing regulatory T cells. J. Immunol. 2005, 174, 1841–1850. [Google Scholar] [CrossRef]
- Paschos, N.K.; Brown, W.E.; Eswaramoorthy, R.; Hu, J.C.; Athanasiou, K.A. Advances in tissue engineering through stem cell-based co-culture. J. Tissue Eng. Regen. Med. 2015, 9, 488–503. [Google Scholar] [CrossRef]
- Schmidt, S.I.; Bogetofte, H.; Ritter, L.; Agergaard, J.B.; Hammerich, D.; Kabiljagic, A.A.; Wlodarczyk, A.; Lopez, S.G.; Sørensen, M.D.; Jørgensen, M.L.; et al. Microglia-Secreted Factors Enhance Dopaminergic Differentiation of Tissue- and iPSC-Derived Human Neural Stem Cells. Stem Cell Rep. 2021, 16, 281–294. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Long, Z.; Zeng, L.; Wu, Y. Protoplasmic astrocytes enhance the ability of neural stem cells to differentiate into neurons in vitro. PLoS ONE 2012, 7, e38243. [Google Scholar] [CrossRef]
- Kempuraj, D.; Khan, M.M.; Thangavel, R.; Xiong, Z.; Yang, E.; Zaheer, A. Glia maturation factor induces interleukin-33 release from astrocytes: Implications for neurodegenerative diseases. J. Neuroimmune Pharmacol. 2013, 8, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Abdelfattah-Hassan, A.; Swelum, A.A. Feeder Cell Type Affects the Growth of In Vitro Cultured Bovine Trophoblast Cells. Biomed. Res. Int. 2017, 2017, 1061589. [Google Scholar] [CrossRef] [PubMed]
- Le-Bel, G.; Cortez Ghio, S.; Guérin, L.P.; Bisson, F.; Germain, L.; Guérin, S.L. Irradiated Human Fibroblasts as a Substitute Feeder Layer to Irradiated Mouse 3T3 for the Culture of Human Corneal Epithelial Cells: Impact on the Stability of the Transcription Factors Sp1 and NFI. Int. J. Mol. Sci. 2019, 20, 6296. [Google Scholar] [CrossRef] [PubMed]
- Bongso, T.A.; Fong, C.Y.; Ng, S.C.; Ratnam, S.S. Human ampullary co-cultures for blastocyst transfer in assisted reproduction. Ann. Acad. Med. Singap. 1992, 21, 571–575. [Google Scholar] [PubMed]
- Trettner, S.; Findeisen, A.; Taube, S.; Horn, P.A.; Sasaki, E.; zur Nieden, N.I. Osteogenic induction from marmoset embryonic stem cells cultured in feeder-dependent and feeder-independent conditions. Osteoporos. Int. 2014, 25, 1255–1266. [Google Scholar] [CrossRef]
- López-Fagundo, C.; Livi, L.L.; Ramchal, T.; Darling, E.M.; Hoffman-Kim, D. A biomimetic synthetic feeder layer supports the proliferation and self-renewal of mouse embryonic stem cells. Acta Biomater. 2016, 39, 55–64. [Google Scholar] [CrossRef]
- Rathinam, M.L.; Watts, L.T.; Narasimhan, M.; Riar, A.K.; Mahimainathan, L.; Henderson, G.I. Astrocyte mediated protection of fetal cerebral cortical neurons from rotenone and paraquat. Environ. Toxicol. Pharmacol. 2012, 33, 353–360. [Google Scholar] [CrossRef]
- Guo, H.; Ma, J.; Tong, Y.; Qu, Y.; Mu, D.Z.; Mao, M. A comparative study on three models of co-culture of neurons and astrocytes. Chin. J. Contemp. Pediatr. 2010, 12, 984–987. [Google Scholar]
- Appelt-Menzel, A.; Cubukova, A.; Metzger, M. Establishment of a Human Blood-Brain Barrier Co-Culture Model Mimicking the Neurovascular Unit Using Induced Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2018, 47, e62. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell. Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.T.; Daly, K.A.; Brennan-Pierce, E.P.; Johnson, S.A.; Carruthers, C.A.; D′Amore, A.; Nagarkar, S.P.; Velankar, S.S.; Badylak, S.F. A hydrogel derived from decellularized dermal extracellular matrix. Biomaterials 2012, 33, 7028–7038. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, C. Composition and Mechanism of Three-Dimensional Hydrogel System in Regulating Stem Cell Fate. Tissue Eng. Part B Rev. 2020, 26, 498–518. [Google Scholar] [CrossRef]
- Absalan, F.; Pasandi, M.S.; Ghasemi Hamidabadi, H.; Saeednia, S.; Bojnordi, M.N.; Zahiri, M.; Alizadeh, R.; Bagher, Z. Matrigel enhances differentiation of human adipose tissue-derived stem cells into dopaminergic neuron. Neurosci. Lett. 2021, 760, 136070. [Google Scholar] [CrossRef]
- Yan, W.; Liu, W.; Qi, J.; Fang, Q.; Fan, Z.; Sun, G.; Han, Y.; Zhang, D.; Xu, L.; Wang, M.; et al. A Three-Dimensional Culture System with Matrigel Promotes Purified Spiral Ganglion Neuron Survival and Function In Vitro. Mol. Neurobiol. 2018, 55, 2070–2084. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, Y.; Zhang, H.; Zhang, Q.; Zhao, Y.; Xiao, Z.; Liu, W.; Chen, B.; Gao, L.; Sun, Z.; et al. Small molecules combined with collagen hydrogel direct neurogenesis and migration of neural stem cells after spinal cord injury. Biomaterials 2021, 269, 120479. [Google Scholar] [CrossRef]
- Moxon, S.R.; Corbett, N.J.; Fisher, K.; Potjewyd, G.; Domingos, M.; Hooper, N.M. Blended alginate/collagen hydrogels promote neurogenesis and neuronal maturation. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 104, 109904. [Google Scholar] [CrossRef]
- Alvarez-Primo, F.; Anil Kumar, S.; Manciu, F.S.; Joddar, B. Fabrication of Surfactant-Dispersed HiPco Single-Walled Carbon Nanotube-Based Alginate Hydrogel Composites as Cellular Products. Int. J. Mol. Sci. 2019, 20, 4802. [Google Scholar] [CrossRef]
- Ye, L.; Ji, H.; Liu, J.; Tu, C.H.; Kappl, M.; Koynov, K.; Vogt, J.; Butt, H.J. Carbon Nanotube-Hydrogel Composites Facilitate Neuronal Differentiation While Maintaining Homeostasis of Network Activity. Adv. Mater. 2021, 33, e2102981. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, J.; Liu, C.; Zhang, S.; Gao, F.; Guo, W.; Sun, X.; Zhang, C.; Li, H.; Rao, Z.; et al. Understanding the role of tissue-specific decellularized spinal cord matrix hydrogel for neural stem/progenitor cell microenvironment reconstruction and spinal cord injury. Biomaterials 2021, 268, 120596. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Xu, P.; Guan, J.; Zhang, C.; Chang, J.; Yang, F.; Xiao, H.; Sun, H.; Zhang, Z.; Wang, M.; et al. Promoting 3D neuronal differentiation in hydrogel for spinal cord regeneration. Colloids Surf. B: Biointerfaces 2020, 194, 111214. [Google Scholar] [CrossRef] [PubMed]
- Stoppini, L.; Buchs, P.A.; Muller, D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 1991, 37, 173–182. [Google Scholar] [CrossRef]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell. Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell. Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Involvement of Rac and Rho signaling in cancer cell motility in 3D substrates. Oncogene 2009, 28, 1570–1583. [Google Scholar] [CrossRef]
- Hirata, E.; Yukinaga, H.; Kamioka, Y.; Arakawa, Y.; Miyamoto, S.; Okada, T.; Sahai, E.; Matsuda, M. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell. Sci. 2012, 125, 858–868. [Google Scholar] [CrossRef]
- Guilluy, C.; Garcia-Mata, R.; Burridge, K. Rho protein crosstalk: Another social network? Trends Cell. Biol. 2011, 21, 718–726. [Google Scholar] [CrossRef]
- Humpel, C. Organotypic brain slice cultures: A review. Neuroscience 2015, 305, 86–98. [Google Scholar] [CrossRef]
- Wang, S.; Wang, B.; Pan, N.; Fu, L.; Wang, C.; Song, G.; An, J.; Liu, Z.; Zhu, W.; Guan, Y.; et al. Differentiation of human induced pluripotent stem cells to mature functional Purkinje neurons. Sci. Rep. 2015, 5, 9232. [Google Scholar] [CrossRef]
- Xiao, L.; Ide, R.; Saiki, C.; Kumazawa, Y.; Okamura, H. Human Dental Pulp Cells Differentiate toward Neuronal Cells and Promote Neuroregeneration in Adult Organotypic Hippocampal Slices In Vitro. Int. J. Mol. Sci. 2017, 18, 1745. [Google Scholar] [CrossRef]
- Gonmanee, T.; Sritanaudomchai, H.; Vongsavan, K.; Faisaikarm, T.; Songsaad, A.; White, K.L.; Thonabulsombat, C. Neuronal differentiation of dental pulp stem cells from human permanent and deciduous teeth following coculture with rat auditory brainstem slices. Anat. Rec. (Hoboken) 2020, 303, 2931–2946. [Google Scholar] [CrossRef] [PubMed]
- Lassiter, C.M.; Gal, J.S.; Becker, S.; Hartman, N.W.; Grabel, L. Embryonic stem cell-derived neural progenitors transplanted to the hippocampus migrate on host vasculature. Stem Cell Res. 2016, 16, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, S.; Sypecka, J.; Jablonska, A.; Strojek, L.; Wielgos, M.; Domanska-Janik, K.; Sarnowska, A. Neuroprotective Potential and Paracrine Activity of Stromal Vs. Culture-Expanded hMSC Derived from Wharton Jelly under Co-Cultured with Hippocampal Organotypic Slices. Mol. Neurobiol. 2018, 55, 6021–6036. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Saiki, C.; Okamura, H. Oxidative Stress-Tolerant Stem Cells from Human Exfoliated Deciduous Teeth Decrease Hydrogen Peroxide-Induced Damage in Organotypic Brain Slice Cultures from Adult Mice. Int. J. Mol. Sci. 2019, 20, 1858. [Google Scholar] [CrossRef] [PubMed]
- Marques-Torrejon, M.A.; Gangoso, E.; Pollard, S.M. Modelling glioblastoma tumour-host cell interactions using adult brain organotypic slice co-culture. Dis. Model. Mech. 2018, 11, dmm031435. [Google Scholar] [CrossRef]
- Baudouin, L.; Adès, N.; Kanté, K.; Czarnecki, A.; Bachelin, C.; Baskaran, A.; Langui, D.; Millécamps, A.; Gurchenkov, B.; Velut, Y.; et al. Co-culture of exogenous oligodendrocytes with unmyelinated cerebella: Revisiting ex vivo models and new tools to study myelination. Glia 2021, 69, 1916–1931. [Google Scholar] [CrossRef]
- Sekizar, S.; Williams, A. Ex Vivo Slice Cultures to Study Myelination, Demyelination, and Remyelination in Mouse Brain and Spinal Cord. Methods Mol. Biol. 2019, 1936, 169–183. [Google Scholar]
- Pinkernelle, J.; Fansa, H.; Ebmeyer, U.; Keilhoff, G. Prolonged minocycline treatment impairs motor neuronal survival and glial function in organotypic rat spinal cord cultures. PLoS ONE 2013, 8, e73422. [Google Scholar] [CrossRef]
- Mikhailova, M.M.; Panteleyev, A.A., Jr.; Paltsev, M.A.; Panteleyev, A.A. Spinal cord tissue affects sprouting from aortic fragments in ex vivo co-culture. Cell Biol. Int. 2019, 43, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, H.; Ming, G.L. Brain organoids: Advances, applications and challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.G. Congenital Myasthenic Syndromes in 2018. Curr. Neurol. Neurosci. Rep. 2018, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Nicolau, S.; Kao, J.C.; Liewluck, T. Trouble at the junction: When myopathy and myasthenia overlap. Muscle Nerve 2019, 60, 648–657. [Google Scholar] [CrossRef]
- Gonzalez-Freire, M.; de Cabo, R.; Studenski, S.A.; Ferrucci, L. The Neuromuscular Junction: Aging at the Crossroad between Nerves and Muscle. Front. Aging Neurosci. 2014, 6, 208. [Google Scholar] [CrossRef]
- Faustino Martins, J.M.; Fischer, C.; Urzi, A.; Vidal, R.; Kunz, S.; Ruffault, P.L.; Kabuss, L.; Hube, I.; Gazzerro, E.; Birchmeier, C.; et al. Self-Organizing 3D Human Trunk Neuromuscular Organoids. Cell Stem Cell 2020, 26, 172–186.e6. [Google Scholar] [CrossRef]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 2018, 4, eaat5847. [Google Scholar] [CrossRef]
- Afshar Bakooshli, M.; Lippmann, E.S.; Mulcahy, B.; Iyer, N.; Nguyen, C.T.; Tung, K.; Stewart, B.A.; van den Dorpel, H.; Fuehrmann, T.; Shoichet, M.; et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. eLife 2019, 8, e44530. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Cakir, B.; Patterson, B.; Kim, K.Y.; Sun, P.; Kang, Y.J.; Zhong, M.; Liu, X.; Patra, P.; et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 2019, 24, 487–497.e7. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Patterson, B.; Kang, Y.J.; Govindaiah, G.; Roselaar, N.; Cakir, B.; Kim, K.Y.; Lombroso, A.P.; Hwang, S.M.; et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21, 383–398.e7. [Google Scholar] [CrossRef] [PubMed]
- Giandomenico, S.L.; Mierau, S.B.; Gibbons, G.M.; Wenger, L.M.D.; Masullo, L.; Sit, T.; Sutcliffe, M.; Boulanger, J.; Tripodi, M.; Derivery, E.; et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019, 22, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Park, S.J.; Ha, T.; Lee, S.N.; Cho, H.Y.; Choi, J.W. Electrophysiological Monitoring of Neurochemical-Based Neural Signal Transmission in a Human Brain-Spinal Cord Assembloid. ACS Sens. 2022, 7, 409–414. [Google Scholar] [CrossRef]
- de Jongh, R.; Spijkers, X.M.; Pasteuning-Vuhman, S.; Vulto, P.; Pasterkamp, R.J. Neuromuscular junction-on-a-chip: ALS disease modeling and read-out development in microfluidic devices. J. Neurochem. 2021, 157, 393–412. [Google Scholar] [CrossRef]
- Natarajan, A.; Sethumadhavan, A.; Krishnan, U.M. Toward Building the Neuromuscular Junction: In Vitro Models to Study Synaptogenesis and Neurodegeneration. ACS Omega 2019, 4, 12969–12977. [Google Scholar] [CrossRef]
- Habibey, R.; Rojo Arias, J.E.; Striebel, J.; Busskamp, V. Microfluidics for Neuronal Cell and Circuit Engineering. Chem Rev. 2022, 122, 14842–14880. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef]
- Taylor, A.M.; Dieterich, D.C.; Ito, H.T.; Kim, S.A.; Schuman, E.M. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron 2010, 66, 57–68. [Google Scholar] [CrossRef]
- De Vitis, E.; La Pesa, V.; Gervaso, F.; Romano, A.; Quattrini, A.; Gigli, G.; Moroni, L.; Polini, A. A microfabricated multi-compartment device for neuron and Schwann cell differentiation. Sci. Rep. 2021, 11, 7019. [Google Scholar] [CrossRef]
- Adriani, G.; Ma, D.; Pavesi, A.; Kamm, R.D.; Goh, E.L. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab Chip 2017, 17, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Zahavi, E.E.; Ionescu, A.; Gluska, S.; Gradus, T.; Ben-Yaakov, K.; Perlson, E. A compartmentalized microfluidic neuromuscular co-culture system reveals spatial aspects of GDNF functions. J. Cell. Sci. 2015, 128, 1241–1252. [Google Scholar] [PubMed]
- Kunze, A.; Meissner, R.; Brando, S.; Renaud, P. Co-pathological connected primary neurons in a microfluidic device for Alzheimer studies. Biotechnol. Bioeng. 2011, 108, 2241–2245. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Wetzel, I.; Marriott, I.; Dréau, D.; D’Avanzo, C.; Kim, D.Y.; Tanzi, R.E.; Cho, H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018, 21, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Berdichevsky, Y.; Staley, K.J.; Yarmush, M.L. Building and manipulating neural pathways with microfluidics. Lab Chip 2010, 10, 999–1004. [Google Scholar] [CrossRef]
- Musso, D.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef]
- Garcez, P.P.; Loiola, E.C.; Madeiro da Costa, R.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef]
- Bullard-Feibelman, K.M.; Govero, J.; Zhu, Z.; Salazar, V.; Veselinovic, M.; Diamond, M.S.; Geiss, B.J. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017, 137, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Xie, J.; Ding, C.; Li, J.; Wang, Y.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C.; Jin, T.; Gao, Y.; et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J. Med. Virol. 2020, 92, 2004–2010. [Google Scholar] [CrossRef]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27, 951–961.e5. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.H.; Sun, A.; Je, H.S.; Tan, E.K. Unravelling Pathophysiology of Neurological and Psychiatric Complications of COVID-19 Using Brain Organoids. Neuroscientist 2021. [Google Scholar] [CrossRef]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell. Res. 2020, 30, 928–931. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, Y.; Qi, H.; Ma, Q.; Xu, L.; Chen, W.; Chen, G.; Xu, X. A novel brain neurovascular unit model with neurons, astrocytes and microvascular endothelial cells of rat. Int. J. Biol. Sci. 2013, 9, 174–189. [Google Scholar] [CrossRef]
- Ikemoto, S. Dopamine reward circuitry: Two projection systems from the ventral midbrain to the nucleus accumbens-olfactory tubercle complex. Brain Res. Rev. 2007, 56, 27–78. [Google Scholar] [CrossRef]
- Liu, S.; Tian, Z.; Yin, F.; Zhao, Q.; Fan, M. Generation of dopaminergic neurons from human fetal mesencephalic progenitors after co-culture with striatal-conditioned media and exposure to lowered oxygen. Brain Res. Bull. 2009, 80, 62–68. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, T.T.; Wu, C.X.; Lan, X.; Du, G.H. Targeting the neurovascular unit: Development of a new model and consideration for novel strategy for Alzheimer’s disease. Brain Res. Bull. 2011, 86, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Tohda, C.; Kuboyama, T. Current and future therapeutic strategies for functional repair of spinal cord injury. Pharmacol. Ther. 2011, 132, 57–71. [Google Scholar] [CrossRef] [PubMed]

| Co-Cultured Cell Type | 3D Construction | Effect on Cell-Interaction | Investigations | Refs |
|---|---|---|---|---|
| Human adult MSCs | Matrigel | Matrigel promotes cell growth and differentiation. | Cell survival rate. | [25] |
| SGNs | Matrigel | Matrigel promotes the survival of purified SGNs in vitro and maintained their morphological structure and function. | Neurite length. | [26] |
| NSCs | collagen hydrogel | Collagen hydrogel increases neuronal differentiation of NSCs and induces their migration. | Number of different types of cells, electrophysiological evaluation, and behavioral assessments. | [27] |
| iPSCs | Blended alginate/collagen hydrogels | The hydrogel matrix promoted neuronal differentiation and maturation. | Cell morphology and synaptophysin density. | [28] |
| NSCs and bone MSCs | gelatin methacryloyl | The gelatin methacryloyl promoted the generation of neurons and oligodendrocytes. | The percentage of live cells. | [32] |
| NSC | CNT | CNT could facilitate neuronal differentiation while maintaining neuronal homeostasis. | Cell viability and calcium imaging. | [30] |
| NSCs/NPCs | DTM | DSCM-gel promotes NSCs/NPCs proliferation, migration, neuron-like differentiation, and synapse formation. | Number of different types of cells and number of synapses in different periods. | [31] |
| System Composition | Features | Objectives | Advantages | Refs |
|---|---|---|---|---|
| Rat hippocampal neurons | Consists of two main chambers connected by multiple parallel microgrooves. | To visualize synapses. | Synapses originating from cell bodies in one compartment can be identified. | [70] |
| Neuroblastoma cell and primary SCs | Consists of three perfusable compartments with distinct inlets and outlets, interconnected through a series of narrow and parallel microgrooves. | To perform cell differentiation on the chip. | Up to three different cell populations can be cultured in a fluidically independent circuit. | [71] |
| Primary rat astrocytes and neurons and human cerebral microvascular endothelial cells. | Consists of two central 3D hydrogel regions and two media channels. | To assess the influence of the neurovascular microfluidic system on neural cell growth and functionality. | Supports the addition of other cell types present in the neurovasculature such as pericytes and microglia. | [72] |
| Motor neurons and muscle cells. | Consists of two main compartments or channels that are connected by parallel grooves. | To form a functional NMJ in the microfluidic chamber. | Allows an independent manipulation of neuronal or muscle cell populations. | [73] |
| Cortical neurons | Consists of two lateral cell culture channels, 24 junction channels, and the main channel. | To study interactions between healthy and diseased neurons in AD. | Connecting healthy and diseased neurons through local perfusion therapy. | [74] |
| Neuron, astrocyte, and microglia. | Consists of a central chamber and an angular chamber. | To model neurodegeneration and neuroinflammation in AD. | Microglia recruitment was achieved in the microfluidic system. | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Meng, X.; Yu, X.; Wang, G.; Dong, Z.; Zhou, Z.; Qi, M.; Yu, X.; Ji, T.; Wang, F. From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction. Int. J. Mol. Sci. 2022, 23, 13116. https://doi.org/10.3390/ijms232113116
Liu R, Meng X, Yu X, Wang G, Dong Z, Zhou Z, Qi M, Yu X, Ji T, Wang F. From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction. International Journal of Molecular Sciences. 2022; 23(21):13116. https://doi.org/10.3390/ijms232113116
Chicago/Turabian StyleLiu, Rongrong, Xiaoting Meng, Xiyao Yu, Guoqiang Wang, Zhiyong Dong, Zhengjie Zhou, Mingran Qi, Xiao Yu, Tong Ji, and Fang Wang. 2022. "From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction" International Journal of Molecular Sciences 23, no. 21: 13116. https://doi.org/10.3390/ijms232113116
APA StyleLiu, R., Meng, X., Yu, X., Wang, G., Dong, Z., Zhou, Z., Qi, M., Yu, X., Ji, T., & Wang, F. (2022). From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction. International Journal of Molecular Sciences, 23(21), 13116. https://doi.org/10.3390/ijms232113116






