HLGNN-MDA: Heuristic Learning Based on Graph Neural Networks for miRNA–Disease Association Prediction
Abstract
1. Introduction
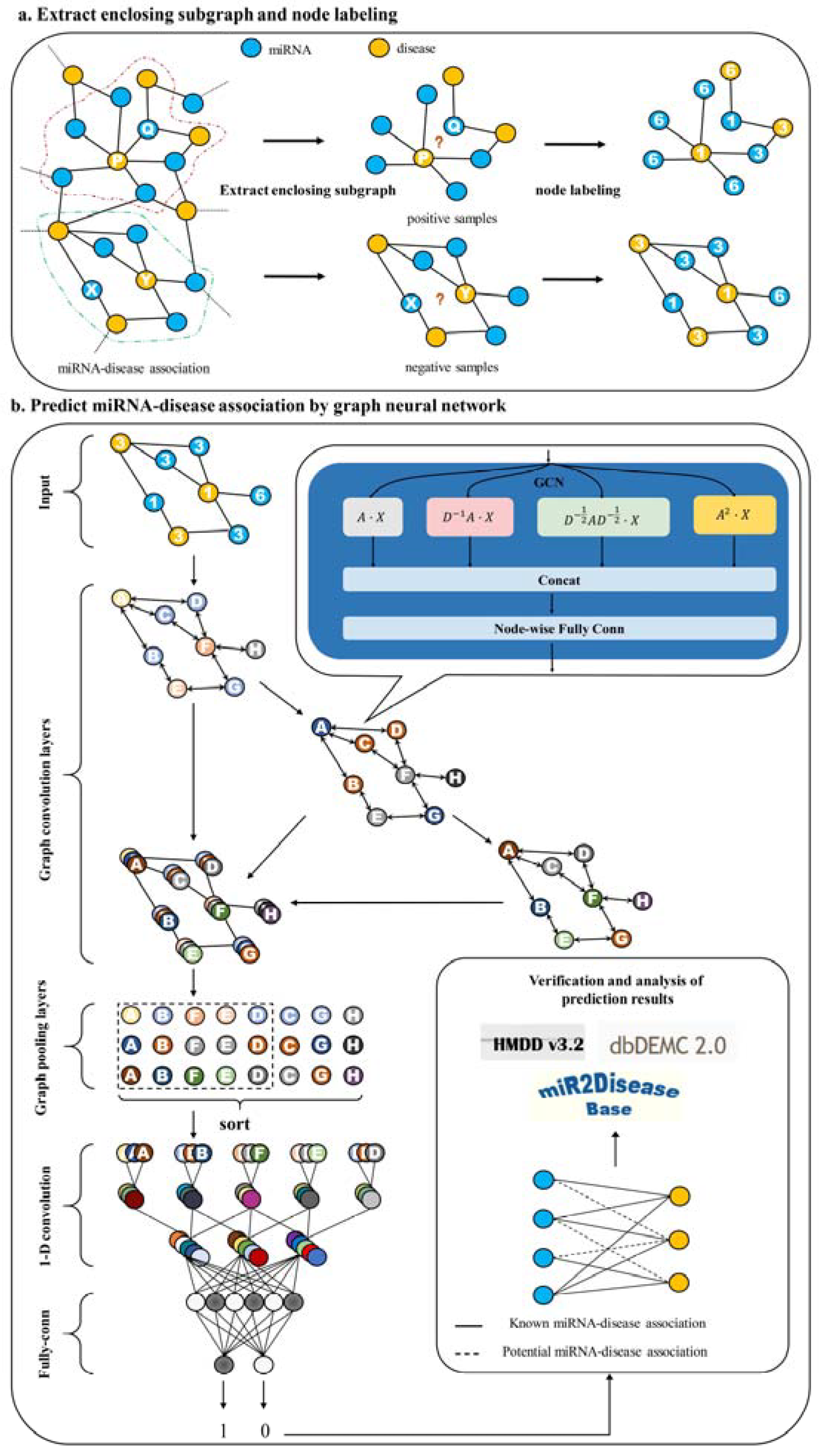
2. Results and Discussion
2.1. Performance Analysis of HLGNN-MDA Mode
2.2. Influence of Different Hops in the Enclosing Subgraph
2.3. Analysis of the Improved Graph Convolutional Layer
2.4. Validation of Prediction Results
2.5. Case Study
2.5.1. Breast Cancer
2.5.2. Hepatocellular Carcinoma
2.5.3. Renal Cell Carcinoma
3. Materials and Methods
3.1. Data Resources
3.2. Methods
3.2.1. Extraction of the Enclosing Subgraph of Node Pair
3.2.2. Label Nodes
3.2.3. Construct Graph Neural Network
3.2.4. Evaluation Metrics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense com-plementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Perez-Rodriguez, D.; Lopez-Fernandez, H.; Agis-Balboa, R.C. Application of miRNA-seq in neuropsychiatry: A method-ological perspective. Comput. Biol. Med. 2021, 135, 104603. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, M.; Zou, Q. Computational biology and chemistry Special section editorial: Computational analyses for miRNA. Comput. Biol. Chem. 2021, 91, 107448. [Google Scholar] [CrossRef]
- Shaker, F.; Nikravesh, A.; Arezumand, R.; Aghaee-Bakhtiari, S.H. Web-based tools for miRNA studies analysis. Comput. Biol. Med. 2020, 127, 104060. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Teng, M.; Zhang, D.; Li, L.; Liu, Y. Signal transducers and activators of transcription-1 (STAT1) regulates microRNA transcription in interferon gamma-stimulated HeLa cells. PLoS ONE 2010, 5, e11794. [Google Scholar]
- Zhao, Y.; Wang, F.; Juan, L. MicroRNA Promoter Identification in Arabidopsis using Multiple Histone Markers. BioMed Res. Int. 2015, 2015, 861402. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, F.; Chen, S.; Wan, J.; Wang, G. Methods of MicroRNA Promoter Prediction and Transcription Factor Mediated Regulatory Network. BioMed Res. Int. 2017, 2017, 7049406. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Liu, J.; Wang, G. The stacking strategy-based hybrid framework for identifying non-coding RNAs. Brief. Bioinform. 2021, 22, bbab023. [Google Scholar] [CrossRef]
- Tétreault, N.; De Guire, V. miRNAs: Their discovery, biogenesis and mechanism of action. Clin. Biochem. 2013, 46, 842–845. [Google Scholar] [CrossRef]
- Tian, L.; Wang, S.L. Exploring miRNA Sponge Networks of Breast Cancer by Combining miRNA-disease-lncRNA and miRNA-target Networks. Curr. Bioinform. 2021, 16, 385–394. [Google Scholar] [CrossRef]
- Han, W.; Lu, D.; Wang, C.; Cui, M.; Lu, K. Identification of Key mRNAs, miRNAs, and mRNA-miRNA Network Involved in Papillary Thyroid Carcinoma. Curr. Bioinform. 2021, 16, 146–153. [Google Scholar] [CrossRef]
- Sarkar, J.P.; Saha, I.; Sarkar, A.; Maulik, U. Machine learning integrated ensemble of feature selection methods followed by survival analysis for predicting breast cancer subtype specific miRNA biomarkers. Comput. Biol. Med. 2021, 131, 104244. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Li, D.; Wang, X.; Li, L.; Zou, Q. Cancer Diagnosis Through IsomiR Expression with Machine Learning Method. Curr. Bioinform. 2018, 13, 57–63. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Zahra, A.; Mumtaz, S.; Fatmi, M.Q.; Khan, M.J. Integrated In-silico Analysis to Study the Role of microRNAs in the Detection of Chronic Kidney Diseases. Curr. Bioinform. 2020, 15, 144–154. [Google Scholar] [CrossRef]
- Porta, C.; Figlin, R.A. MiR-193a-3p and miR-224 mediate renal cell carcinoma progression by targeting al-pha-2,3-sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol. Carcinog. 2019, 58, 1926–1927. [Google Scholar]
- Zhao, Z.; Zhang, C.; Li, M.; Yu, X.; Liu, H.; Chen, Q.; Wang, J.; Shen, S.; Jiang, J. Integrative Analysis of miRNA-mediated Competing Endogenous RNA Network Reveals the lncRNAs-mRNAs Interaction in Glioblastoma Stem Cell Differentiation. Curr. Bioinform. 2020, 15, 1187–1196. [Google Scholar] [CrossRef]
- Zhu, Q.; Fan, Y.; Pan, X. Fusing Multiple Biological Networks to Effectively Predict miRNA-disease Associations. Curr. Bioinform. 2021, 16, 371–384. [Google Scholar] [CrossRef]
- Li, J.Q.; Rong, Z.H.; Chen, X.; Yan, G.Y.; You, Z.H. MCMDA: Matrix completion for MiRNA-disease association prediction. Oncotarget 2017, 8, 21187–21199. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Peng, M.; Liao, B.; Huang, G.; Liang, W.; Li, K. Improved low-rank matrix recovery method for predicting miRNA-disease association. Sci. Rep. 2017, 7, 6007. [Google Scholar] [CrossRef] [PubMed]
- Ha, J. MDMF: Predicting miRNA-Disease Association Based on Matrix Factorization with Disease Similarity Constraint. J. Pers. Med. 2022, 12, 885. [Google Scholar] [CrossRef]
- Chen, X.; Yin, J.; Qu, J.; Huang, L. MDHGI: Matrix Decomposition and Heterogeneous Graph Inference for miRNA-disease association prediction. PLoS Comput. Biol. 2018, 14, e1006418. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Park, C.; Park, S. IMIPMF: Inferring miRNA-disease interactions using probabilistic matrix factorization. J. Biomed. Inform. 2020, 102, 103358. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H.; Deng, L.; Liu, Y.; Zhang, Y.; Dai, Q. WBSMDA: Within and Between Score for MiRNA-Disease Association prediction. Sci. Rep. 2016, 6, 21106. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Park, C. MLMD, MLMD: Metric Learning for predicting miRNA-Disease associations. IEEE Access 2021, 9, 78847–78858. [Google Scholar] [CrossRef]
- Chen, X.; Li, T.H.; Zhao, Y.; Wang, C.C.; Zhu, C.C. Deep-belief network for predicting potential miRNA-disease associations. Brief. Bioinform. 2021, 22, bbaa186. [Google Scholar]
- Zhang, Z.-Y.; Sun, Z.-J.; Yang, Y.-H.; Lin, H. Towards a better prediction of subcellular location of long non-coding RNA. Front. Comput. Sci. 2022, 16, 165903. [Google Scholar] [CrossRef]
- Yang, H.; Luo, Y.; Ren, X.; Wu, M.; He, X.; Peng, B.; Deng, K.; Yan, D.; Tang, H.; Lin, H. Risk Prediction of Diabetes: Big data mining with fusion of multi-farious physical examination indicators. Inf. Fusion 2021, 75, 140–149. [Google Scholar] [CrossRef]
- Lu, X.; Gao, Y.; Zhu, Z.; Ding, L.; Wang, X.; Liu, F.; Li, J. A Constrained Probabilistic Matrix Decomposition Method for Predicting miRNA-disease Associations. Curr. Bioinform. 2021, 16, 524–533. [Google Scholar] [CrossRef]
- Liu, Z.P. Predicting lncRNA-protein Interactions by Machine Learning Methods: A Review. Curr. Bioinform. 2020, 15, 831–840. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, G.; Yan, C.; Yi, H.; Wu, F.X.; Wang, J. MDAPlatform: A Component-based Platform for Constructing and Assessing miRNA-disease Association Prediction Methods. Curr. Bioinform. 2021, 16, 710–721. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Q.; Rodriguez-Paton, A.; Zeng, X. Meta-Path Methods for Prioritizing Candidate Disease miRNAs. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Liu, L.; Lü, L.; Zou, Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics 2018, 34, 2425–2432. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Chu, Y.; Li, Z.; Zhao, Y.; Mao, X.; Wang, Y.; Xiong, Y.; Wei, D.Q. MDA-CF: Predicting MiRNA-Disease associations based on a cascade forest model by fusing multi-source information. Comput. Biol. Med. 2021, 136, 104706. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, G.; Jin, S.; Li, Y.; Wang, Y. Predicting human microRNA-disease associations based on support vector machine. Int. J. Data Min. Bioinform. 2013, 8, 282–293. [Google Scholar] [CrossRef]
- Chen, X.; Clarence Yan, C.; Zhang, X.; Li, Z.; Deng, L.; Zhang, Y.; Dai, Q. RBMMMDA: Predicting multiple types of disease-microRNA associations. Sci. Rep. 2015, 5, 13877. [Google Scholar] [CrossRef]
- Phan, A.V.; Le Nguyen, M.; Nguyen, Y.L.; Bui, L.T. Dgcnn: A convolutional neural network over large-scale labeled graphs. Neural. Netw. 2018, 108, 533–543. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, J.; Chen, S.; Gong, M.; Gao, D.; Zhu, M.; Gan, W. Review of the Applications of Deep Learning in Bioinformatics. Curr. Bioinform. 2021, 15, 898–911. [Google Scholar] [CrossRef]
- Ayachit, G.; Shaikh, I.; Pandya, H.; Das, J. Salient Features, Data and Algorithms for MicroRNA Screening from Plants: A Review on the Gains and Pitfalls of Machine Learning Techniques. Curr. Bioinform. 2021, 15, 1091–1103. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, J.-P. Identification of Carcinogenic Chemicals with Network Embedding and Deep Learning Methods. Curr. Bioinform. 2021, 15, 1017–1026. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Z.; Jiang, Y.; Mao, Z.; Wang, D.; Lin, H.; Xu, D. DM3Loc: Multi-label mRNA subcellular localization prediction and analysis based on multi-head self-attention mechanism. Nucleic Acids Res. 2021, 49, e46. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Dao, F.Y.; Zulfiqar, H.; Lin, H. DeepIPs: Comprehensive assessment and computational identification of phosphorylation sites of SARS-CoV-2 infection using a deep learning-based approach. Brief Bioinform. 2021, 22, bbab244. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Dao, F.-Y.; Zulfiqar, H.; Su, W.; Ding, H.; Liu, L.; Lin, H. A sequence-based deep learning approach to predict CTCF-mediated chromatin loop. Brief. Bioinform. 2021, 22, bbab031. [Google Scholar] [CrossRef]
- Fu, L.; Peng, Q. A deep ensemble model to predict miRNA-disease association. Sci. Rep. 2017, 7, 14482. [Google Scholar] [CrossRef]
- Geete, K.; Pandey, M. Robust Transcription Factor Binding Site Prediction Using Deep Neural Networks. Curr. Bioinform. 2020, 15, 1137–1152. [Google Scholar] [CrossRef]
- Peng, J.; Hui, W.; Li, Q.; Chen, B.; Hao, J.; Jiang, Q.; Shang, X.; Wei, Z. A learning-based framework for miRNA-disease association identification using neural networks. Bioinformatics 2019, 35, 4364–4371. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, H.; Lin, L.; Wang, H.; Gao, Y.; Zhao, L.; Chen, Y.-P.P.; Chen, R.; Gu, L. A Similarity Searching System for Biological Phenotype Images Using Deep Convolutional Encoder-decoder Architecture. Curr. Bioinform. 2019, 14, 628–639. [Google Scholar] [CrossRef]
- Chu, Y.; Wang, X.; Dai, Q.; Wang, Y.; Wang, Q.; Peng, S.; Wei, X.; Qiu, J.; Salahub, D.R.; Xiong, Y.; et al. MDA-GCNFTG: Identifying miRNA-disease associations based on graph convolutional networks via graph sampling through the feature and topology graph. Brief. Bioinform. 2021, 22, bbab165. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, J.-Y.; Yin, J. Predicting microRNA-disease associations using bipartite local models and hubness-aware regression. RNA Biol. 2018, 15, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, L.; Qu, J.; Guan, N.N.; Li, J.Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics 2018, 34, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Che, K.; Guo, M.; Wang, C.; Liu, X.; Chen, X. Predicting MiRNA-Disease Association by Latent Feature Extraction with Positive Samples. Genes 2019, 10, 80. [Google Scholar] [CrossRef]
- Chen, X.; Niu, Y.W.; Wang, G.H.; Yan, G.Y. MKRMDA: Multiple kernel learning-based Kronecker regularized least squares for MiRNA-disease association prediction. J. Transl. Med. 2017, 15, 251. [Google Scholar] [CrossRef]
- Zhang, M.; Cui, Z.; Neumann, M.; Chen, Y. An End-to-End Deep Learning Architecture for Graph Classification. Proc. Conf. AAAI Artif. Intell. 2018, 32, 4438–4445. [Google Scholar] [CrossRef]
- Du, X.; Yao, Y. ConvsPPIS: Identifying Protein-protein Interaction Sites by an Ensemble Convolutional Neural Network with Feature Graph. Curr. Bioinform. 2020, 15, 368–378. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, L.; Wang, A.; Tang, W.; Zhao, Y.; Zhao, H.; Teschendorff, A.E. dbDEMC 2.0: Updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2016, 45, D812–D818. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef]
- Tao, Z.; Shi, A.; Lu, C.; Song, T.; Zhang, Z.; Zhao, J. Breast Cancer: Epidemiology and Etiology. Cell Biophys. 2014, 72, 333–338. [Google Scholar] [CrossRef]
- Assiri, A.A.; Mourad, N.; Shao, M.; Kiel, P.; Liu, W.; Skaar, T.C.; Overholser, B.R. MicroRNA 362-3p Reduces hERG-related Current and Inhibits Breast Cancer Cells Proliferation. Cancer Genom. Proteom. 2019, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- NI, F.; Gui, Z.; Guo, Q.; Hu, Z.; Wang, X.; Chen, D.; Wang, S. Downregulation of miR-362-5p inhibits proliferation, migration and invasion of human breast cancer MCF7 cells. Oncol. Lett. 2015, 11, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, B.H.; Rudolph, L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Jeyaram, C.; Philip, M.; Perumal, R.C.; Benny, J.; Jayakumari, J.M.; Ramasamy, M.S. A Computational Approach to Identify Novel Potential Precursor miRNAs and their Targets from Hepatocellular Carcinoma Cells. Curr. Bioinform. 2019, 14, 24–32. [Google Scholar] [CrossRef]
- Ye, Y.; Zhuang, J.; Wang, G.; He, S.; Zhang, S.; Wang, G.; Ni, J.; Wang, J.; Xia, W. MicroRNA-495 suppresses cell proliferation and invasion of hepatocellular carcinoma by directly targeting insulin-like growth factor receptor-1. Exp. Ther. Med. 2018, 15, 1150–1158. [Google Scholar]
- Ljungberg, B.; Campbell, S.C.; Cho, H.Y.; Jacqmin, D.; Lee, J.E.; Weikert, S.; Kiemeney, L.A. The Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2011, 60, 1317. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human mi-croRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y. Link prediction based on graph neural networks. In Advances in Neural Information Processing Systems 31, Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS 2018), Montreal, QC, Canada, 3–8 December 2018; Neural Information Processing Systems Foundation, Inc.: San Diego, CA, USA, 2018. [Google Scholar]
- Thomas, N.; Kipf, M.W. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Advances in Neural Information Processing Systems 30, Proceedings of the 31st Annual Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Neural Information Processing Systems Foundation, Inc.: San Diego, CA, USA, 2017; pp. 1025–1035. [Google Scholar]
- Dehmamy, N.; Barabási, A.L.; Yu, R. Understanding the Representation Power of Graph Neural Networks in Learning Graph Topology. In Advances in Neural Information Processing Systems 32, Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Neural Information Processing Systems Foundation, Inc.: San Diego, CA, USA, 2019. [Google Scholar]
- Lan, Y.; Li, Q. Supervised Learning in Spiking Neural Networks with Synaptic Delay Plasticity: An Overview. Curr. Bioinform. 2020, 15, 854–865. [Google Scholar] [CrossRef]








| Model | ACC | Precision | Recall | AUROC | AUPR | MCC |
|---|---|---|---|---|---|---|
| BNPMDA | 0.79088 | 0.87069 | 0.68324 | 0.85648 | 0.88275 | 0.59574 |
| IMCMDA | 0.77274 | 0.80102 | 0.72578 | 0.84004 | 0.84989 | 0.54791 |
| LFEMDA | 0.84751 | 0.85590 | 0.83573 | 0.90039 | 0.91289 | 0.69522 |
| BLHARMDA | 0.85442 | 0.85619 | 0.85193 | 0.92838 | 0.92699 | 0.70885 |
| MKRMDA | 0.84549 | 0.87610 | 0.80479 | 0.89658 | 0.91971 | 0.69328 |
| HLGNN-MDA-hop1 | 0.85442 | 0.86263 | 0.84309 | 0.92974 | 0.92779 | 0.70902 |
| HLGNN-MDA-hop2 | 0.85976 | 0.85917 | 0.86059 | 0.92833 | 0.92927 | 0.71952 |
| HLGNN-MDA-hop3 | 0.85635 | 0.86745 | 0.84125 | 0.92863 | 0.93007 | 0.71303 |
| HLGNN-MDA-hop4 | 0.85912 | 0.86709 | 0.84825 | 0.93086 | 0.93247 | 0.71840 |
| Model | ACC | Precision | Recall | AUROC | AUPR | MCC |
|---|---|---|---|---|---|---|
| HLGNN-MDA-hop1 | 0.88122 | 0.90430 | 0.85267 | 0.93535 | 0.93281 | 0.76368 |
| HLGNN-MDA-hop2 | 0.92726 | 0.93939 | 0.91344 | 0.97212 | 0.97564 | 0.85484 |
| HLGNN-MDA-hop3 | 0.93831 | 0.95946 | 0.91529 | 0.97266 | 0.97744 | 0.87754 |
| HLGNN-MDA-hop4 | 0.96869 | 0.96690 | 0.97053 | 0.99178 | 0.99332 | 0.93739 |
| Model | ACC | Precision | Recall | AUROC | AUPR | MCC |
|---|---|---|---|---|---|---|
| HLGNN-MDA-a-hop1 | 0.85820 | 0.91121 | 0.79374 | 0.92795 | 0.92303 | 0.72242 |
| HLGNN-MDA-a-hop2 | 0.90055 | 0.90503 | 0.89503 | 0.94681 | 0.94629 | 0.80115 |
| HLGNN-MDA-a-hop3 | 0.94843 | 0.94516 | 0.95212 | 0.98538 | 0.98626 | 0.89689 |
| HLGNN-MDA-a-hop4 | 0.93186 | 0.95183 | 0.90976 | 0.97535 | 0.97945 | 0.86456 |
| HLGNN-MDA-b-hop1 | 0.86096 | 0.90164 | 0.81031 | 0.93369 | 0.93412 | 0.72565 |
| HLGNN-MDA-b-hop2 | 0.87845 | 0.92371 | 0.82505 | 0.93721 | 0.94106 | 0.76126 |
| HLGNN-MDA-b-hop3 | 0.89042 | 0.91569 | 0.86004 | 0.94265 | 0.94575 | 0.78229 |
| HLGNN-MDA-b-hop4 | 0.88858 | 0.90734 | 0.86556 | 0.94257 | 0.94877 | 0.77799 |
| HLGNN-MDA-c-hop1 | 0.85635 | 0.84127 | 0.87845 | 0.92729 | 0.92241 | 0.71340 |
| HLGNN-MDA-c-hop2 | 0.92265 | 0.91652 | 0.93002 | 0.96673 | 0.96729 | 0.84540 |
| HLGNN-MDA-c-hop3 | 0.88398 | 0.92464 | 0.83610 | 0.93691 | 0.94300 | 0.77150 |
| HLGNN-MDA-c-hop4 | 0.93923 | 0.96311 | 0.91344 | 0.97610 | 0.97901 | 0.87962 |
| HLGNN-MDA-d-hop1 | 0.86280 | 0.89879 | 0.81768 | 0.92999 | 0.92586 | 0.72857 |
| HLGNN-MDA-d-hop2 | 0.93831 | 0.94238 | 0.93370 | 0.98012 | 0.98081 | 0.87665 |
| HLGNN-MDA-d-hop3 | 0.87569 | 0.92324 | 0.81952 | 0.93266 | 0.94060 | 0.75617 |
| HLGNN-MDA-d-hop4 | 0.94015 | 0.95437 | 0.92449 | 0.98585 | 0.98702 | 0.88073 |
| DGCNN-hop1 | 0.85820 | 0.88822 | 0.81952 | 0.92889 | 0.92889 | 0.71854 |
| DGCNN-hop2 | 0.87201 | 0.90400 | 0.83241 | 0.93509 | 0.93831 | 0.74636 |
| DGCNN-hop3 | 0.88582 | 0.90522 | 0.86188 | 0.94241 | 0.94083 | 0.88302 |
| DGCNN-hop4 | 0.89411 | 0.91961 | 0.86372 | 0.95250 | 0.95707 | 0.89079 |
| Rank | MicroRNA | Validation | Rank | MicroRNA | Validation |
|---|---|---|---|---|---|
| 1 | hsa-mir-211 | yes <H, D> | 26 | hsa-mir-30e | yes <H, D> |
| 2 | hsa-mir-186 | yes <D> | 27 | hsa-mir-494 | yes <H, D> |
| 3 | hsa-mir-744 | yes <H, D> | 28 | hsa-mir-421 | yes <H, D> |
| 4 | hsa-mir-138 | yes <H, D> | 29 | hsa-mir-501 | yes <H, D> |
| 5 | hsa-mir-154 | yes <D> | 30 | hsa-mir-99b | yes <H, D> |
| 6 | hsa-mir-216b | yes <H, D> | 31 | hsa-mir-196b | yes <H, D> |
| 7 | hsa-mir-106a | yes <H, D> | 32 | hsa-mir-185 | yes <H, D> |
| 8 | hsa-mir-432 | yes <H, D> | 33 | hsa-mir-484 | yes <H, D> |
| 9 | hsa-mir-32 | yes <H, D> | 34 | hsa-mir-144 | yes <H, D> |
| 10 | hsa-mir-381 | yes <H, D> | 35 | hsa-mir-592 | yes <H, D> |
| 11 | hsa-mir-142 | yes <H, D> | 36 | hsa-mir-130a | yes <H, D> |
| 12 | hsa-mir-150 | yes <H, D> | 37 | hsa-mir-542 | yes <H, D> |
| 13 | hsa-mir-491 | yes <H, D> | 38 | hsa-mir-1224 | yes <H, D> |
| 14 | hsa-mir-449a | yes <H, D> | 39 | hsa-mir-376a | yes <H, D> |
| 15 | hsa-mir-362 | no | 40 | hsa-mir-451 | yes <H, D, M> |
| 16 | hsa-mir-28 | yes <H, D> | 41 | hsa-mir-433 | yes <H, D> |
| 17 | hsa-mir-378a | yes <H, D> | 42 | hsa-mir-483 | yes <H, D> |
| 18 | hsa-mir-212 | yes <H, D> | 43 | hsa-mir-1207 | yes <H, D> |
| 19 | hsa-mir-98 | yes <H, D, M> | 44 | hsa-mir-33b | yes <H, D> |
| 20 | hsa-mir-92b | yes <H, D> | 45 | hsa-mir-15b | yes <H, D> |
| 21 | hsa-mir-455 | yes <H, D> | 46 | hsa-mir-630 | yes <H, D> |
| 22 | hsa-mir-590 | yes <H, D> | 47 | hsa-mir-622 | yes <H, D> |
| 23 | hsa-mir-330 | yes <H, D> | 48 | hsa-mir-1271 | yes <H, D> |
| 24 | hsa-mir-675 | yes <H, D> | 49 | hsa-mir-424 | yes <H, D> |
| 25 | hsa-mir-217 | yes <H, D> | 50 | hsa-mir-95 | yes <H, D> |
| Rank | MicroRNA | Validation | Rank | MicroRNA | Validation |
|---|---|---|---|---|---|
| 1 | hsa-mir-143 | yes <H, D, M> | 26 | hsa-mir-23b | yes <H, D, M> |
| 2 | hsa-mir-196b | yes <H, D> | 27 | hsa-mir-574 | yes <H, D> |
| 3 | hsa-mir-137 | yes <H, D, M> | 28 | hsa-mir-26b | yes <H, D, M> |
| 4 | hsa-mir-520c | yes <H, D> | 29 | hsa-mir-495 | no |
| 5 | hsa-mir-376c | yes <H, D> | 30 | hsa-mir-328 | yes <H, D, M> |
| 6 | hsa-mir-184 | yes <H, D> | 31 | hsa-mir-452 | yes <H, D> |
| 7 | hsa-mir-215 | yes <H, D, M> | 32 | hsa-mir-204 | yes <H, D> |
| 8 | hsa-mir-302a | yes <H, D> | 33 | hsa-mir-135b | yes <H, D> |
| 9 | hsa-mir-34b | yes <H, D> | 34 | hsa-mir-95 | yes <H, D> |
| 10 | hsa-mir-339 | yes <H, D> | 35 | hsa-mir-185 | yes <H, D, M> |
| 11 | hsa-mir-708 | yes <H, D> | 36 | hsa-mir-206 | yes <H, D> |
| 12 | hsa-mir-193 | yes <H, D> | 37 | hsa-mir-449a | yes <H, D> |
| 13 | hsa-mir-30e | yes <H, D, M> | 38 | hsa-mir-520a | yes <H, D> |
| 14 | hsa-mir-488 | yes <H, D> | 39 | hsa-mir-194 | yes <H, D, M> |
| 15 | hsa-mir-200 | yes <H, M> | 40 | hsa-mir-451 | yes <H, D> |
| 16 | hsa-mir-342 | yes <H, D> | 41 | hsa-mir-149 | yes <H, D> |
| 17 | hsa-mir-367 | yes <H, D> | 42 | hsa-mir-153 | yes <H, D> |
| 18 | hsa-mir-302d | yes <H, D> | 43 | hsa-mir-299 | yes <H, D> |
| 19 | hsa-mir-494 | yes <H, D> | 44 | hsa-mir133a | yes <H, D, M> |
| 20 | hsa-mir-128 | yes <H, D, M> | 45 | hsa-mir-633 | yes <D> |
| 21 | hsa-mir-340 | yes <H, D> | 46 | hsa-mir-132 | yes <H, D, M> |
| 22 | hsa-mir-33b | yes <H, D> | 47 | hsa-mir-27b | yes <H, D> |
| 23 | hsa-mir-625 | yes <H, D> | 48 | hsa-mir-935 | yes <H, D> |
| 24 | hsa-mir-424 | yes <H, D> | 49 | hsa-mir-32 | yes <H, D> |
| 25 | hsa-mir-151b | yes <H, D> | 50 | hsa-mir-186 | yes <H, D, M> |
| Rank | MicroRNA | Validation | Rank | MicroRNA | Validation |
|---|---|---|---|---|---|
| 1 | hsa-mir-20a | yes <H, D, M> | 26 | hsa-mir-181a | yes <H, D> |
| 2 | hsa-mir-17 | yes <H, D, M> | 27 | hsa-mir-192 | yes <H, D> |
| 3 | hsa-mir-27b | yes <H, D> | 28 | hsa-mir-22 | yes <H, D> |
| 4 | hsa-mir-221 | yes <H, D, M> | 29 | hsa-mir-182 | yes <H, D, M> |
| 5 | hsa-mir-223 | yes <H, D, M> | 30 | hsa-mir-29b | yes <H, D, M> |
| 6 | hsa-mir-31 | yes <H, D> | 31 | hsa-mir-15a | yes <H, D, M> |
| 7 | hsa-mir-29a | yes <H, D, M> | 32 | hsa-mir-375 | yes <H, D> |
| 8 | hsa-mir-125b | yes <H, D> | 33 | hsa-mir-486 | yes <D> |
| 9 | hsa-mir-133a | yes <H, D, M> | 34 | hsa-mir-15b | yes <H, D> |
| 10 | hsa-mir-125a | yes <H, D> | 35 | hsa-mir-107 | yes <H, D> |
| 11 | hsa-mir-18a | yes <H, D> | 36 | hsa-mir-328 | yes <D> |
| 12 | hsa-mir-1 | yes <H, D> | 37 | hsa-mir-23a | yes <D> |
| 13 | hsa-mir-30a | yes <H, D, M> | 38 | hsa-mir-194 | yes <H, D> |
| 14 | hsa-mir-181b | yes <H, D> | 39 | hsa-mir-193b | yes <H, D> |
| 15 | hsa-mir-19b | yes <H, D, M> | 40 | hsa-mir-196b | yes <D> |
| 16 | hsa-mir-214 | yes <H, D, M> | 41 | hsa-mir-137 | yes <H, D> |
| 17 | hsa-mir-130a | yes <H, D> | 42 | hsa-mir-191 | yes <H, D, M> |
| 18 | hsa-mir-222 | yes <H, D> | 43 | hsa-mir-302a | no |
| 19 | hsa-mir-148a | yes <H, D> | 44 | hsa-mir-135b | yes <D> |
| 20 | hsa-mir-25 | yes <D> | 45 | hsa-mir-451b | no |
| 21 | hsa-mir-133b | yes <H, D> | 46 | hsa-mir-342 | yes <D, M> |
| 22 | hsa-mir-183 | yes <H, D> | 47 | hsa-mir-30b | yes <H, D> |
| 23 | hsa-mir-106a | yes <H, D, M> | 48 | hsa-mir-373 | no |
| 24 | hsa-mir-24 | yes <D> | 49 | hsa-mir-212 | yes <D> |
| 25 | hsa-mir-132 | yes <D> | 50 | hsa-mir-193a | yes <H, D> |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Ju, B.; Ren, S. HLGNN-MDA: Heuristic Learning Based on Graph Neural Networks for miRNA–Disease Association Prediction. Int. J. Mol. Sci. 2022, 23, 13155. https://doi.org/10.3390/ijms232113155
Yu L, Ju B, Ren S. HLGNN-MDA: Heuristic Learning Based on Graph Neural Networks for miRNA–Disease Association Prediction. International Journal of Molecular Sciences. 2022; 23(21):13155. https://doi.org/10.3390/ijms232113155
Chicago/Turabian StyleYu, Liang, Bingyi Ju, and Shujie Ren. 2022. "HLGNN-MDA: Heuristic Learning Based on Graph Neural Networks for miRNA–Disease Association Prediction" International Journal of Molecular Sciences 23, no. 21: 13155. https://doi.org/10.3390/ijms232113155
APA StyleYu, L., Ju, B., & Ren, S. (2022). HLGNN-MDA: Heuristic Learning Based on Graph Neural Networks for miRNA–Disease Association Prediction. International Journal of Molecular Sciences, 23(21), 13155. https://doi.org/10.3390/ijms232113155







