Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins
Abstract
1. Introduction
2. Results
2.1. Salt Stress Hinders Soybean Root and Shoot Growth
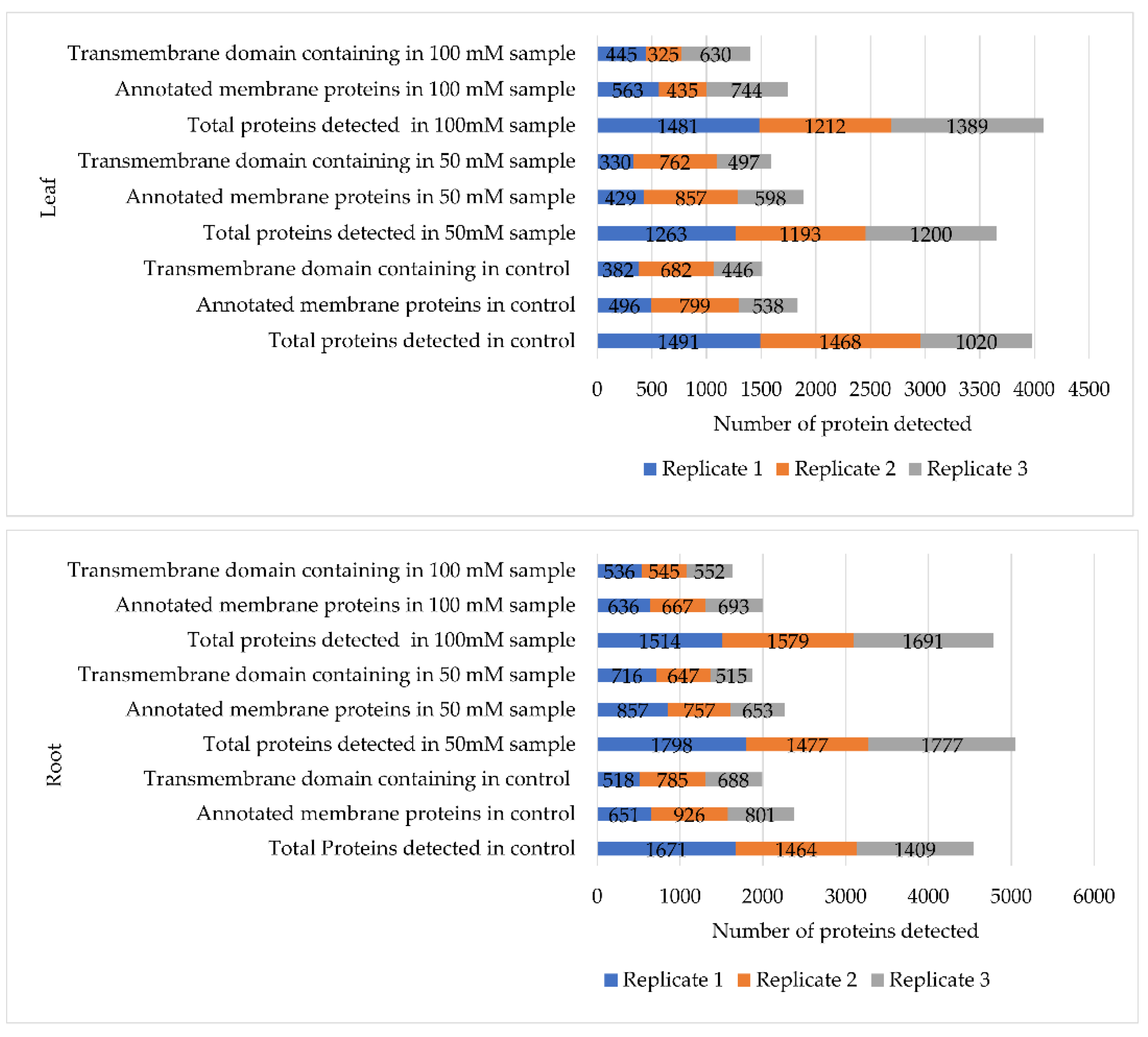
2.2. Leaf and Root Membrane Proteome under Salt Stress Revealed by Orbitrap
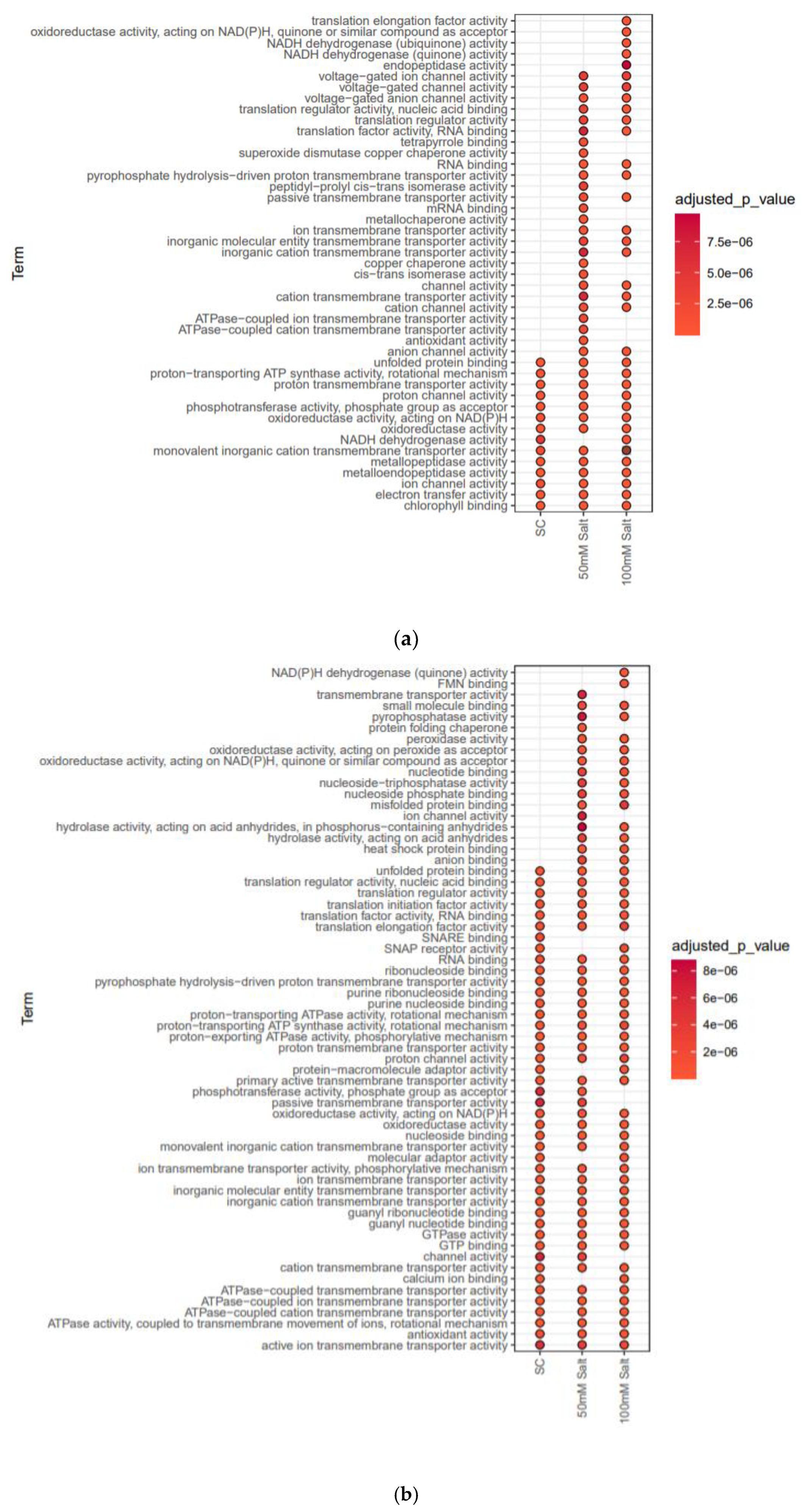
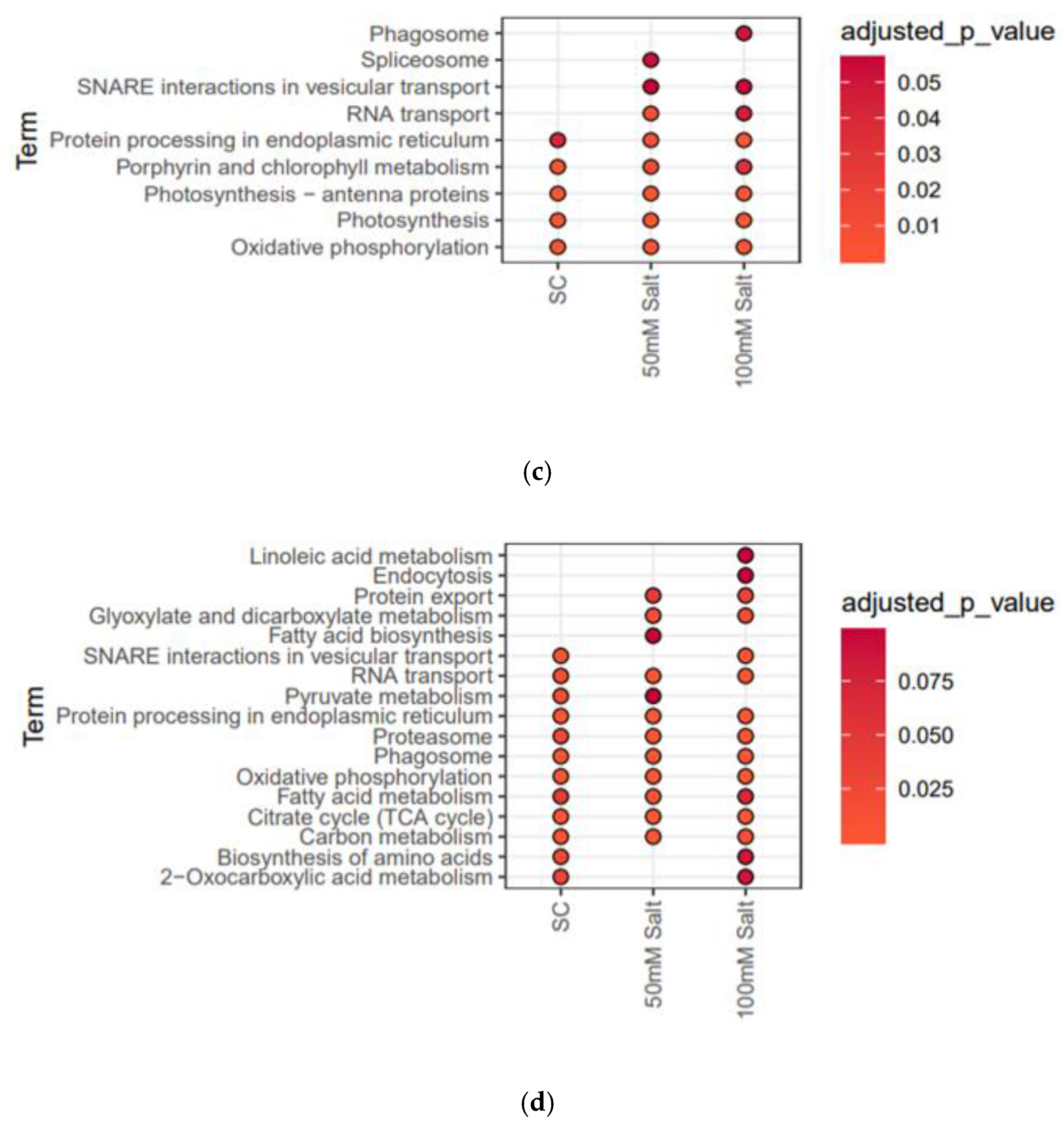
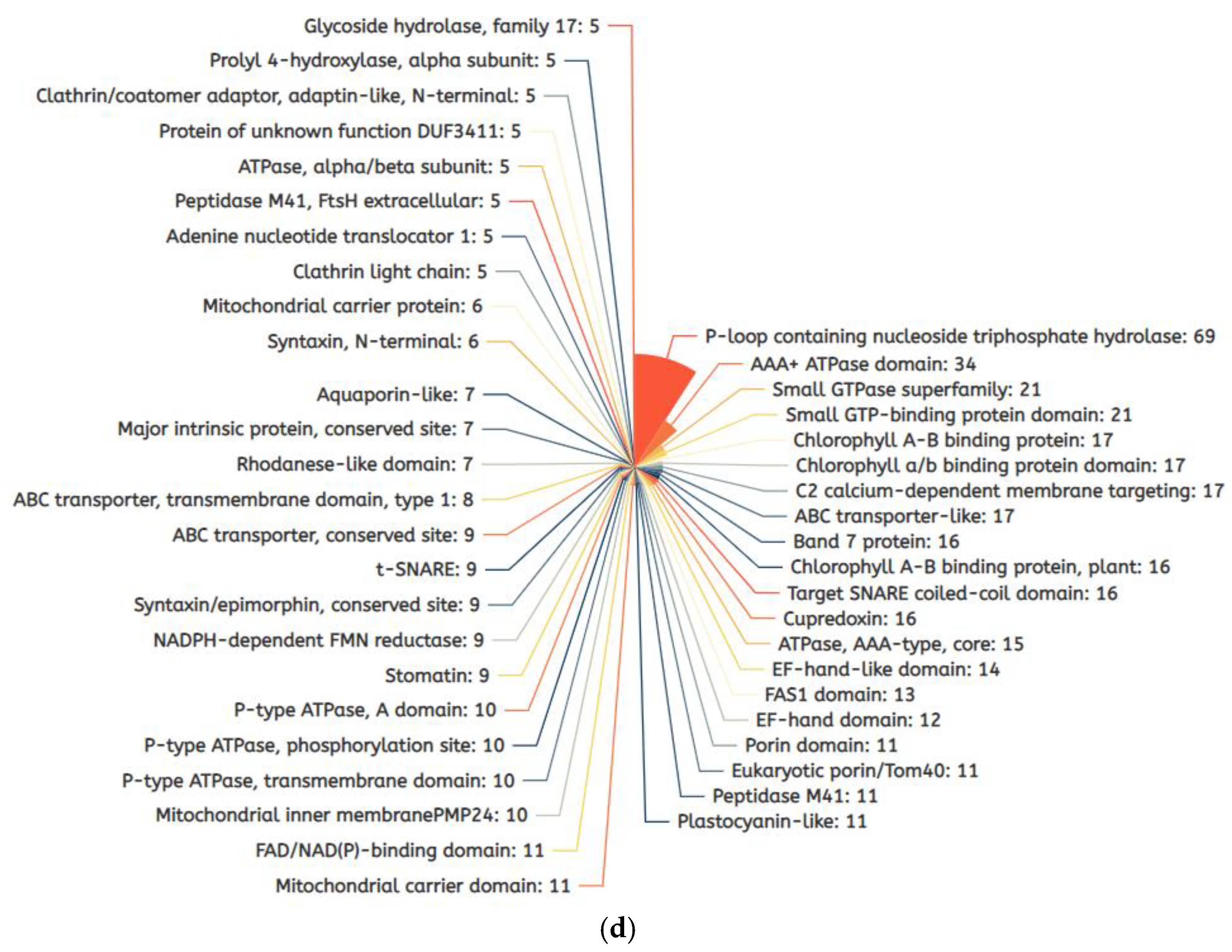
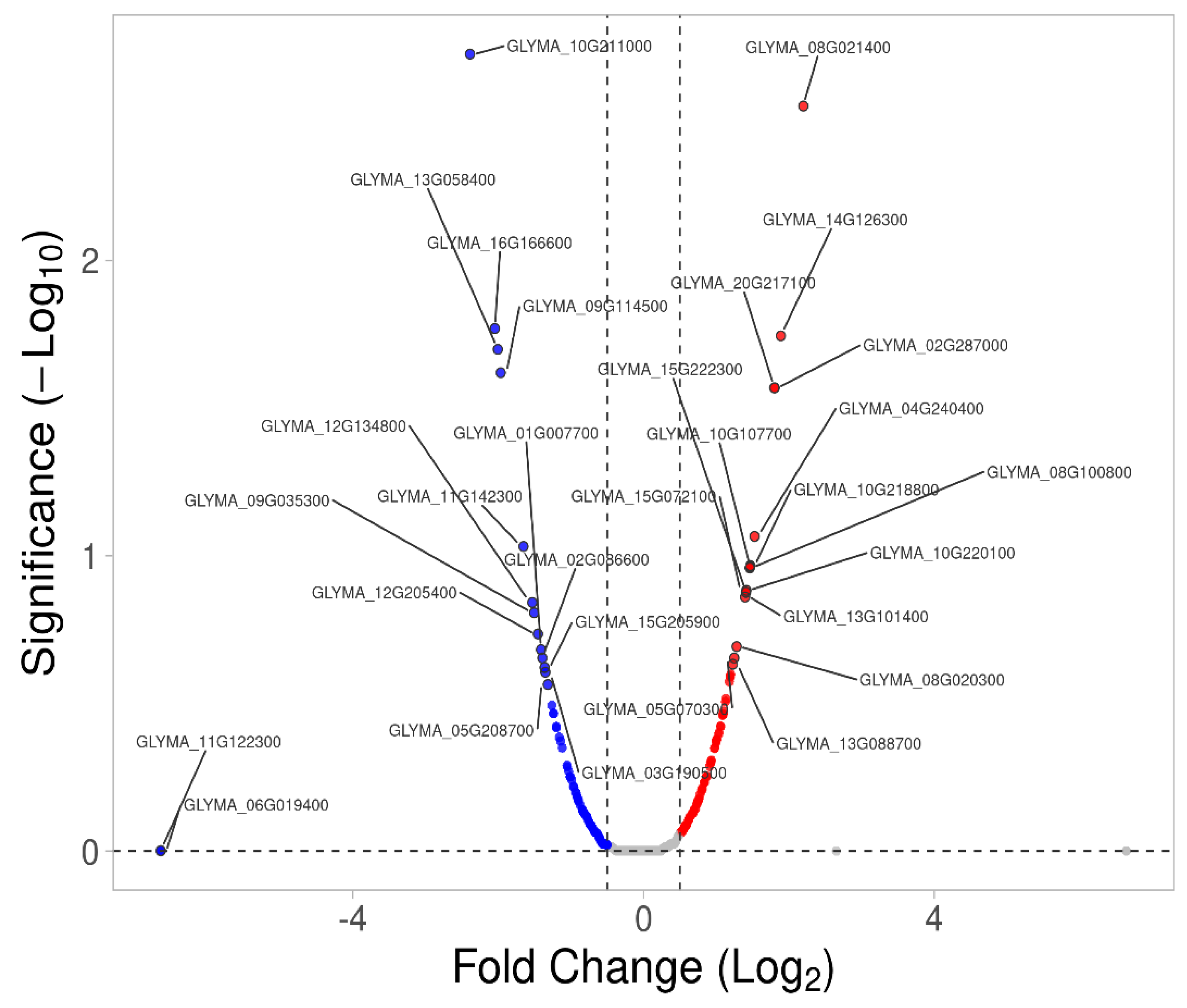
2.3. Differentially Expressed Membrane Proteins (DEMPs) in Soybean Leaves and Roots under Salt Stress
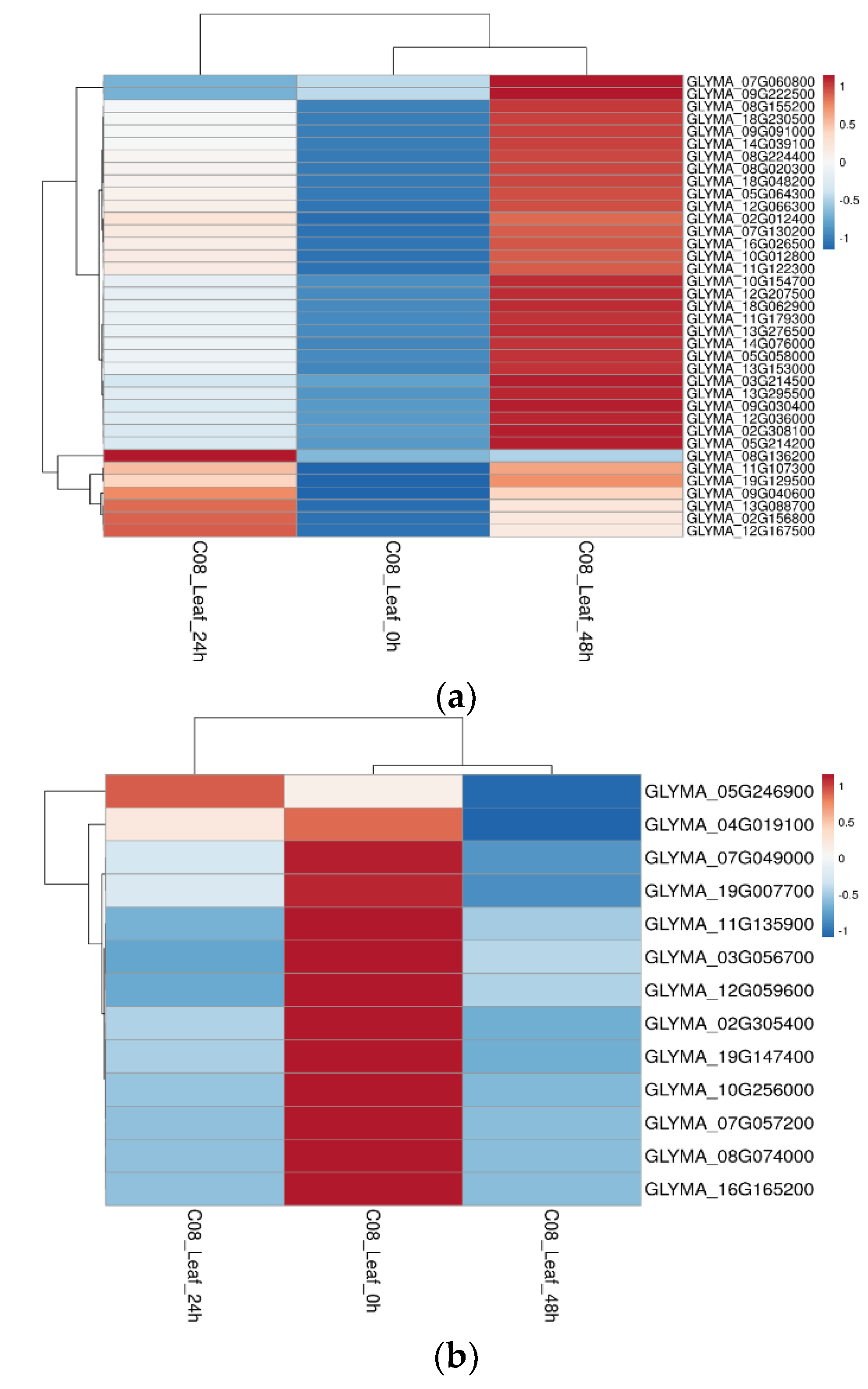
2.4. Transcriptomic Verifications of DEMPs in Root and Leaf Tissues under Salt Stress
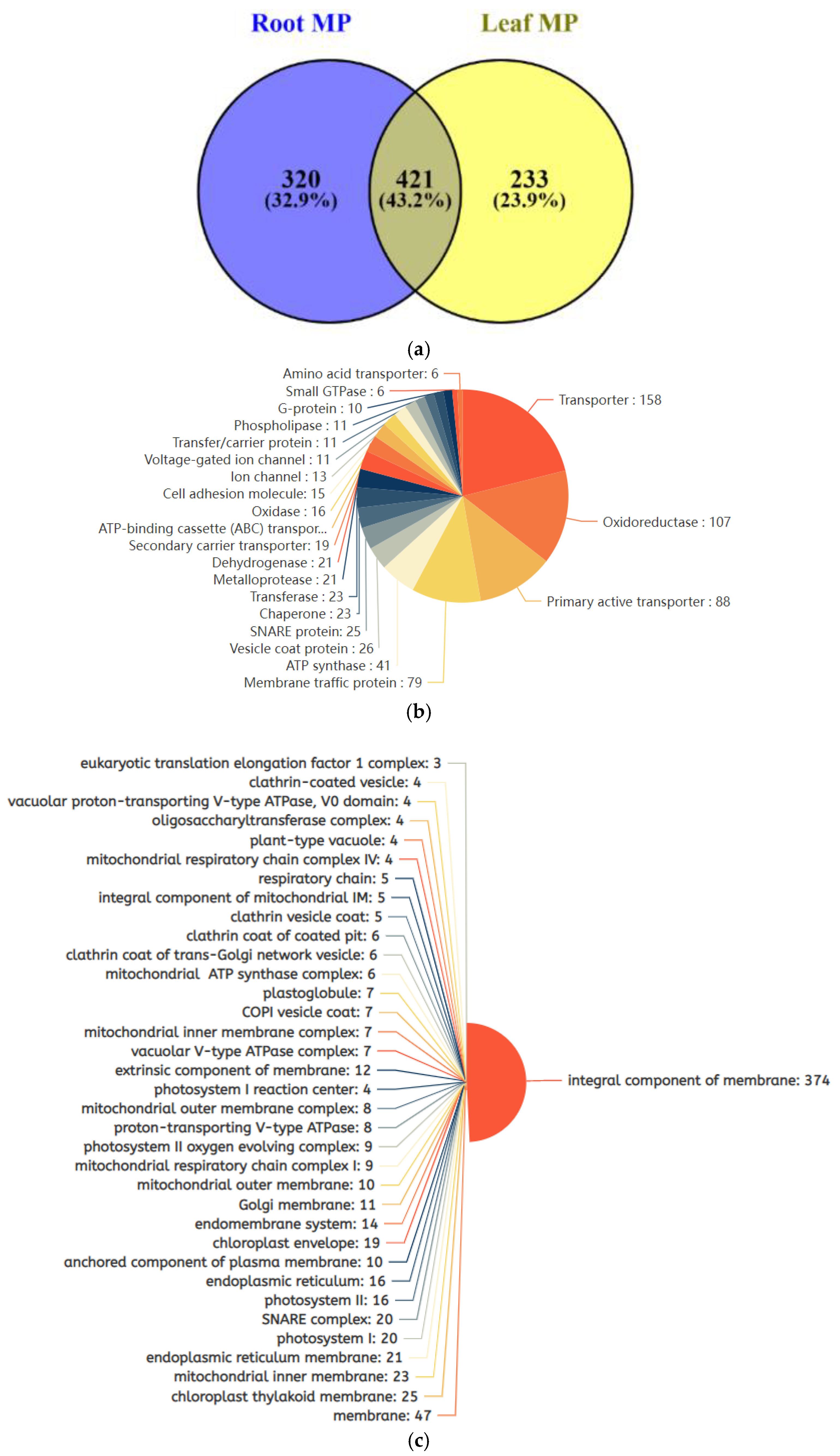
2.5. Salt-Stress-Induced Membrane Proteins from Soybean Leaf and Root Tissues
3. Discussion
4. Materials and Methods
4.1. Soybean Stress Treatment
4.2. Extraction of Membrane Proteins and Trypsin Digestion
4.3. Orbitrap-Based Liquid Chromatography-Tandem MS (LC-MS/MS) Analysis
4.4. Data Analysis and Interpretation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R. Comparative Physiology of Salt and Water Stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J.-K. Understanding and Improving Salt Tolerance in Plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.; Dai, S. Proteomics-Based Investigation of Salt-Responsive Mechanisms in Plant Roots. J. Proteom. 2013, 82, 230–253. [Google Scholar] [CrossRef]
- Kongpracha, P.; Wiriyasermkul, P.; Isozumi, N.; Moriyama, S.; Kanai, Y.; Nagamori, S. Simple but Efficacious Enrichment of Integral Membrane Proteins and Their Interactions for In-Depth Membrane Proteomics. Mol. Cell. Proteom. 2022, 21, 100206. [Google Scholar] [CrossRef]
- César-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X.; Reithmeier, R.A.; Hepworth, D.; Hediger, M.A.; Edwards, A.M.; et al. A Call for Systematic Research on Solute Carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, L.; Merchan, F.; Laporte, P.; Thompson, R.; Clarke, J.; Sousa, C.; Crespi, M. A Novel Plant Leucine-Rich Repeat Receptor Kinase Regulates the Response of Medicago Truncatula Roots to Salt Stress. Plant Cell 2009, 21, 668–680. [Google Scholar] [CrossRef]
- Várady, G.; Cserepes, J.; Németh, A.; Szabó, E.; Sarkadi, B. Cell Surface Membrane Proteins as Personalized Biomarkers: Where We Stand and Where We Are Headed. Biomark. Med. 2013, 7, 803–819. [Google Scholar] [CrossRef]
- Sobhanian, H.; Razavizadeh, R.; Nanjo, Y.; Ehsanpour, A.A.; Jazii, F.R.; Motamed, N.; Komatsu, S. Proteome Analysis of Soybean Leaves, Hypocotyls and Roots under Salt Stress. Proteome Sci. 2010, 8, 19. [Google Scholar] [CrossRef]
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y.; et al. Mechanisms of Soybean Roots’ Tolerances to Salinity Revealed by Proteomic and Phosphoproteomic Comparisons Between Two Cultivars. Mol. Cell. Proteom. MCP 2016, 15, 266–288. [Google Scholar] [CrossRef]
- Ma, H.; Song, L.; Shu, Y.; Wang, S.; Niu, J.; Wang, Z.; Yu, T.; Gu, W.; Ma, H. Comparative Proteomic Analysis of Seedling Leaves of Different Salt Tolerant Soybean Genotypes. J. Proteom. 2012, 75, 1529–1546. [Google Scholar] [CrossRef]
- Chen, H.; Chen, X.; Gu, H.; Wu, B.; Zhang, H.; Yuan, X.; Cui, X. GmHKT1;4, a Novel Soybean Gene Regulating Na+/K+ Ratio in Roots Enhances Salt Tolerance in Transgenic Plants. Plant Growth Regul. 2014, 73, 299–308. [Google Scholar] [CrossRef]
- Wei, P.; Che, B.; Shen, L.; Cui, Y.; Wu, S.; Cheng, C.; Liu, F.; Li, M.-W.; Yu, B.; Lam, H.-M. Identification and Functional Characterization of the Chloride Channel Gene, GsCLC-C2 from Wild Soybean. BMC Plant Biol. 2019, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.-W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.-L.; et al. Identification of a Novel Salt Tolerance Gene in Wild Soybean by Whole-Genome Sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef]
- Luo, G.-Z.; Wang, H.-W.; Huang, J.; Tian, A.-G.; Wang, Y.-J.; Zhang, J.-S.; Chen, S.-Y. A Putative Plasma Membrane Cation/Proton Antiporter from Soybean Confers Salt Tolerance in Arabidopsis. Plant Mol. Biol. 2005, 59, 809–820. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Sun, G.; He, Y.; Zhuang, H.; Sun, T.; Qi, J.; Wu, J. Genome-Wide Identification of Calcium-Dependent Protein Kinases in Soybean and Analyses of Their Transcriptional Responses to Insect Herbivory and Drought Stress. Sci. Rep. 2016, 6, 18973. [Google Scholar] [CrossRef]
- Liu, C.-W.; Chang, T.-S.; Hsu, Y.-K.; Wang, A.Z.; Yen, H.-C.; Wu, Y.-P.; Wang, C.-S.; Lai, C.-C. Comparative Proteomic Analysis of Early Salt Stress Responsive Proteins in Roots and Leaves of Rice. Proteomics 2014, 14, 1759–1775. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Qi, Y.; Chen, F.; Meng, Y.; Luo, X.; Shuai, H.; Zhou, W.; Ding, J.; Du, J.; Liu, J.; et al. Salt Stress Represses Soybean Seed Germination by Negatively Regulating GA Biosynthesis While Positively Mediating ABA Biosynthesis. Front. Plant Sci. 2017, 8, 1372. [Google Scholar] [CrossRef]
- Wu, H. Plant Salt Tolerance and Na+ Sensing and Transport. Crop J. 2018, 6, 215–225. [Google Scholar] [CrossRef]
- UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.-W.; Wong, F.-L.; Yung, W.-S.; Ku, Y.-S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.-Y.; et al. Transcriptomic Reprogramming in Soybean Seedlings under Salt Stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Arif, M.R.; Islam, M.T.; Robin, A.H.K. Salinity Stress Alters Root Morphology and Root Hair Traits in Brassica napus. Plants 2019, 8, 192. [Google Scholar] [CrossRef] [PubMed]
- Otie, V.; Udo, I.; Shao, Y.; Itam, M.O.; Okamoto, H.; An, P.; Eneji, E.A. Salinity Effects on Morpho-Physiological and Yield Traits of Soybean (Glycine max L.) as Mediated by Foliar Spray with Brassinolide. Plants 2021, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, D.-Z.; Zhang, G.-P. Advances in Studies on Ion Transporters Involved in Salt Tolerance and Breeding Crop Cultivars with High Salt Tolerance. J. Zhejiang Univ. Sci. B 2020, 21, 426–441. [Google Scholar] [CrossRef]
- Mansour, M.M.F. Role of Vacuolar Membrane Transport Systems in Plant Salinity Tolerance. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Shomali, A.; Azad, N.; Hassani, B.; Lastochkina, O.; Li, T. Calcium Signaling and Salt Tolerance Are Diversely Entwined in Plants. Plant Signal. Behav. 2019, 14, 1665455. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Zhang, D.Y.; Xu, Z.L.; Xu, L.; Yi, J.X.; He, X.L.; Huang, Y.H.; Liu, X.Q.; Khan, A.A.; Trethowan, R.M.; et al. Uncovering the Salt Response of Soybean by Unraveling Its Wild and Cultivated Functional Genomes Using Tag Sequencing. PLoS ONE 2012, 7, e48819. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimova, U.; Zivcak, M.; Gasparovic, K.; Rastogi, A.; Allakhverdiev, S.I.; Yang, X.; Brestic, M. Electron and Proton Transport in Wheat Exposed to Salt Stress: Is the Increase of the Thylakoid Membrane Proton Conductivity Responsible for Decreasing the Photosynthetic Activity in Sensitive Genotypes? Photosynth. Res. 2021, 150, 195–211. [Google Scholar] [CrossRef]
- Kwon, C.; Lee, J.-H.; Yun, H.S. SNAREs in Plant Biotic and Abiotic Stress Responses. Mol. Cells 2020, 43, 501–508. [Google Scholar] [CrossRef]
- Leterrier, M.; Barroso, J.B.; Valderrama, R.; Palma, J.M.; Corpas, F.J. NADP-Dependent Isocitrate Dehydrogenase from Arabidopsis Roots Contributes in the Mechanism of Defence against the Nitro-Oxidative Stress Induced by Salinity. Sci. World J. 2012, 2012, 694740. [Google Scholar] [CrossRef]
- Cui, P.; Xiong, L. Environmental Stress and Pre-MRNA Splicing. Mol. Plant 2015, 8, 1302–1303. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [PubMed]
- Gogna, M.; Choudhary, A.; Mishra, G.; Kapoor, R.; Bhatla, S.C. Changes in Lipid Composition in Response to Salt Stress and Its Possible Interaction with Intracellular Na+-K+ Ratio in Sunflower (Helianthus annuus L.). Environ. Exp. Bot. 2020, 178, 104147. [Google Scholar] [CrossRef]
- Baral, A.; Irani, N.G.; Fujimoto, M.; Nakano, A.; Mayor, S.; Mathew, M.K. Salt-Induced Remodeling of Spatially Restricted Clathrin-Independent Endocytic Pathways in Arabidopsis Root. Plant Cell 2015, 27, 1297–1315. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Li, C.; Liu, Y.; Yu, J. A Remorin Gene SiREM6, the Target Gene of SiARDP, from Foxtail Millet (Setaria Italica) Promotes High Salt Tolerance in Transgenic Arabidopsis. PLoS ONE 2014, 9, e100772. [Google Scholar] [CrossRef]
- Jia, J.; Liang, Y.; Gou, T.; Hu, Y.; Zhu, Y.; Huo, H.; Guo, J.; Gong, H. The Expression Response of Plasma Membrane Aquaporins to Salt Stress in Tomato Plants. Environ. Exp. Bot. 2020, 178, 104190. [Google Scholar] [CrossRef]
- Chen, L.-M.; Fang, Y.-S.; Zhang, C.-J.; Hao, Q.-N.; Cao, D.; Yuan, S.-L.; Chen, H.-F.; Yang, Z.-L.; Chen, S.-L.; Shan, Z.-H.; et al. GmSYP24, a Putative Syntaxin Gene, Confers Osmotic/Drought, Salt Stress Tolerances and ABA Signal Pathway. Sci. Rep. 2019, 9, 5990. [Google Scholar] [CrossRef]
- Sun, T.-J.; Fan, L.; Yang, J.; Cao, R.-Z.; Yang, C.-Y.; Zhang, J.; Wang, D.-M. A Glycine Max Sodium/Hydrogen Exchanger Enhances Salt Tolerance through Maintaining Higher Na+ Efflux Rate and K+/Na+ Ratio in Arabidopsis. BMC Plant Biol. 2019, 19, 469. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na(+)/H(+) Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Schapire, A.L.; Voigt, B.; Jasik, J.; Rosado, A.; Lopez-Cobollo, R.; Menzel, D.; Salinas, J.; Mancuso, S.; Valpuesta, V.; Baluska, F.; et al. Arabidopsis Synaptotagmin 1 Is Required for the Maintenance of Plasma Membrane Integrity and Cell Viability. Plant Cell 2008, 20, 3374–3388. [Google Scholar] [CrossRef]
- Krausko, M.; Kusá, Z.; Peterková, D.; Labajová, M.; Kumar, A.; Pavlovič, A.; Bačovčinová, M.; Bačkor, M.; Jásik, J. The Absence of the AtSYT1 Function Elevates the Adverse Effect of Salt Stress on Photosynthesis in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1751. [Google Scholar] [CrossRef]
- Hasegawa, P.M. Sodium (Na+) Homeostasis and Salt Tolerance of Plants. Environ. Exp. Bot. 2013, 92, 19–31. [Google Scholar] [CrossRef]
- Beamer, Z.G.; Routray, P.; Choi, W.-G.; Spangler, M.K.; Lokdarshi, A.; Roberts, D.M. Aquaporin Family Lactic Acid Channel NIP2;1 Promotes Plant Survival under Low Oxygen Stress in Arabidopsis. Plant Physiol. 2021, 187, 2262–2278. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zhao, Q.; David, L.; Chen, S.; Dai, S. Salinity Response in Chloroplasts: Insights from Gene Characterization. Int. J. Mol. Sci. 2017, 18, 1011. [Google Scholar] [CrossRef]
- Batelli, G.; Verslues, P.E.; Agius, F.; Qiu, Q.; Fujii, H.; Pan, S.; Schumaker, K.S.; Grillo, S.; Zhu, J.-K. SOS2 Promotes Salt Tolerance in Part by Interacting with the Vacuolar H+-ATPase and Upregulating Its Transport Activity. Mol. Cell. Biol. 2007, 27, 7781–7790. [Google Scholar] [CrossRef] [PubMed]
- Gévaudant, F.; Duby, G.; von Stedingk, E.; Zhao, R.; Morsomme, P.; Boutry, M. Expression of a Constitutively Activated Plasma Membrane H+-ATPase Alters Plant Development and Increases Salt Tolerance. Plant Physiol. 2007, 144, 1763–1776. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.-H. Potassium Transport and Signaling in Higher Plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef]
- Huda, K.M.K.; Banu, M.S.A.; Tuteja, R.; Tuteja, N. Global Calcium Transducer P-Type Ca2+-ATPases Open New Avenues for Agriculture by Regulating Stress Signalling. J. Exp. Bot. 2013, 64, 3099–3109. [Google Scholar] [CrossRef]
- Bu, Y.; Takano, T.; Liu, S. The Role of Ammonium Transporter (AMT) against Salt Stress in Plants. Plant Signal. Behav. 2019, 14, 1625696. [Google Scholar] [CrossRef]
- Meco, V.; Egea, I.; Ortíz-Atienza, A.; Drevensek, S.; Esch, E.; Yuste-Lisbona, F.J.; Barneche, F.; Vriezen, W.; Bolarin, M.C.; Lozano, R.; et al. The Salt Sensitivity Induced by Disruption of Cell Wall-Associated Kinase 1 (SlWAK1) Tomato Gene Is Linked to Altered Osmotic and Metabolic Homeostasis. Int. J. Mol. Sci. 2020, 21, 6308. [Google Scholar] [CrossRef]
- Grossi, C.E.M.; Santin, F.; Quintana, S.A.; Fantino, E.; Ulloa, R.M. Calcium-Dependent Protein Kinase 2 Plays a Positive Role in the Salt Stress Response in Potato. Plant Cell Rep. 2022, 41, 535–548. [Google Scholar] [CrossRef]
- Ueda, M.; Tsutsumi, N.; Fujimoto, M. Salt Stress Induces Internalization of Plasma Membrane Aquaporin into the Vacuole in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2016, 474, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. Salt Stress Responses in Arabidopsis Utilize a Signal Transduction Pathway Related to Endoplasmic Reticulum Stress Signaling. Plant J. Cell Mol. Biol. 2007, 51, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y.H.; Song, M.; Wang, Y.; Xu, W.; Wu, L.; Wang, H.; Ma, Z. Overexpression of a Triticum Aestivum Calreticulin Gene (TaCRT1) Improves Salinity Tolerance in Tobacco. PLoS ONE 2015, 10, e0140591. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Karim, S.; Zhang, H.; Aronsson, H. Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS). Int. J. Mol. Sci. 2017, 18, 309. [Google Scholar] [CrossRef] [PubMed]
- Inaba, T.; Alvarez-Huerta, M.; Li, M.; Bauer, J.; Ewers, C.; Kessler, F.; Schnell, D.J. Arabidopsis Tic110 Is Essential for the Assembly and Function of the Protein Import Machinery of Plastids. Plant Cell 2005, 17, 1482–1496. [Google Scholar] [CrossRef]
- Wojtyla, Ł.; Kosmala, A.; Garnczarska, M. Lupine Embryo Axes under Salinity Stress. II. Mitochondrial Proteome Response. Acta Physiol. Plant. 2013, 35, 2383–2392. [Google Scholar] [CrossRef][Green Version]
- Luo, Q.; Peng, M.; Zhang, X.; Lei, P.; Ji, X.; Chow, W.; Meng, F.; Sun, G. Comparative Mitochondrial Proteomic, Physiological, Biochemical and Ultrastructural Profiling Reveal Factors Underpinning Salt Tolerance in Tetraploid Black Locust (Robinia pseudoacacia L.). BMC Genom. 2017, 18, 648. [Google Scholar] [CrossRef]
- Li, W.; Zhao, F.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of Early Salt Stress Responsive Proteins in Seedling Roots of Upland Cotton (Gossypium hirsutum L.) Employing ITRAQ-Based Proteomic Technique. Front. Plant Sci. 2015, 6, 732. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A Magnesium Transporter OsMGT1 Plays a Critical Role in Salt Tolerance in Rice. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A Putative Plasma Membrane Na(+)/H(+) Antiporter GmSOS1 Is Critical for Salt Stress Tolerance in Glycine Max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef]
- Cao, M.-J.; Wang, Z.; Wirtz, M.; Hell, R.; Oliver, D.J.; Xiang, C.-B. SULTR3;1 Is a Chloroplast-Localized Sulfate Transporter in Arabidopsis Thaliana. Plant J. Cell Mol. Biol. 2013, 73, 607–616. [Google Scholar] [CrossRef] [PubMed]




| Ensemble Protein ID | Primary Protein Name | Arabidopsis Identifier | No. of Peptides | No. of PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_09G040600 | Abscisic acid receptor PYL12 | PYL12_ARATH | 12 | 39 | 16.5 | 4.83 |
| GLYMA_11G096400 | COP1-interactive protein 1 | CIP1_ARATH | 20 | 58 | 153.2 | 4.7 |
| GLYMA_09G222500 | Cation/H(+) antiporter 8 | CHX8_ARATH | 2 | 12 | 5.7 | 10.17 |
| GLYMA_12G036000 | Cation/H(+) antiporter 8 | CHX8_ARATH | 2 | 10 | 5.6 | 10.17 |
| GLYMA_08G155200 | ATP synthase subunit O, mitochondrial | ATPO_ARATH | 15 | 80 | 26.9 | 9.52 |
| GLYMA_09G091000 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | NDUA6_ARATH | 6 | 45 | 17.1 | 7.81 |
| GLYMA_05G058000 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial | NDUB8_ARATH | 3 | 9 | 13.5 | 7.97 |
| GLYMA_11G107300 | Synaptotagmin-1 | SYT1_ARATH | 5 | 8 | 61.7 | 6.73 |
| GLYMA_05G064300 | Ras-related protein RABD2c (AtRABD2c) | RAD2C_ARATH | 9 | 30 | 22.6 | 5.71 |
| GLYMA_02G308100 | Transmembrane emp24 domain-containing protein p24beta2 | P24B2_ARATH | 2 | 7 | 25.4 | 6.47 |
| GLYMA_09G030400 | Coatomer subunit epsilon-1 | COPE1_ARATH | 1 | 4 | 32.3 | 5.57 |
| GLYMA_08G224400 | V-type proton ATPase catalytic subunit A (V-ATPase subunit A) | VATA_ARATH | 34 | 276 | 68.7 | 5.67 |
| GLYMA_05G214200 | V-type proton ATPase subunit E3 (V-ATPase subunit E3) | VATE3_ARATH | 22 | 123 | 27.6 | 6.8 |
| GLYMA_08G020300 | V-type proton ATPase subunit E1 (V-ATPase subunit E1) | VATE1_ARATH | 21 | 141 | 26.8 | 6.25 |
| GLYMA_14G076000 | Outer envelope pore protein 24B, chloroplastic | OP24B_ARATH | 14 | 83 | 23.7 | 9.26 |
| GLYMA_14G214700 | Probable inactive receptor kinase At1g48480 | Y1848_ARATH | 2 | 2 | 68.1 | 7.2 |
| GLYMA_09G271500 | Probable receptor-like protein kinase At5g24010 | Y5241_ARATH | 3 | 13 | 35.4 | 7.84 |
| GLYMA_12G167500 | Uncharacterized protein At5g49945 | Y5994_ARATH | 7 | 17 | 50.8 | 6.34 |
| GLYMA_08G136200 | Annexin D2 | ANXD2_ARATH | 14 | 31 | 35.6 | 6.46 |
| GLYMA_10G154700 | Mitochondrial Rho GTPase 1 (AtMIRO1) | MIRO1_ARATH | 1 | 4 | 71.9 | 5.59 |
| GLYMA_11G120200 | Probable NAD(P)H dehydrogenase (quinone) FQR1-like 2 | FQRL2_ARATH | 9 | 37 | 27.3 | 6 |
| GLYMA_02G028200 | Metalloendoproteinase 5-MMP (At5-MMP) | 5MMP_ARATH | 2 | 3 | 43.8 | 9.09 |
| GLYMA_13G276500 | Proton pump-interactor 1 | PPI1_ARATH | 36 | 173 | 69.3 | 9 |
| GLYMA_12G207500 | Prohibitin-2, mitochondrial (Atphb2) | PHB2_ARATH | 20 | 105 | 37.1 | 9.63 |
| GLYMA_18G230500 | Uncharacterized protein At3g61260 | Y3126_ARATH | 12 | 40 | 20.1 | 8.7 |
| GLYMA_09G271800 | Protein RETICULATA-RELATED 3, chloroplastic | RER3_ARATH | 4 | 4 | 36.9 | 8.27 |
| GLYMA_12G207600 | Fasciclin-like arabinogalactan protein 9 | FLA9_ARATH | 17 | 159 | 26.3 | 9.11 |
| GLYMA_19G180700 | Fasciclin-like arabinogalactan protein 11 | FLA11_ARATH | 1 | 6 | 26.3 | 5.85 |
| GLYMA_02G012400 | Cytochrome c oxidase subunit 5C-2 | CX5C2_ARATH | 7 | 41 | 7.1 | 8.59 |
| GLYMA_10G012800 | Cytochrome c oxidase subunit 5C-2 | CX5C2_ARATH | 7 | 47 | 7.1 | 8.59 |
| GLYMA_01G064700 | Protein trichome birefringence-like 37 | TBL37_ARATH | 1 | 9 | 21.8 | 6.64 |
| GLYMA_11G179300 | Alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial | NDC1_ARATH | 31 | 238 | 39.3 | 9.42 |
| GLYMA_18G062900 | Peptidyl-prolyl cis-trans isomerase CYP19-4 (PPIase CYP19-4) | CP19D_ARATH | 7 | 28 | 21.9 | 9.17 |
| GLYMA_19G129500 | Rhomboid-like protein 20 (AtRBL20) | RBL20_ARATH | 2 | 6 | 36.6 | 10.02 |
| GLYMA_15G016300 | Photosystem I reaction center subunit VI-2, chloroplastic | PSAH2_ARATH | 6 | 28 | 14.9 | 9.99 |
| GLYMA_01G181700 | Probable serine/threonine-protein kinase PBL19 | PBL19_ARATH | 1 | 1 | 41.9 | 8.73 |
| GLYMA_02G272600 | Protein RESTRICTED TEV MOVEMENT 2 | RTM2_ARATH | 7 | 21 | 28.9 | 8.02 |
| GLYMA_14G039100 | Elongation factor 1-beta 1 (EF-1-beta 1) | EF1B1_ARATH | 7 | 15 | 24.2 | 4.68 |
| GLYMA_13G153000 | Ras-related protein RABF2a (AtRABF2a) | RAF2A_ARATH | 3 | 13 | 25.6 | 7.3 |
| GLYMA_18G048200 | Protein TIC 40, chloroplastic | TIC40_ARATH | 8 | 26 | 47.3 | 6.44 |
| GLYMA_11G122300 | Mitochondrial import receptor subunit TOM6 homolog | TOM6_ARATH | 2 | 4 | 6.2 | 9.61 |
| GLYMA_13G088700 | Annexin D1 | ANXD1_ARATH | 13 | 52 | 35.9 | 6.96 |
| GLYMA_07G060800 | Sugar transport protein 7 | STP7_ARATH | 4 | 12 | 27.2 | 10.08 |
| GLYMA_04G015000 | Monosaccharide-sensing protein 2 | MSSP2_ARATH | 2 | 7 | 78.7 | 5.72 |
| GLYMA_07G130200 | Protein DETOXIFICATION 50 (AtDTX50) | DTX50_ARATH | 3 | 16 | 8.7 | 8.53 |
| GLYMA_13G295500 | ADP, ATP carrier protein 3, mitochondrial | ADT3_ARATH | 21 | 129 | 39.6 | 9.85 |
| GLYMA_12G066300 | Aquaporin NIP2-1 | NIP21_ARATH | 6 | 29 | 7.8 | 10.61 |
| GLYMA_12G095500 | Putative purine permease 20 (AtPUP20) | PUP20_ARATH | 4 | 9 | 27 | 9.39 |
| GLYMA_16G026500 | Succinate dehydrogenase subunit 8, mitochondrial | SDH8_ARATH | 2 | 7 | 5.7 | 8.62 |
| GLYMA_05G067900 | RING-H2 finger protein ATL28 | ATL28_ARATH | 1 | 1 | 6.4 | 8.69 |
| Ensemble Protein IDs | Primary Protein Name | Arbidopsis Identifier | Peptides | PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_02G147200 | RAN GTPase-activating protein 1 (AtRanGAP1; RanGAP1) | RAGP1_ARATH | 3 | 3 | 57.7 | 4.78 |
| GLYMA_02G305400 | Chlorophyll a-b binding protein 2.1, chloroplastic | CB21_ARATH | 8 | 40 | 28.6 | 5.66 |
| GLYMA_03G056700 | Temperature-sensitive sn-2 acyl-lipid omega-3 desaturase (ferredoxin), chloroplastic | FAD3D_ARATH | 2 | 8 | 51.3 | 7.71 |
| GLYMA_03G179500 | Syntaxin-71 (AtSYP71) | SYP71_ARATH | 3 | 9 | 29.7 | 5.94 |
| GLYMA_04G019100 | ATP-dependent zinc metalloprotease FTSH 5, chloroplastic (AtFTSH5) | FTSH5_ARATH | 18 | 90 | 74.1 | 6.19 |
| GLYMA_04G107400 | Mitochondrial import inner membrane translocase subunit TIM44-2 | TI442_ARATH | 1 | 2 | 54.4 | 8.82 |
| GLYMA_05G147700 | Non-specific lipid transfer protein GPI-anchored 1 (AtLTPG-1; Protein LTP-GPI-ANCHORED 1) | LTPG1_ARATH | 2 | 9 | 19 | 8.24 |
| GLYMA_05G246900 | PRA1 family protein B5 (AtPRA1.B5) | PR1B5_ARATH | 1 | 4 | 22.6 | 8.66 |
| GLYMA_07G049000 | Protein CURVATURE THYLAKOID 1B, chloroplastic | CUT1B_ARATH | 1 | 3 | 18.1 | 8.76 |
| GLYMA_07G057200 | Rhodanese-like domain-containing protein 4, chloroplastic | STR4_ARATH | 14 | 43 | 46.4 | 6.23 |
| GLYMA_08G074000 | Photosystem I chlorophyll a/b-binding protein 5, chloroplastic (Lhca5) | LHCA5_ARATH | 2 | 5 | 27.4 | 7.49 |
| GLYMA_09G114500 | Exocyst complex component SEC3A (AtSec3a) | SEC3A_ARATH | 1 | 2 | 101 | 5.87 |
| GLYMA_09G134500 | Protein SRC2 homolog (AtSRC2) | SRC2_ARATH | 5 | 18 | 30.3 | 5.03 |
| GLYMA_10G256000 | Photosystem I reaction center subunit N, chloroplastic (PSI-N) | PSAN_ARATH | 6 | 20 | 15.4 | 9.67 |
| GLYMA_11G054900 | PI-PLC X domain-containing protein At5g67130 | Y5713_ARATH | 7 | 24 | 46.1 | 7.5 |
| GLYMA_11G135900 | Protein PLASTID TRANSCRIPTIONALLY ACTIVE 16, chloroplastic (pTAC16) | PTA16_ARATH | 35 | 136 | 55.5 | 8.65 |
| GLYMA_12G059600 | Protein PLASTID TRANSCRIPTIONALLY ACTIVE 16, chloroplastic (pTAC16) | PTA16_ARATH | 32 | 131 | 55 | 8.03 |
| GLYMA_13G028200 | Photosystem II protein D1 (PSII D1 protein) | PSBA_ARATH | 3 | 43 | 29.8 | 5.14 |
| GLYMA_16G165200 | Chlorophyll a-b binding protein 1, chloroplastic | CB1C_ARATH | 14 | 61 | 28 | 5.41 |
| GLYMA_19G007700 | Beta carbonic anhydrase 1, chloroplastic (AtbCA1; AtbetaCA1) | BCA1_ARATH | 3 | 4 | 36.7 | 7.43 |
| GLYMA_19G147400 | Delta(12)-fatty-acid desaturase | FAD6E_ARATH | 2 | 5 | 43.8 | 8.46 |
| Ensembl Protein ID | Primary Protein Name | Arabidopsis Identifier | No. of Peptides | No. of PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_01G181700 | Probable serine/threonine-protein kinase PBL19 | PBL19_ARATH | 1 | 1 | 41.9 | 8.73 |
| GLYMA_02G287000 | Peroxisomal and mitochondrial division factor 2 | PMD2_ARATH | 16 | 49 | 34.9 | 5.1 |
| GLYMA_04G174800 | ATPase 11, plasma membrane-type | PMA11_ARATH | 19 | 62 | 105.2 | 6.73 |
| GLYMA_04G240400 | Vacuolar protein sorting-associated protein 45 homolog (AtVPS45) | VPS45_ARATH | 1 | 2 | 64.9 | 6.77 |
| GLYMA_05G078000 | Wall-associated receptor kinase-like 6 | WAKLF_ARATH | 1 | 1 | 25.2 | 9.41 |
| GLYMA_05G215100 | COP1-interactive protein 1 | CIP1_ARATH | 8 | 13 | 139.4 | 4.94 |
| GLYMA_06G161400 | Bidirectional sugar transporter SWEET15 (AtSWEET15) | SWT15_ARATH | 4 | 8 | 68.7 | 5.67 |
| GLYMA_08G020300 | V-type proton ATPase subunit E1 (V-ATPase subunit E1) | VATE1_ARATH | 21 | 141 | 26.8 | 6.25 |
| GLYMA_08G021400 | COP1-interactive protein 1 | CIP1_ARATH | 1 | 1 | 162.5 | 5.01 |
| GLYMA_08G100800 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Y1677_ARATH | 1 | 2 | 104.3 | 6.68 |
| GLYMA_08G348200 | Alpha-mannosidase I MNS4 | MNS4_ARATH | 2 | 22 | 6.8 | 9.2 |
| GLYMA_10G024000 | Phospholipid:diacylglycerol acyltransferase 1 (AtPDAT) | PDAT1_ARATH | 1 | 2 | 21.6 | 10.18 |
| GLYMA_10G107700 | Calcium-transporting ATPase 4, plasma membrane-type | ACA4_ARATH | 8 | 30 | 113.9 | 6.37 |
| GLYMA_10G132300 | Ammonium transporter 1 member 1 (AtAMT1;1) | AMT11_ARATH | 1 | 5 | 53.3 | 7.3 |
| GLYMA_10G218800 | Somatic embryogenesis receptor kinase 1 (AtSERK1) | SERK1_ARATH | 2 | 8 | 68.9 | 5.73 |
| GLYMA_10G220100 | Ras-related protein RABG3a (AtRABG3a) | RAG3A_ARATH | 3 | 5 | 23.1 | 5.68 |
| GLYMA_11G146900 | Receptor-like protein 4 (AtRLP4) | RLP4_ARATH | 1 | 2 | 16.4 | 9.6 |
| GLYMA_11G250100 | Mitochondrial import receptor subunit TOM20-2 | TO202_ARATH | 2 | 9 | 22.5 | 5.21 |
| GLYMA_12G028300 | Delta(24)-sterol reductase | DIM_ARATH | 11 | 81 | 66 | 8.22 |
| GLYMA_13G101400 | Caffeoylshikimate esterase | CSE_ARATH | 1 | 1 | 36.7 | 7.06 |
| GLYMA_14G126300 | Cytochrome b-c1 complex subunit 7-2 | QCR72_ARATH | 16 | 170 | 14.6 | 9.61 |
| GLYMA_15G072100 | Clathrin light chain 1 | CLC1_ARATH | 31 | 139 | 35.3 | 5.19 |
| GLYMA_15G222300 | Calcium-dependent protein kinase 2 | CDPK2_ARATH | 1 | 1 | 58.8 | 5.77 |
| GLYMA_16G154200 | Putative syntaxin-131 (AtSYP131) | SY131_ARATH | 2 | 4 | 34.8 | 7.94 |
| GLYMA_18G141900 | High affinity nitrate transporter 2.5 (AtNRT2:5) | NRT25_ARATH | 1 | 10 | 55.5 | 9.2 |
| GLYMA_20G217100 | Early nodulin-like protein 1 | ENL1_ARATH | 1 | 3 | 21.5 | 7.46 |
| Ensembl Protein ID | Primary Protein Name | Arabidopsis Identifier | No. of Peptides | No. of PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_01G007700 | Mitochondrial import inner membrane translocase subunit TIM17-2 | TI172_ARATH | 4 | 14 | 20.9 | 5.12 |
| GLYMA_02G086600 | Binding partner of ACD11 1 | BPA1_ARATH | 2 | 2 | 33.7 | 5.45 |
| GLYMA_02G307700 | Fasciclin-like arabinogalactan protein 7 | FLA7_ARATH | 2 | 6 | 27.1 | 5.29 |
| GLYMA_02G309400 | Transmembrane 9 superfamily member 12 | TMN12_ARATH | 2 | 4 | 75.1 | 6.86 |
| GLYMA_03G190500 | Bifunctional enolase 2/transcriptional activator | ENO2_ARATH | 10 | 45 | 47.6 | 5.69 |
| GLYMA_04G095900 | Tetratricopeptide repeat domain-containing protein PYG7, chloroplastic | PYG7_ARATH | 7 | 20 | 24.1 | 8.7 |
| GLYMA_04G181000 | Probable cyclic nucleotide-gated ion channel 16 | CNG16_ARATH | 1 | 1 | 79.4 | 9.16 |
| GLYMA_04G220400 | Probable inactive receptor kinase At5g10020 | Y5020_ARATH | 1 | 1 | 113.6 | 7.11 |
| GLYMA_04G222800 | Probable LRR receptor-like serine/threonine-protein kinase IRK | IRK_ARATH | 1 | 1 | 104.7 | 6.61 |
| GLYMA_05G016100 | CHAPERONE-LIKE PROTEIN OF POR1, chloroplastic (AtCPP1) | CPP1_ARATH | 1 | 1 | 28.3 | 9.88 |
| GLYMA_05G199200 | Calnexin homolog 1 | CALX1_ARATH | 4 | 14 | 61.6 | 4.91 |
| GLYMA_06G019400 | ATP-dependent zinc metalloprotease FTSH 5, chloroplastic (AtFTSH5) | FTSH5_ARATH | 6 | 13 | 74.3 | 6.24 |
| GLYMA_06G066400 | ER membrane protein complex subunit 7 homolog | Y4213_ARATH | 11 | 47 | 23.3 | 8.76 |
| GLYMA_07G014400 | Probable mitochondrial-processing peptidase subunit alpha-1, mitochondrial | MPPA1_ARATH | 17 | 76 | 54.4 | 6.27 |
| GLYMA_07G059400 | SUN domain-containing protein 1 (AtSUN1) | SUN1_ARATH | 1 | 7 | 8.5 | 6.54 |
| GLYMA_08G348000 | Galacturonosyltransferase 8 | GAUT8_ARATH | 1 | 1 | 64.1 | 9.16 |
| GLYMA_09G035300 | ABC transporter G family member 19 (ABC transporter ABCG.19; AtABCG19) | AB19G_ARATH | 9 | 47 | 12.2 | 5.07 |
| GLYMA_09G114500 | Exocyst complex component SEC3A (AtSec3a) | SEC3A_ARATH | 1 | 2 | 101 | 5.87 |
| GLYMA_09G243700 | Subtilisin-like protease SBT5.4 | SBT54_ARATH | 5 | 10 | 84.3 | 9.14 |
| GLYMA_10G208300 | Ras-related protein RABF1 (AtRABF1) | RABF1_ARATH | 1 | 4 | 21.7 | 7.14 |
| GLYMA_10G211000 | Aquaporin PIP2-2 | PIP22_ARATH | 1 | 2 | 31.7 | 7.85 |
| GLYMA_11G122300 | Mitochondrial import receptor subunit TOM6 homolog | TOM6_ARATH | 2 | 4 | 6.2 | 9.61 |
| GLYMA_11G142300 | Ras-related protein RABE1c (AtRABE1c) | RAE1C_ARATH | 10 | 35 | 23.6 | 7.83 |
| GLYMA_12G134800 | Early nodulin-like protein 1 | ENL1_ARATH | 2 | 7 | 22.8 | 6.92 |
| GLYMA_12G205400 | ADP, ATP carrier protein 3, mitochondrial | ADT3_ARATH | 19 | 123 | 39.4 | 9.79 |
| GLYMA_13G058400 | Membrane-anchored ubiquitin-fold protein 2 (AtMUB2) | MUB2_ARATH | 3 | 7 | 12.6 | 6.09 |
| GLYMA_14G076000 | Outer envelope pore protein 24B, chloroplastic | OP24B_ARATH | 14 | 83 | 23.7 | 9.26 |
| GLYMA_14G201500 | Protein TIC110, chloroplastic | TI110_ARATH | 10 | 35 | 109.8 | 6.19 |
| GLYMA_15G205900 | Temperature-induced lipocalin-1 (AtTIL1) | TIL_ARATH | 3 | 9 | 21.2 | 8.07 |
| GLYMA_16G166600 | 2-succinyl-6-hydroxy-2,4- cyclohexadiene-1-carboxylate synthase | PHYLO_ARATH | 3 | 9 | 12.6 | 6.86 |
| GLYMA_17G021200 | Probable prolyl 4-hydroxylase 4 (AtP4H4) | P4H4_ARATH | 11 | 44 | 33 | 7.24 |
| GLYMA_20G006500 | Proton pump-interactor 1 | PPI1_ARATH | 3 | 4 | 161.1 | 4.51 |
| Ensembl Protein ID | Primary Protein Name | Arabidopsis Identifier | No. of Peptides | No. of PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_05G019400 | ABC transporter B family member 27 (ABC transporter ABCB.27; AtABCB27) | AB27B_ARATH | 1 | 1 | 68.5 | 8.46 |
| GLYMA_01G194500 | ADP-ribosylation factor-like protein 8a (AtARL8a) | ARL8A_ARATH | 4 | 5 | 20.5 | 8.15 |
| GLYMA_16G076500 | AP-4 complex subunit epsilon | AP4E_ARATH | 1 | 1 | 107.4 | 5.92 |
| GLYMA_12G172500 | Aquaporin PIP2-2 | PIP22_ARATH | 1 | 2 | 30.8 | 8.13 |
| GLYMA_09G056300 | ATPase 2, plasma membrane-type | PMA2_ARATH | 24 | 44 | 112.4 | 6.42 |
| GLYMA_04G203800 | Bidirectional sugar transporter SWEET15 (AtSWEET15) | SWT15_ARATH | 2 | 2 | 67.1 | 6.11 |
| GLYMA_06G324300 | Cellulose synthase-like protein G1 (AtCslG1) | CSLG1_ARATH | 4 | 8 | 79.5 | 8.57 |
| GLYMA_12G095700 | Chaperone protein dnaJ 2 (AtDjA2) | DNAJ2_ARATH | 5 | 7 | 46.3 | 6.28 |
| GLYMA_17G030700 | COBRA-like protein 7 | COBL7_ARATH | 1 | 1 | 70.4 | 6.89 |
| GLYMA_07G202300 | Cytochrome P450 705A20 | C75AK_ARATH | 2 | 2 | 58.9 | 9.01 |
| GLYMA_07G189900 | External alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial | NDB2_ARATH | 2 | 4 | 65.2 | 7.65 |
| GLYMA_09G267700 | Fasciclin-like arabinogalactan protein 10 | FLA10_ARATH | 14 | 41 | 42.5 | 5.97 |
| GLYMA_12G069300 | Fasciclin-like arabinogalactan protein 11 | FLA11_ARATH | 1 | 2 | 27.8 | 8.88 |
| GLYMA_02G308700 | Fasciclin-like arabinogalactan protein 2 | FLA2_ARATH | 9 | 23 | 43.5 | 6.87 |
| GLYMA_02G307700 | Fasciclin-like arabinogalactan protein 7 | FLA7_ARATH | 2 | 2 | 27.1 | 5.29 |
| GLYMA_05G032500 | GDP-mannose transporter GONST2 | GONS2_ARATH | 1 | 1 | 6.1 | 7.24 |
| GLYMA_11G118500 | Guanine nucleotide-binding protein subunit beta | GBB_ARATH | 1 | 4 | 41 | 7.27 |
| GLYMA_06G188900 | HIPL1 protein | HIPL1_ARATH | 1 | 2 | 75 | 5.17 |
| GLYMA_05G029800 | Hypersensitive-induced response protein 1 (AtHIR1) | HIR1_ARATH | 11 | 18 | 31.3 | 7.01 |
| GLYMA_15G248900 | Interactor of constitutive active ROPs 1 | ICR1_ARATH | 1 | 1 | 42 | 5.76 |
| GLYMA_02G079400 | Light-harvesting complex-like protein OHP2, chloroplastic | OHP2_ARATH | 5 | 12 | 19.8 | 8.87 |
| GLYMA_14G099800 | Mitochondrial carrier protein CoAc1 | COAC1_ARATH | 1 | 2 | 29.6 | 10.04 |
| GLYMA_08G115600 | Mitochondrial dicarboxylate/tricarboxylate transporter DTC | DTC_ARATH | 8 | 20 | 32 | 9.35 |
| GLYMA_13G096000 | Mitochondrial outer membrane protein porin 4 | VDAC4_ARATH | 8 | 22 | 29.7 | 8.57 |
| GLYMA_19G101100 | Mitochondrial phosphate carrier protein 3, mitochondrial | MPCP3_ARATH | 6 | 14 | 39.8 | 9.26 |
| GLYMA_09G114000 | Outer envelope pore protein 24B, chloroplastic | OP24B_ARATH | 17 | 69 | 23.7 | 9.26 |
| GLYMA_18G297700 | Piezo-type mechanosensitive ion channel homolog | PIEZO_ARATH | 2 | 2 | 21.1 | 9.85 |
| GLYMA_06G020400 | Plastocyanin minor isoform, chloroplastic | PLAS1_ARATH | 1 | 2 | 16.8 | 5.2 |
| GLYMA_20G141900 | Probable dolichyl-diphosphooligosaccharide-subunit 3B | OST3B_ARATH | 2 | 2 | 37.8 | 9.25 |
| GLYMA_15G064700 | Probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 | FQRL1_ARATH | 17 | 37 | 21.7 | 6.95 |
| GLYMA_13G212700 | Probable prolyl 4-hydroxylase 4 (AtP4H4) | P4H4_ARATH | 9 | 21 | 33.3 | 5.81 |
| GLYMA_08G288900 | SUPPRESSOR OF QUENCHING 1, chloroplastic | SOQ1_ARATH | 2 | 3 | 44.1 | 8.41 |
| GLYMA_13G307600 | Putative syntaxin-131 (AtSYP131) | SY131_ARATH | 6 | 8 | 34.5 | 7.49 |
| GLYMA_07G028500 | Pyrophosphate-energized vacuolar membrane proton pump 1 | AVP1_ARATH | 2 | 5 | 80.7 | 5.58 |
| GLYMA_11G118800 | Ras-related protein RABG3f (AtRABG3f) | RAG3F_ARATH | 2 | 3 | 23 | 5.19 |
| GLYMA_11G146900 | Receptor-like protein 4 (AtRLP4) | RLP4_ARATH | 1 | 1 | 16.4 | 9.6 |
| GLYMA_07G072100 | Serine/threonine-protein kinase BSK3 | BSK3_ARATH | 1 | 1 | 55.2 | 6.1 |
| GLYMA_02G275500 | SNF1-related protein kinase regulatory subunit beta-1 (AKIN subunit beta-1; AKINB1; AKINbeta1) | KINB1_ARATH | 1 | 1 | 30.8 | 6.32 |
| GLYMA_04G246100 | Sucrose transport protein SUC9 | SUC9_ARATH | 1 | 1 | 14.8 | 9.25 |
| GLYMA_17G152900 | Tobamovirus multiplication protein 2A (AtTOM2A) | TOM2A_ARATH | 2 | 6 | 31.5 | 5.36 |
| GLYMA_10G243500 | Transmembrane emp24 domain-containing protein p24beta3 | P24B3_ARATH | 2 | 2 | 23.9 | 7.09 |
| GLYMA_19G093900 | Triacylglycerol lipase SDP1 | SDP1_ARATH | 1 | 4 | 8.5 | 9.47 |
| GLYMA_03G214700 | UDP-glucuronic acid decarboxylase 2 | UXS2_ARATH | 10 | 29 | 48.1 | 7.97 |
| GLYMA_14G035500 | Vesicle-associated membrane protein 713 (AtVAMP713) | VA713_ARATH | 3 | 4 | 25 | 9.42 |
| GLYMA_20G014300 | Auxin efflux carrier component 3 (AtPIN3) | PIN3_ARATH | 1 | 1 | 72.2 | 7.64 |
| GLYMA_16G038800 | Binding partner of ACD11 1 | BPA1_ARATH | 2 | 4 | 29.9 | 9.11 |
| GLYMA_05G204300 | Gamma carbonic anhydrase-like 1, mitochondrial (AtCAL1; GAMMA CAL1) | GCAL1_ARATH | 9 | 42 | 27.7 | 8.18 |
| GLYMA_09G009300 | Mitochondrial pyruvate carrier 4 | MPC4_ARATH | 1 | 4 | 10.5 | 10.36 |
| GLYMA_09G271600 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 | NDUA1_ARATH | 4 | 27 | 7.3 | 8.88 |
| GLYMA_13G212700 | Probable prolyl 4-hydroxylase 4 (AtP4H4) | P4H4_ARATH | 6 | 11 | 33.3 | 5.81 |
| GLYMA_05G150300 | Protein TIC 22-like, chloroplastic | TI22L_ARATH | 2 | 2 | 28.2 | 8.4 |
| GLYMA_01G036200 | RAN GTPase-activating protein 1 (AtRanGAP1; RanGAP1) | RAGP1_ARATH | 1 | 1 | 65.1 | 8.07 |
| GLYMA_16G090700 | RPM1-interacting protein 4 | RIN4_ARATH | 1 | 1 | 27.1 | 8.29 |
| GLYMA_19G093900 | Triacylglycerol lipase SDP1 | SDP1_ARATH | 1 | 3 | 8.5 | 9.47 |
| GLYMA_07G008600 | UDP-glucuronate 4-epimerase 4 | GAE4_ARATH | 1 | 1 | 14.6 | 11.84 |
| GLYMA_14G043500 | Very-long-chain enoyl-CoA reductase | TECR_ARATH | 3 | 9 | 35.9 | 9.63 |
| GLYMA_05G214200 | V-type proton ATPase subunit E3 (V-ATPase subunit E3) | VATE3_ARATH | 10 | 22 | 27.6 | 6.8 |
| Ensembl Protein ID | Primary Protein Name | Arabidopsis Identifier | No. of Peptides | No. of PSMs | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| GLYMA_13G220000 | ABC transporter C family member 2 (ABC transporter ABCC.2; AtABCC2) | AB2C_ARATH | 2 | 2 | 181.7 | 6.61 |
| GLYMA_11G098800 | ABC transporter G family member 8 (ABC transporter ABCG.8; AtABCG8) | AB8G_ARATH | 1 | 4 | 21.2 | 9.44 |
| GLYMA_12G094000 | Alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial | NDC1_ARATH | 4 | 29 | 39.5 | 8.19 |
| GLYMA_10G167800 | Ammonium transporter 1 member 2 (AtAMT1;2) | AMT12_ARATH | 1 | 2 | 53.6 | 7.44 |
| GLYMA_11G119800 | Arabinosyltransferase RRA3 | RRA3_ARATH | 1 | 1 | 49.4 | 8.78 |
| GLYMA_04G162800 | ATP synthase subunit gamma, mitochondrial | ATPG3_ARATH | 9 | 42 | 35.2 | 9.29 |
| GLYMA_06G019400 | ATP-dependent zinc metalloprotease FTSH 5, chloroplastic (AtFTSH5) | FTSH5_ARATH | 3 | 4 | 74.3 | 6.24 |
| GLYMA_16G204600 | Bifunctional enolase 2/transcriptional activator | ENO2_ARATH | 4 | 17 | 48 | 6.47 |
| GLYMA_16G038800 | Binding partner of ACD11 1 | BPA1_ARATH | 2 | 4 | 29.9 | 9.11 |
| GLYMA_08G180800 | AtBAK1; BRI1-associated receptor kinase 1 | BAK1_ARATH | 1 | 2 | 67.9 | 5.59 |
| GLYMA_12G095700 | Chaperone protein dnaJ 2 (AtDjA2) | DNAJ2_ARATH | 5 | 15 | 46.3 | 6.28 |
| GLYMA_12G040500 | Coatomer subunit delta | COPD_ARATH | 1 | 2 | 58.4 | 5.59 |
| GLYMA_11G118600 | Coatomer subunit gamma | COPG_ARATH | 4 | 7 | 98.7 | 5.15 |
| GLYMA_11G063400 | Cystinosin homolog | CTNS_ARATH | 1 | 2 | 42.9 | 8.12 |
| GLYMA_05G110100 | Cytochrome c oxidase subunit 6a, mitochondrial (AtCOX6a) | COX6A_ARATH | 5 | 41 | 11.5 | 9.09 |
| GLYMA_17G157000 | Cytochrome c oxidase subunit 6a, mitochondrial (AtCOX6a) | COX6A_ARATH | 5 | 41 | 11.6 | 9.35 |
| GLYMA_06G176200 | Cytochrome P450 71A22 | C71AM_ARATH | 1 | 1 | 58.5 | 7.36 |
| GLYMA_10G286200 | Dihydroorotate dehydrogenase (quinone), mitochondrial (DHOdehase) | PYRD_ARATH | 1 | 2 | 48.3 | 9.31 |
| GLYMA_13G171700 | Dolichol-phosphate mannosyltransferase subunit 1 | DPM1_ARATH | 1 | 1 | 29.3 | 9.54 |
| GLYMA_09G129900 | DUF21 domain-containing protein At5g52790 | Y5279_ARATH | 1 | 1 | 39.1 | 8.59 |
| GLYMA_07G170200 | E3 ubiquitin protein ligase RIE1 | RIE1_ARATH | 1 | 1 | 39.6 | 6.34 |
| GLYMA_16G071500 | EIN2-CEND | EIN2_ARATH | 1 | 1 | 486.8 | 4.31 |
| GLYMA_02G276600 | Elongation factor 1-beta 1 (EF-1-beta 1) | EF1B1_ARATH | 11 | 43 | 24.3 | 4.68 |
| GLYMA_20G194100 | Exocyst complex component EXO70A1 (AtExo70a1) | E70A1_ARATH | 1 | 3 | 72.9 | 7.2 |
| GLYMA_17G260400 | Expansin-A11 (AtEXPA11) | EXP11_ARATH | 1 | 2 | 27.5 | 8.84 |
| GLYMA_08G239300 | FT-interacting protein 1 | FTIP1_ARATH | 1 | 2 | 117.1 | 8.57 |
| GLYMA_04G013100 | Guanine nucleotide-binding protein subunit beta | GBB_ARATH | 1 | 1 | 41 | 7.44 |
| GLYMA_17G124900 | High-affinity nitrate transporter 3.1 | NRT31_ARATH | 1 | 1 | 23.2 | 9.09 |
| GLYMA_05G133700 | LysM domain-containing GPI-anchored protein 2 | LYM2_ARATH | 1 | 3 | 40.2 | 7.24 |
| GLYMA_10G057000 | Membrane magnesium transporter | MMGT_ARATH | 1 | 1 | 11.9 | 7.03 |
| GLYMA_10G049900 | Mitochondrial import inner membrane translocase subunit TIM14-1 | TI141_ARATH | 1 | 2 | 12.2 | 10.13 |
| GLYMA_07G193700 | Mitochondrial inner membrane protein OXA1 | OXA1_ARATH | 1 | 3 | 47.1 | 9.35 |
| GLYMA_01G157300 | Monosaccharide-sensing protein 3 | MSSP3_ARATH | 2 | 3 | 79.4 | 6.18 |
| GLYMA_03G114600 | Oxygen-evolving enhancer protein 3-2, chloroplastic (OEE3) | PSBQ2_ARATH | 2 | 4 | 24.8 | 9.85 |
| GLYMA_12G001900 | Patellin-3 | PATL3_ARATH | 2 | 6 | 48.9 | 5.44 |
| GLYMA_14G199000 | Sorting nexin 2B | SNX2B_ARATH | 3 | 5 | 62.3 | 5.5 |
| GLYMA_08G180400 | Sulfate transporter 1.3 | SUT13_ARATH | 1 | 3 | 71.9 | 9.32 |
| GLYMA_10G195800 | Sulfoquinovosyl transferase SQD2 | SQD2_ARATH | 1 | 1 | 55.4 | 6.83 |
| GLYMA_13G185300 | SUN domain-containing protein 1 (AtSUN1) | SUN1_ARATH | 1 | 1 | 50.5 | 8.19 |
| GLYMA_12G032400 | Synaptotagmin-2 | SYT2_ARATH | 3 | 11 | 61.6 | 6.65 |
| GLYMA_18G215900 | Thioredoxin H9 (AtTrxh9) | TRXH9_ARATH | 1 | 2 | 15.7 | 5.17 |
| GLYMA_07G010300 | Transmembrane 9 superfamily member 7 | TMN7_ARATH | 2 | 4 | 73.7 | 8.32 |
| GLYMA_08G193400 | Transmembrane 9 superfamily member 7 | TMN7_ARATH | 2 | 4 | 73.6 | 8.22 |
| GLYMA_16G054600 | Triacylglycerol lipase SDP1 | SDP1_ARATH | 2 | 4 | 8.7 | 9.7 |
| GLYMA_03G214700 | UDP-glucuronic acid decarboxylase 2 | UXS2_ARATH | 10 | 37 | 48.1 | 7.97 |
| GLYMA_07G119500 | Vacuolar protein-sorting-associated protein 11 homolog (AtVPS11) | VPS11_ARATH | 1 | 2 | 109.4 | 6 |
| GLYMA_15G044700 | V-type proton ATPase subunit a1 (V-ATPase subunit a1) | VHAA1_ARATH | 2 | 7 | 94.2 | 6.4 |
| GLYMA_20G106500 | V-type proton ATPase subunit B2 (V-ATPase subunit B2) | VATB2_ARATH | 33 | 210 | 54.2 | 5.02 |
| GLYMA_02G065500 | V-type proton ATPase subunit C (V-ATPase subunit C) | VATC_ARATH | 13 | 50 | 42.5 | 5.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, H.M.; Chen, S.; Zhang, S.; Khalid, M.; Uzair, M.; Wilmarth, P.A.; Ahmad, S.; Lam, H.-M. Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins. Int. J. Mol. Sci. 2022, 23, 13270. https://doi.org/10.3390/ijms232113270
Rehman HM, Chen S, Zhang S, Khalid M, Uzair M, Wilmarth PA, Ahmad S, Lam H-M. Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins. International Journal of Molecular Sciences. 2022; 23(21):13270. https://doi.org/10.3390/ijms232113270
Chicago/Turabian StyleRehman, Hafiz Mamoon, Shengjie Chen, Shoudong Zhang, Memoona Khalid, Muhammad Uzair, Phillip A. Wilmarth, Shakeel Ahmad, and Hon-Ming Lam. 2022. "Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins" International Journal of Molecular Sciences 23, no. 21: 13270. https://doi.org/10.3390/ijms232113270
APA StyleRehman, H. M., Chen, S., Zhang, S., Khalid, M., Uzair, M., Wilmarth, P. A., Ahmad, S., & Lam, H.-M. (2022). Membrane Proteomic Profiling of Soybean Leaf and Root Tissues Uncovers Salt-Stress-Responsive Membrane Proteins. International Journal of Molecular Sciences, 23(21), 13270. https://doi.org/10.3390/ijms232113270







