Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression
Abstract
1. Introduction
2. Results
2.1. DSF/Copper Suppresses the Cell Proliferation of DTC Cell Lines
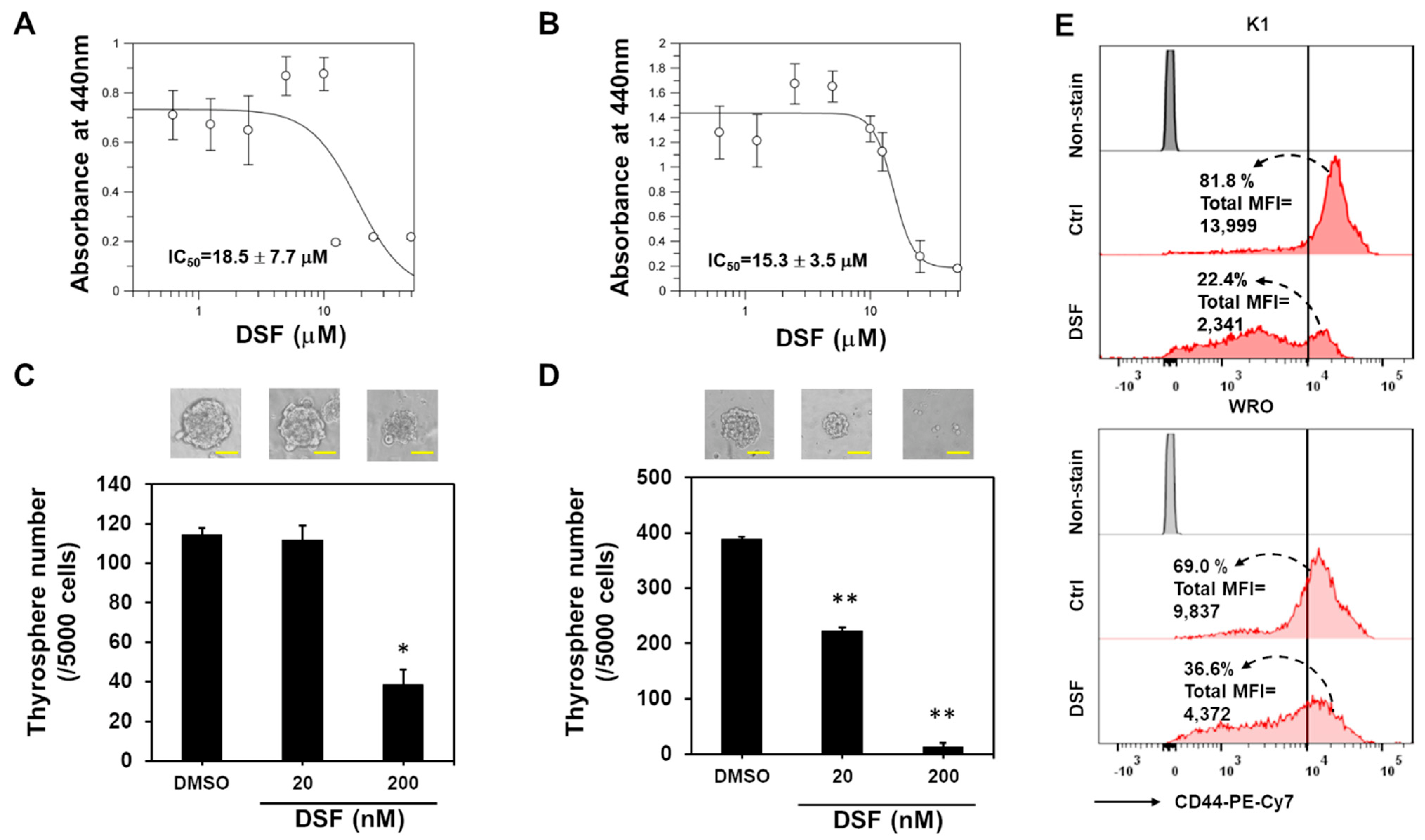
2.2. BMI1 Downregulation Is Responsible for the Anti-CSC Activity of DSF/Copper in DTC Cell Lines
2.3. DSF/Copper Inhibits BMI1 Expression through c-Myc and E2F1 Downregulation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Chemicals and Reagents
4.3. WST-1 Based Cell Proliferation Assay
4.4. Thyrosphere Cultivation
4.5. Determination of Cell Proliferation by the Incorporation of Atto-488 Conjugated dUTP
4.6. Flow Cytometrical Analyses of Cell Cycle Distributions or CD44 Expression
4.7. Quantitative RT-PCR
4.8. Manipulation of BMI1 Expression Levels in K1 Cells
4.9. Western Blot Analysis
4.10. Chromatin Immunoprecipitation
4.11. c-Myc Reporter Assay
4.12. Overexpression of c-Myc or E2F1
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hussain, F.; Iqbal, S.; Mehmood, A.; Bazarbashi, S.; ElHassan, T.; Chaudhri, N. Incidence of thyroid cancer in the Kingdom of Saudi Arabia, 2000–2010. Hematol. Oncol. Stem. Cell Ther. 2013, 6, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- Carling, T.; Udelsman, R. Thyroid cancer. Annu. Rev. Med. 2014, 65, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Devesa, S.S.; Sosa, J.A.; Check, D.; Kitahara, C.M. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA 2017, 317, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, J.; Park, S.Y.; Kim, T.H.; Kim, S.W.; Chung, J.H. Clinical course from diagnosis to death in patients with well-differentiated thyroid cancer. Cancers 2020, 12, 2323. [Google Scholar] [CrossRef]
- Aboelnaga, E.M.; Ahmed, R.A. Difference between papillary and follicular thyroid carcinoma outcomes: An experience from Egyptian institution. Cancer Biol. Med. 2015, 12, 53–59. [Google Scholar] [CrossRef]
- Araque, K.A.; Gubbi, S.; Klubo-Gwiezdzinska, J. Updates on the management of thyroid cancer. Horm. Metab. Res 2020, 52, 562–577. [Google Scholar] [CrossRef]
- Lee, S.L. Complications of radioactive iodine treatment of thyroid carcinoma. J. Natl. Compr. Cancer Netw. 2010, 8, 1277–1286. [Google Scholar] [CrossRef]
- Ahn, S.H.; Henderson, Y.C.; Williams, M.D.; Lai, S.Y.; Clayman, G.L. Detection of thyroid cancer stem cells in papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014, 99, 536–544. [Google Scholar] [CrossRef]
- Zito, G.; Richiusa, P.; Bommarito, A.; Carissimi, E.; Russo, L.; Coppola, A.; Zerilli, M.; Rodolico, V.; Criscimanna, A.; Amato, M.; et al. In vitro identification and characterization of CD133(pos) cancer stem-like cells in anaplastic thyroid carcinoma cell lines. PLoS ONE 2008, 3, e3544. [Google Scholar] [CrossRef]
- Shimamura, M.; Nagayama, Y.; Matsuse, M.; Yamashita, S.; Mitsutake, N. Analysis of multiple markers for cancer stem-like cells in human thyroid carcinoma cell lines. Endocr. J. 2014, 61, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Zito, G.; Coppola, A.; Pizzolanti, G.; Giordano, C. Heterogeneity of stem cells in the thyroid. Adv. Exp. Med. Biol. 2019, 1169, 81–93. [Google Scholar] [CrossRef]
- Khan, A.Q.; Mohamed, E.A.N.; Hakeem, I.; Nazeer, A.; Kuttikrishnan, S.; Prabhu, K.S.; Siveen, K.S.; Nawaz, Z.; Ahmad, A.; Zayed, H.; et al. Sanguinarine induces apoptosis in papillary thyroid cancer cells via generation of reactive oxygen species. Molecules 2020, 25, 1229. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Moore, R.D. Disulfiram treatment of alcoholism. Am. J. Med. 1990, 88, 647–655. [Google Scholar] [CrossRef]
- Xu, X.; Chai, S.; Wang, P.; Zhang, C.; Yang, Y.; Yang, Y.; Wang, K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015, 369, 50–57. [Google Scholar] [CrossRef]
- Chen, D.; Cui, Q.C.; Yang, H.; Dou, Q.P. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006, 66, 10425–10433. [Google Scholar] [CrossRef]
- Shimamura, M.; Kurashige, T.; Mitsutake, N.; Nagayama, Y. Aldehyde dehydrogenase activity plays no functional role in stem cell-like properties in anaplastic thyroid cancer cell lines. Endocrine 2017, 55, 934–943. [Google Scholar] [CrossRef]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent Advances in Repurposing Disulfiram and Disulfiram Derivatives as Copper-Dependent Anticancer Agents. Front. Mol. Biosci. 2021, 8, 741316. [Google Scholar] [CrossRef]
- Zhou, H.M.; Zhang, J.G.; Zhang, X.; Li, Q. Targeting cancer stem cells for reversing therapy resistance: Mechanism, signaling, and prospective agents. Signal Transduct. Target. Ther. 2021, 6, 62. [Google Scholar] [CrossRef]
- Hardin, H.; Zhang, R.; Helein, H.; Buehler, D.; Guo, Z.; Lloyd, R.V. The evolving concept of cancer stem-like cells in thyroid cancer and other solid tumors. Lab. Investig. 2017, 97, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Hajian, M.; Esmaeili, A.; Talebi, A. Comparative evaluation of BMI-1 proto-oncogene expression in normal tissue, adenoma and papillary carcinoma of human thyroid in pathology samples. BMC Res. Notes 2021, 14, 369. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Liu, G.H.; Zhang, H.; Xing, S.; Hu, L.J.; Zhao, W.F.; Xie, B.; Li, M.Z.; Zeng, B.H.; Li, Y.; et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 2013, 280, 2929–2944. [Google Scholar] [CrossRef]
- Lee, Y.C.; Chang, W.W.; Chen, Y.Y.; Tsai, Y.H.; Chou, Y.H.; Tseng, H.C.; Chen, H.L.; Wu, C.C.; Chang-Chien, J.; Lee, H.T.; et al. Hsp90alpha mediates BMI1 expression in breast cancer stem/progenitor cells through facilitating nuclear translocation of c-Myc and EZH2. Int. J. Mol. Sci. 2017, 18, 1986. [Google Scholar] [CrossRef]
- Nowak, K.; Kerl, K.; Fehr, D.; Kramps, C.; Gessner, C.; Killmer, K.; Samans, B.; Berwanger, B.; Christiansen, H.; Lutz, W. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 2006, 34, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Hu, C.; Du, P.; Huang, G. E2F1 maintains gastric cancer stemness properties by regulating stemness-associated genes. J. Oncol. 2021, 2021, 6611327. [Google Scholar] [CrossRef] [PubMed]
- Javadirad, S.M.; Mokhtari, M.; Esfandiarpour, G.; Kolahdouzan, M. The pseudogene problem and RT-qPCR data normalization; SYMPK: A suitable reference gene for papillary thyroid carcinoma. Sci. Rep. 2020, 10, 18408. [Google Scholar] [CrossRef]
- Durante, C.; Haddy, N.; Baudin, E.; Leboulleux, S.; Hartl, D.; Travagli, J.P.; Caillou, B.; Ricard, M.; Lumbroso, J.D.; De Vathaire, F.; et al. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: Benefits and limits of radioiodine therapy. J. Clin. Endocrinol. Metab. 2006, 91, 2892–2899. [Google Scholar] [CrossRef]
- Facchino, S.; Abdouh, M.; Chatoo, W.; Bernier, G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010, 30, 10096–10111. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Luo, C.W.; Huang, W.L.; Wu, C.C.; Chou, C.L.; Chen, C.I.; Chang, S.J.; Chai, C.Y.; Wang, H.C.; Chen, T.Y.; et al. BMI1-KLF4 axis deficiency improves responses to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Radiother. Oncol. 2020, 149, 249–258. [Google Scholar] [CrossRef]
- Schlumberger, M.; Tahara, M.; Wirth, L.J.; Robinson, B.; Brose, M.S.; Elisei, R.; Habra, M.A.; Newbold, K.; Shah, M.H.; Hoff, A.O.; et al. Lenvatinib versus placebo in radioiodine-refractory thyroid cancer. N. Engl. J. Med. 2015, 372, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Shi, Y.; Lv, Y.; Yuan, S.; Ramirez, C.F.A.; Lieftink, C.; Wang, L.; Wang, S.; Wang, C.; Dias, M.H.; et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 2021, 595, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Ngo, M.T.; Peng, S.W.; Kuo, Y.C.; Lin, C.Y.; Wu, M.H.; Chuang, C.H.; Kao, C.X.; Jeng, H.Y.; Lin, G.W.; Ling, T.Y.; et al. A Yes-Associated Protein (YAP) and Insulin-Like Growth Factor 1 Receptor (IGF-1R) signaling loop is involved in sorafenib resistance in hepatocellular carcinoma. Cancers 2021, 13, 3812. [Google Scholar] [CrossRef] [PubMed]
- Creighton, C.J.; Hilger, A.M.; Murthy, S.; Rae, J.M.; Chinnaiyan, A.M.; El-Ashry, D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006, 66, 3903–3911. [Google Scholar] [CrossRef]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 2018, 6, 282–300.e282. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal. Transduct. Target Ther. 2018, 3, 5. [Google Scholar] [CrossRef]
- Huang, C.L.; Liu, D.; Nakano, J.; Yokomise, H.; Ueno, M.; Kadota, K.; Wada, H. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 6938–6946. [Google Scholar] [CrossRef]
- Liu, X.; Hu, C. Novel potential therapeutic target for E2F1 and prognostic factors of E2F1/2/3/5/7/8 in human gastric cancer. Mol. Ther. Methods. Clin. Dev. 2020, 18, 824–838. [Google Scholar] [CrossRef]
- Onda, M.; Nagai, H.; Yoshida, A.; Miyamoto, S.; Asaka, S.I.; Akaishi, J.; Takatsu, K.; Nagahama, M.; Ito, K.; Shimizu, K.; et al. Up-regulation of transcriptional factor E2F1 in papillary and anaplastic thyroid cancers. J. Hum. Genet. 2004, 49, 312–318. [Google Scholar] [CrossRef]
- Chen, C.L.; Uthaya Kumar, D.B.; Punj, V.; Xu, J.; Sher, L.; Tahara, S.M.; Hess, S.; Machida, K. NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016, 23, 206–219. [Google Scholar] [CrossRef]
- Riverso, M.; Montagnani, V.; Stecca, B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene 2017, 36, 3322–3333. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.; Bertino, D.B. E2F-1 as an anticancer drug target. Oncol. Rev. 2009, 3, 207–214. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Skrott, Z.; Mistrik, M.; Andersen, K.K.; Friis, S.; Majera, D.; Gursky, J.; Ozdian, T.; Bartkova, J.; Turi, Z.; Moudry, P.; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Cvek, B. Nonprofit drugs as the salvation of the world’s healthcare systems: The case of Antabuse (disulfiram). Drug Discov. Today 2012, 17, 409–412. [Google Scholar] [CrossRef]
- Wang, K.; Michelakos, T.; Wang, B.; Shang, Z.; DeLeo, A.B.; Duan, Z.; Hornicek, F.J.; Schwab, J.H.; Wang, X. Targeting cancer stem cells by disulfiram and copper sensitizes radioresistant chondrosarcoma to radiation. Cancer Lett. 2021, 505, 37–48. [Google Scholar] [CrossRef]
- Shen, H.T.; Chien, P.J.; Chen, S.H.; Sheu, G.T.; Jan, M.S.; Wang, B.Y.; Chang, W.W. BMI1-mediated pemetrexed resistance in non-small cell lung cancer cells is associated with increased SP1 activation and cancer stemness. Cancers 2020, 12, 2069. [Google Scholar] [CrossRef]
- Wang, W.L.; Hong, G.C.; Chien, P.J.; Huang, Y.H.; Lee, H.T.; Wang, P.H.; Lee, Y.C.; Chang, W.W. Tribbles pseudokinase 3 contributes to cancer stemness of endometrial cancer cells by regulating beta-catenin expression. Cancers 2020, 12, 3785. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Y.-L.; Chien, P.-J.; Hsieh, H.-C.; Shen, H.-T.; Lee, H.-T.; Chen, S.-M.; Chang, W.-W. Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression. Int. J. Mol. Sci. 2022, 23, 13276. https://doi.org/10.3390/ijms232113276
Ni Y-L, Chien P-J, Hsieh H-C, Shen H-T, Lee H-T, Chen S-M, Chang W-W. Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression. International Journal of Molecular Sciences. 2022; 23(21):13276. https://doi.org/10.3390/ijms232113276
Chicago/Turabian StyleNi, Yung-Lun, Peng-Ju Chien, Hung-Chia Hsieh, Huan-Ting Shen, Hsueh-Te Lee, Shih-Ming Chen, and Wen-Wei Chang. 2022. "Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression" International Journal of Molecular Sciences 23, no. 21: 13276. https://doi.org/10.3390/ijms232113276
APA StyleNi, Y.-L., Chien, P.-J., Hsieh, H.-C., Shen, H.-T., Lee, H.-T., Chen, S.-M., & Chang, W.-W. (2022). Disulfiram/Copper Suppresses Cancer Stem Cell Activity in Differentiated Thyroid Cancer Cells by Inhibiting BMI1 Expression. International Journal of Molecular Sciences, 23(21), 13276. https://doi.org/10.3390/ijms232113276





