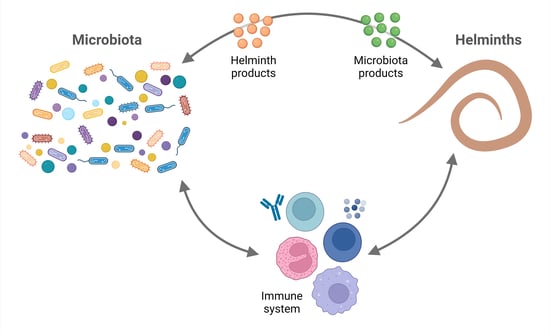
Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”
Abstract
:1. Introduction
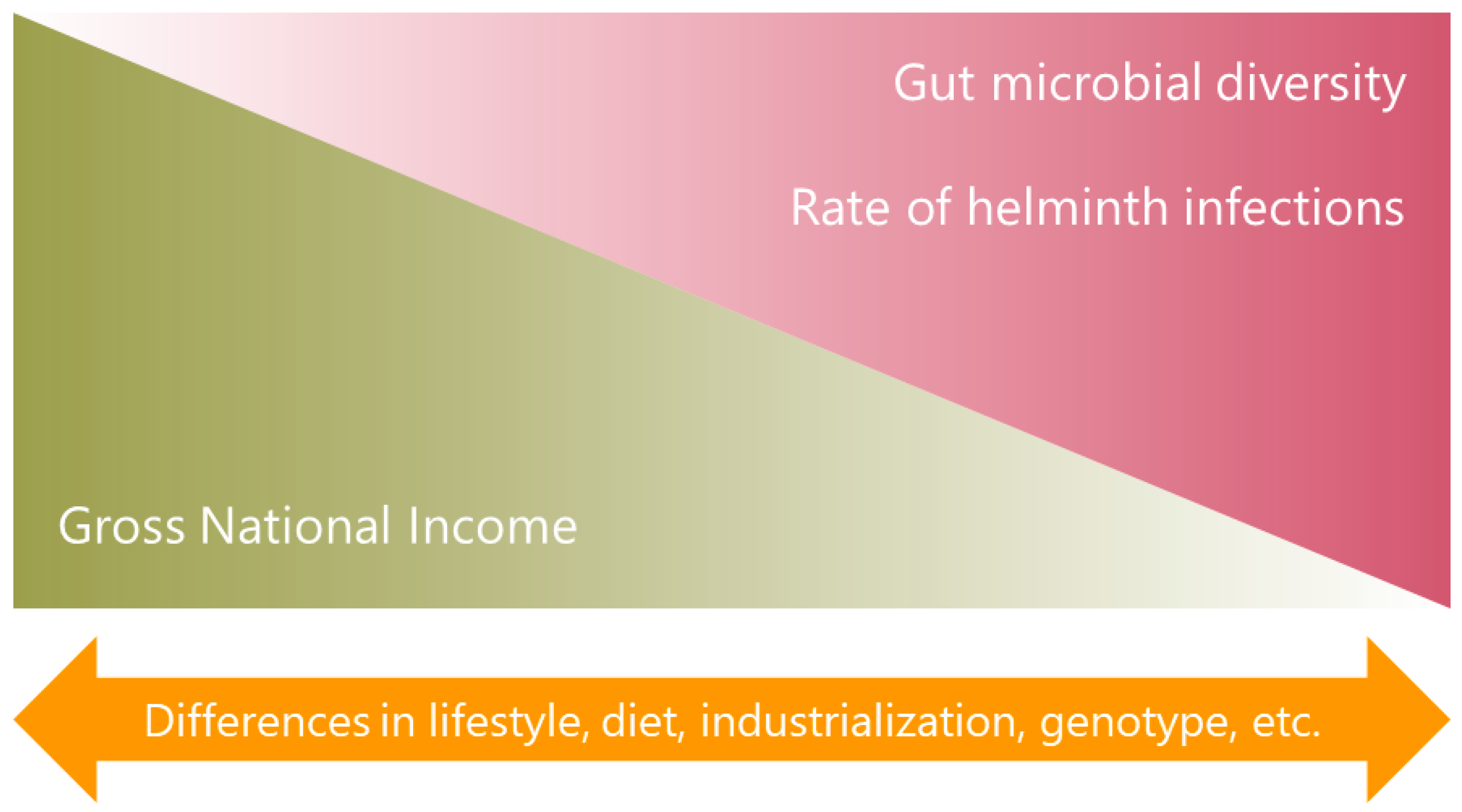
2. Characteristics of the Microbiota in Individuals from Low- and Middle-Income Countries
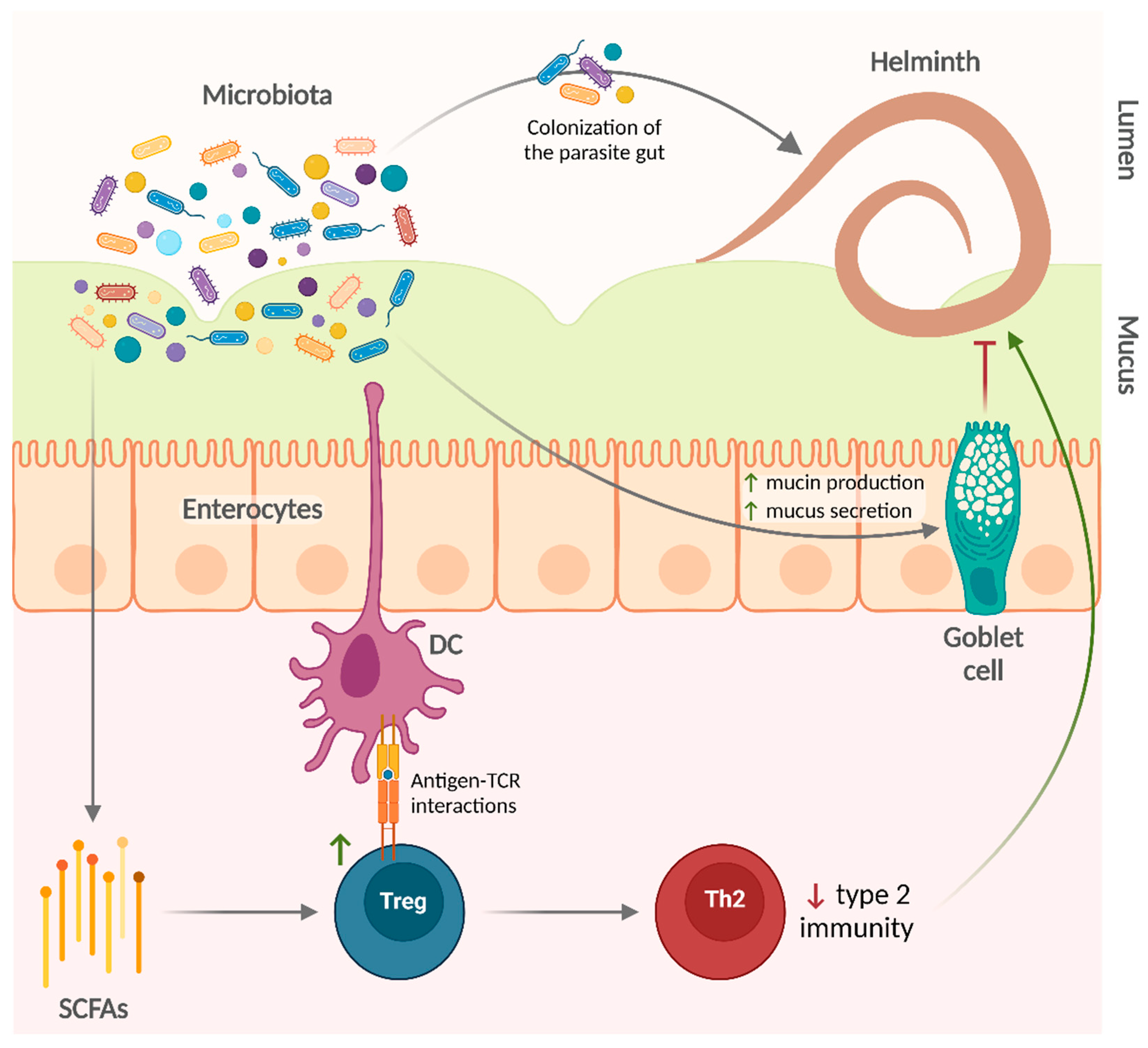
3. Effects of the Microbiota on Helminth Infections
3.1. Animal Models
3.2. Humans
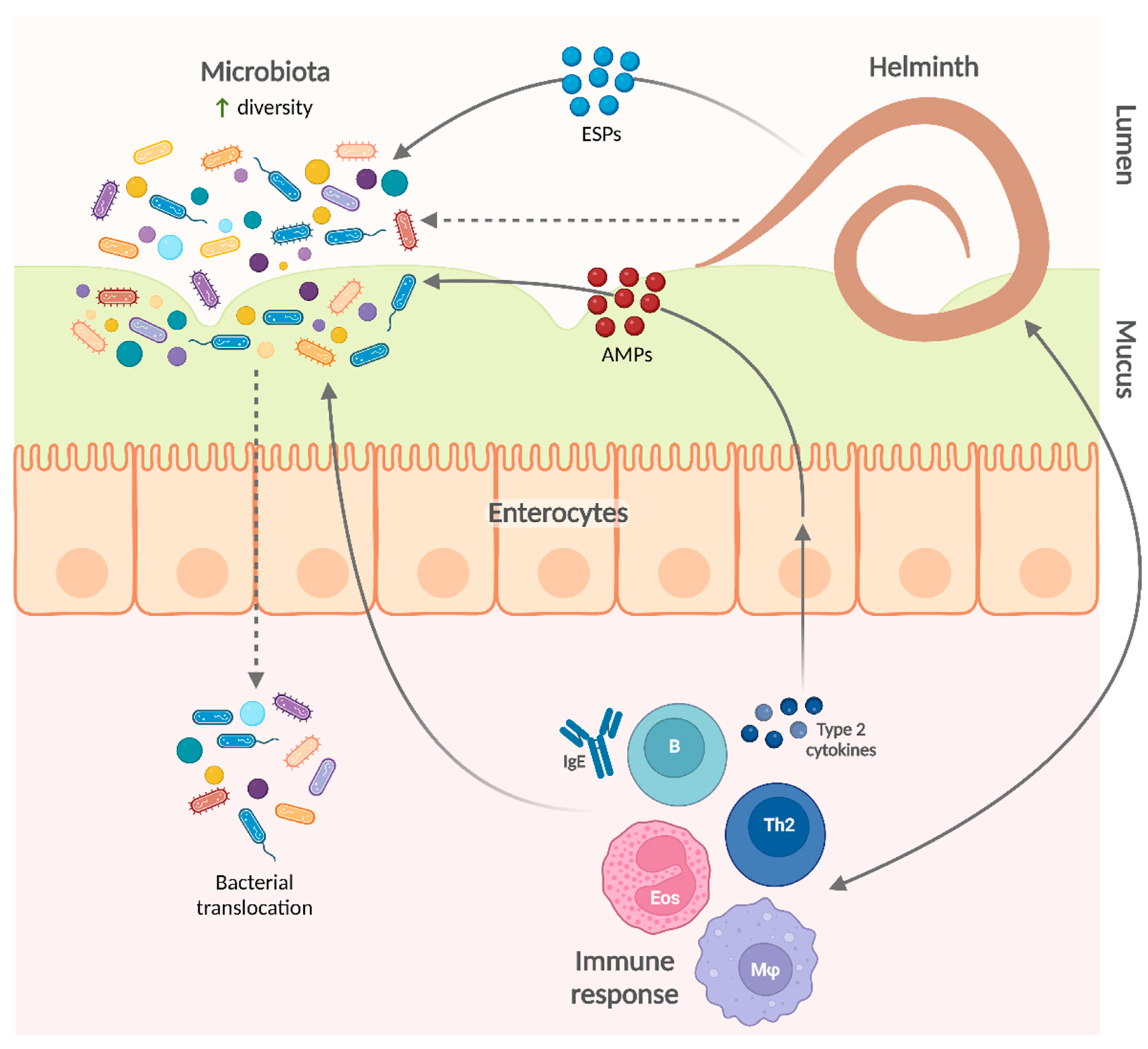
4. Influence of Helminth Infections on the Microbiota
4.1. Animal Models
4.2. Humans
5. Biological and Clinical Consequences of the Modifications Exerted by Helminth Infections on the Microbiota
5.1. Animal Models
5.2. Humans
6. Discussion
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turpin, W.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Kevans, D.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Panaccione, R.; Otley, A.; et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 2016, 48, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.A.; Bacigalupe, R.; Wang, J.; Rühlemann, M.C.; Tito, R.Y.; Falony, G.; Joossens, M.; Vieira-Silva, S.; Henckaerts, L.; Rymenans, L.; et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol. 2020, 5, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A.W. 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin. Exp. Immunol. 2010, 160, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, R.P. Microbiome data dominated by wealthy countries. Science 2022, 375, 709. [Google Scholar] [CrossRef]
- Hosang, L.; Canals, R.C.; van der Flier, F.J.; Hollensteiner, J.; Daniel, R.; Flugel, A.; Odoardi, F. The lung microbiome regulates brain autoimmunity. Nature 2022, 603, 138–144. [Google Scholar] [CrossRef]
- Zakzuk, J.; Casadiego, S.; Mercado, A.; Alvis-Guzman, N.; Caraballo, L. Ascaris lumbricoides infection induces both, reduction and increase of asthma symptoms in a rural community. Acta Trop. 2018, 187, 1–4. [Google Scholar] [CrossRef]
- Jõgi, N.O.; Kitaba, N.; Storaas, T.; Schlünssen, V.; Triebner, K.; Holloway, J.W.; Horsnell, W.G.C.; Svanes, C.; Bertelsen, R.J. Ascaris exposure and its association with lung function, asthma, and DNA methylation in Northern Europe. J. Allergy Clin. Immunol. 2021, 149, 1960–1969. [Google Scholar] [CrossRef]
- Buendía, E.; Zakzuk, J.; San-Juan-Vergara, H.; Zurek, E.; Ajami, N.J.; Caraballo, L. Gut microbiota components are associated with fixed airway obstruction in asthmatic patients living in the tropics. Sci. Rep. 2018, 8, 9582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The World Bank. World Bank Country and Lending Groups. Available online: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups (accessed on 3 July 2022).
- The World Bank. Population, Total-Low & Middle Income. Available online: https://data.worldbank.org/indicator/SP.POP.TOTL?locations=XO (accessed on 3 July 2022).
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vujkovic-cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [Green Version]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The Gut Microbiota of Rural Papua New Guineans: Composition, Diversity Patterns, and Ecological Processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Tang, M.S.; Lim, Y.A.L.; Choy, S.H.; Kurtz, Z.D.; Cox, L.M.; Gundra, U.M.; Cho, I.; Bonneau, R.; Blaser, M.J.; et al. Helminth Colonization Is Associated with Increased Diversity of the Gut Microbiota. PLoS Negl. Trop. Dis. 2014, 8, e2880. [Google Scholar] [CrossRef] [Green Version]
- Rosa, B.A.; Supali, T.; Gankpala, L.; Djuardi, Y.; Sartono, E.; Zhou, Y.; Fischer, K.; Martin, J.; Tyagi, R.; Bolay, F.K.; et al. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 2018, 6, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E.; et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Jha, A.R.; Davenport, E.R.; Gautam, Y.; Bhandari, D.; Tandukar, S.; Ng, K.M.; Fragiadakis, G.K.; Holmes, S.; Gautam, G.P.; Leach, J.; et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018, 16, e2005396. [Google Scholar] [CrossRef]
- Pehrsson, E.C.; Tsukayama, P.; Patel, S.; Mejía-Bautista, M.; Sosa-Soto, G.; Navarrete, K.M.; Calderon, M.; Cabrera, L.; Hoyos-Arango, W.; Bertoli, M.T.; et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature 2016, 533, 212–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vangay, P.; Johnson, A.J.; Ward, T.L.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Hillmann, B.M.; Lucas, S.K.; Beura, L.K.; Thompson, E.A.; Till, L.M.; et al. US Immigration Westernizes the Human Gut Microbiome. Cell 2018, 175, 962–972.e910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copeland, J.K.; Chao, G.; Vanderhout, S.; Acton, E.; Wang, P.W.; Benchimol, E.I.; El Sohami, A.; Croitoru, K.; Gommerman, J.L.; Guttman, D.S.; et al. The Impact of Migration on the Gut Metagenome of South Asian Canadians. Gut Microbes 2021, 13, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Amaruddin, A.I.; Hamid, F.; Koopman, J.P.R.; Muhammad, M.; Brienen, E.A.T.; van Lieshout, L.; Geelen, A.R.; Wahyuni, S.; Kuijper, E.J.; Sartono, E.; et al. The Bacterial Gut Microbiota of Schoolchildren from High and Low Socioeconomic Status: A Study in an Urban Area of Makassar, Indonesia. Microorganisms 2020, 8, 961. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, B.; Oundo, J.; Hossain, M.A.; Antonio, M.; Tamboura, B.; Walker, A.W.; Paulson, J.N.; Parkhill, J.; Omore, R.; Faruque, A.S.G.; et al. Microbiota That Affect Risk for Shigellosis in Children in Low-Income Countries. Emerg. Infect. Dis. 2015, 21, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pop, M.; Walker, A.W.; Paulson, J.; Lindsay, B.; Antonio, M.; Hossain, M.A.; Oundo, J.; Tamboura, B.; Mai, V.; Astrovskaya, I.; et al. Diarrhea in young children from low-income countries leads to large-scale alterations in wintestinal microbiota composition. Genome Biol. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Rausch, S.; Midha, A.; Kuhring, M.; Affinass, N.; Radonic, A.; Kühl, A.A.; Bleich, A.; Renard, B.Y.; Hartmann, S. Parasitic Nematodes Exert Antimicrobial Activity and Benefit from Microbiota-Driven Support for Host Immune Regulation. Front. Immunol. 2018, 9, 2282. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, L.A.; Smith, K.A.; Filbey, K.J.; Harcus, Y.; Hewitson, J.P.; Redpath, S.A.; Valdez, Y.; Yebra, M.J.; Finlay, B.B.; Maizels, R.M. Commensal-pathogen interactions in the intestinal tract. Gut Microbes 2014, 5, 522–532. [Google Scholar] [CrossRef] [Green Version]
- Hayes, K.S.; Bancroft, A.J.; Goldrick, M.; Portsmouth, C.; Roberts, I.S.; Grencis, R.K. Exploitation of the Intestinal Microflora by the Parasitic Nematode Trichuris muris. Science 2010, 328, 1391–1394. [Google Scholar] [CrossRef] [Green Version]
- White, E.C.; Houlden, A.; Bancroft, A.J.; Hayes, K.S.; Goldrick, M.; Grencis, R.K.; Roberts, I.S. Manipulation of host and parasite microbiotas: Survival strategies during chronic nematode infection. Sci. Adv. 2018, 4, 1–10. [Google Scholar] [CrossRef]
- Dea-Ayuela, M.A.; Rama-Iñiguez, S.; Bolás-Fernandez, F. Enhanced susceptibility to Trichuris muris infection of B10Br mice treated with the probiotic Lactobacillus casei. Int. Immunopharmacol. 2008, 8, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Liu, Y.; Wang, J.; Wang, X.; Tang, B.; Liu, M.; Liu, X. β-Glucan-triggered Akkermansia muciniphila expansion facilitates the expulsion of intestinal helminth via TLR2 in mice. Carbohydr. Polym. 2022, 275, 118719. [Google Scholar] [CrossRef]
- Lee, S.C.; Tang, M.S.; Easton, A.V.; Devlin, J.C.; Chua, L.L.; Cho, I.; Moy, F.M.; Khang, T.F.; Lim, Y.A.L.; Loke, P.N. Linking the effects of helminth infection, diet and the gut microbiota with human whole-blood signatures. PLoS Pathog. 2019, 15, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Pillai, A.; Ueno, S.; Zhang, H.; Lee, J.M.; Kato, Y. Cecropin P1 and novel nematode cecropins: A bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 2005, 390, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abner, S.R.; Parthasarathy, G.; Hill, D.E.; Mansfield, L.S. Trichuris suis: Detection of antibacterial activity in excretory-secretory products from adults. Exp. Parasitol. 2001, 99, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Steinel, N.; Weber, J.; Ma, L.; Smith, C.; Correa, D.; Zhu, B.; Bolnick, D.; Wang, G. The gut microbiota response to helminth infection depends on host sex and genotype. ISME J. 2020, 14, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Kreisinger, J.; Bastien, G.; Hauffe, H.C.; Marchesi, J.; Perkins, S.E. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140295. [Google Scholar] [CrossRef] [Green Version]
- Walk, S.T.; Blum, A.M.; Ewing, S.A.-S.; Weinstock, J.V.; Young, V.B. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm. Bowel Dis. 2010, 16, 1841–1849. [Google Scholar] [CrossRef]
- Fricke, W.F.; Song, Y.; Wang, A.-J.; Smith, A.; Grinchuk, V.; Pei, C.; Ma, B.; Lu, N.; Urban, J.F.; Shea-Donohue, T.; et al. Type 2 immunity-dependent reduction of segmented filamentous bacteria in mice infected with the helminthic parasite Nippostrongylus brasiliensis. Microbiome 2015, 3, 40. [Google Scholar] [CrossRef] [Green Version]
- Rapin, A.; Chuat, A.; Lebon, L.; Zaiss, M.M.; Marsland, B.J.; Harris, N.L. Infection with a small intestinal helminth, Heligmosomoides polygyrus bakeri, consistently alters microbial communities throughout the murine small and large intestine. Int. J. Parasitol. 2020, 50, 35–46. [Google Scholar] [CrossRef]
- Rausch, S.; Held, J.; Fischer, A.; Heimesaat, M.M.; Kühl, A.A.; Bereswill, S.; Hartmann, S. Small Intestinal Nematode Infection of Mice Is Associated with Increased Enterobacterial Loads alongside the Intestinal Tract. PLoS ONE 2013, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, F.; Urban, J.F.; Paerewijck, O.; Geldhof, P.; Li, R.W. Ascaris suum infection was associated with a worm-independent reduction in microbial diversity and altered metabolic potential in the porcine gut microbiome. Int. J. Parasitol. 2019, 49, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.R.; Krych, L.; Ahmad, H.F.; Nejsum, P.; Skovgaard, K.; Nielsen, D.S.; Thamsborg, S.M. A polyphenol-enriched diet and Ascaris suum infection modulate mucosal immune responses and gut microbiota composition in pigs. PLoS ONE 2017, 12, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Rosa, B.A.; Snowden, C.; Martin, J.; Fischer, K.; Kupritz, J.; Beshah, E.; Supali, T.; Gankpala, L.; Fischer, P.U.; Urban, J.F.; et al. Whipworm-Associated Intestinal Microbiome Members Consistent Across Both Human and Mouse Hosts. Front. Cell. Infect. Microbiol. 2021, 11, 637570. [Google Scholar] [CrossRef] [PubMed]
- Houlden, A.; Hayes, K.S.; Bancroft, A.J.; Worthington, J.J.; Wang, P.; Grencis, R.K.; Roberts, I.S. Chronic Trichuris muris infection in C57BL/6 mice causes significant changes in host microbiota and metabolome: Effects reversed by pathogen clearance. PLoS ONE 2015, 10, e0125945. [Google Scholar] [CrossRef]
- Holm, J.B.; Sorobetea, D.; Kiilerich, P.; Ramayo-Caldas, Y.; Estellé, J.; Ma, T.; Madsen, L.; Kristiansen, K.; Svensson-Frej, M. Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of lactobacilli. PLoS ONE 2015, 10, 1–22. [Google Scholar] [CrossRef]
- Li, R.W.; Wu, S.; Li, W.; Navarro, K.; Couch, R.D.; Hill, D.; Urban, J.F. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect. Immun. 2012, 80, 2150–2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Li, R.W.; Li, W.; Beshah, E.; Dawson, H.D.; Urban, J.F. Worm Burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS ONE 2012, 7, e35470. [Google Scholar] [CrossRef]
- Stolzenbach, S.; Myhill, L.J.; Andersen, L.O.B.; Krych, L.; Mejer, H.; Williams, A.R.; Nejsum, P.; Stensvold, C.R.; Nielsen, D.S.; Thamsborg, S.M. Dietary Inulin and Trichuris suis Infection Promote Beneficial Bacteria Throughout the Porcine Gut. Front. Microbiol. 2020, 11, 312. [Google Scholar] [CrossRef] [Green Version]
- Schachter, J.; Alvarinho de Oliveira, D.; da Silva, C.M.; de Barros Alencar, A.C.M.; Duarte, M.; da Silva, M.M.P.; Ignácio, A.C.d.P.R.; Lopes-Torres, E.J. Whipworm Infection Promotes Bacterial Invasion, Intestinal Microbiota Imbalance, and Cellular Immunomodulation. Infect. Immun. 2020, 88, e00642-19. [Google Scholar] [CrossRef]
- Nyangahu, D.D.; Darby, M.; Havyarimana, E.; Brown, B.P.; Horsnell, W.; Jaspan, H.B. Preconception helminth infection alters offspring microbiota and immune subsets in a mouse model. Parasite Immunol. 2020, 42, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kupritz, J.; Angelova, A.; Nutman, T.B.; Gazzinelli-Guimaraes, P.H. Helminth-Induced Human Gastrointestinal Dysbiosis: A Systematic Review and Meta-Analysis Reveals Insights into Altered Taxon Diversity and Microbial Gradient Collapse. mBio 2021, 12, e02890-21. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.A.; Krause, L.; McManus, D.P.; Morrison, M.; Weerakoon, K.G.; Connor, M.C.; Olveda, R.M.; Ross, A.G.; Gobert, G.N. Helminths, polyparasitism, and the gut microbiome in the Philippines. Int. J. Parasitol. 2020, 50, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Toro-Londoño, M.A.; Bedoya-Urrego, K.; Garcia-Montoya, G.M.; Galvan-Diaz, A.L.; Alzate, J.F. Intestinal parasitic infection alters bacterial gut microbiota in children. PeerJ 2019, 2019, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramanan, D.; Bowcutt, R.; Lee, S.C.; Tang, M.S.; Kurtz, Z.D.; Ding, Y.; Honda, K.; Gause, W.C.; Blaser, M.J.; Bonneau, R.A.; et al. Helminth infection promotes colonization resistance via type 2 immunity. Science 2016, 352, 608–612. [Google Scholar] [CrossRef] [Green Version]
- Huwe, T.; Prusty, B.K.; Ray, A.; Lee, S.; Ravindran, B.; Michael, E. Interactions between parasitic infections and the human gut microbiome in Odisha, India. Am. J. Trop. Med. Hyg. 2019, 100, 1486–1489. [Google Scholar] [CrossRef]
- Jenkins, T.P.; Rathnayaka, Y.; Perera, P.K.; Peachey, L.E.; Nolan, M.J.; Krause, L.; Rajakaruna, R.S.; Cantacessi, C. Infections by human gastrointestinal helminths are associated with changes in faecal microbiota diversity and composition. PLoS ONE 2017, 12, 1–18. [Google Scholar] [CrossRef]
- Cooper, P.; Walker, A.W.; Reyes, J.; Chico, M.; Salter, S.J.; Vaca, M.; Parkhill, J. Patent Human Infections with the Whipworm, Trichuris trichiura, Are Not Associated with Alterations in the Faecal Microbiota. PLoS ONE 2013, 8, e76573. [Google Scholar] [CrossRef] [Green Version]
- Pane, S.; Sacco, A.; Iorio, A.; Romani, L.; Putignani, L. Strongyloides stercoralis infestation in a child: How a nematode can affect gut microbiota. Int. J. Mol. Sci. 2021, 22, 2131. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, Z.; Shen, Y.; Rosa, B.A.; Martin, J.; Cao, S.; Zhou, Y.; Mitreva, M.; Cao, J. Alteration of the fecal microbiota in Chinese patients with Schistosoma japonicum infection. Parasite 2021, 28, 1. [Google Scholar] [CrossRef]
- Kay, G.L.; Millard, A.; Sergeant, M.J.; Midzi, N.; Gwisai, R.; Mduluza, T.; Ivens, A.; Nausch, N.; Mutapi, F.; Pallen, M. Differences in the faecal microbiome in schistosoma haematobium infected children vs. uninfected children. PLoS Negl. Trop. Dis. 2015, 9, e0003861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Jiang, Z.; Huang, W.; Yin, J.; Ou, S.; Jiang, Y.; Meng, L.; Cao, S.; Yu, A.; Cao, J.; et al. Altered Gut Microbiota Composition in Subjects Infected with Clonorchis sinensis. Front. Microbiol. 2018, 9, 2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prommi, A.; Prombutara, P.; Watthanakulpanich, D.; Adisakwattana, P.; Kusolsuk, T.; Yoonuan, T.; Poodeepiyasawat, A.; Homsuwan, N.; Prummongkol, S.; Tanita, M.; et al. Intestinal parasites in rural communities in Nan Province, Thailand: Changes in bacterial gut microbiota associated with minute intestinal fluke infection. Parasitology 2020, 147, 972–984. [Google Scholar] [CrossRef]
- Easton, A.V.; Quiñones, M.; Vujkovic-Cvijin, I.; Oliveira, R.G.; Kepha, S.; Odiere, M.R.; Anderson, R.M.; Belkaid, Y.; Nutman, T.B. The impact of anthelmintic treatment on human gut microbiota based on cross-sectional and pre-and postdeworming comparisons in Western Kenya. mBio 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Umesaki, Y.; Setoyama, H.; Matsumoto, S.; Imaoka, A.; Itoh, K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999, 67, 3504–3511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.; Chain, F.; Miquel, S.; Lu, J.; Gratadoux, J.J.; Sokol, H.; Verdu, E.F.; Bercik, P.; Bermudez-Humaran, L.G.; Langella, P. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm. Bowel Dis. 2014, 20, 417–430. [Google Scholar] [CrossRef]
- Ramanan, D.; Tang, M.S.; Bowcutt, R.; Loke, P.n.; Cadwell, K. Bacterial Sensor Nod2 Prevents Inflammation of the Small Intestine by Restricting the Expansion of the Commensal Bacteroides vulgatus. Immunity 2014, 41, 311–324. [Google Scholar] [CrossRef] [Green Version]
- Shute, A.; Callejas, B.E.; Li, S.; Wang, A.; Jayme, T.S.; Ohland, C.; Lewis, I.A.; Layden, B.T.; Buret, A.G.; McKay, D.M. Cooperation between host immunity and the gut bacteria is essential for helminth-evoked suppression of colitis. Microbiome 2021, 9, 186. [Google Scholar] [CrossRef]
- Su, C.; Su, L.; Li, Y.; Long, S.R.; Chang, J.; Zhang, W.; Walker, W.A.; Xavier, R.J.; Cherayil, B.J.; Shi, H.N. Helminth-induced alterations of the gut microbiota exacerbate bacterial colitis. Mucosal Immunol. 2018, 11, 144–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaiss, M.M.; Rapin, A.; Lebon, L.; Dubey, L.K.; Mosconi, I.; Sarter, K.; Piersigilli, A.; Menin, L.; Walker, A.W.; Rougemont, J.; et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity 2015, 43, 998–1010. [Google Scholar] [CrossRef] [Green Version]
- Su, C.W.; Chen, C.Y.; Jiao, L.; Long, S.R.; Mao, T.; Ji, Q.; O’Donnell, S.; Stanton, C.; Zheng, S.; Walker, W.A.; et al. Helminth-Induced and Th2-Dependent Alterations of the Gut Microbiota Attenuate Obesity Caused by High-Fat Diet. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.A.; Choi, J.H.; Baek, K.W.; Lee, D.I.; Jeong, M.J.; Yu, H.S. Trichinella spiralis infection ameliorated diet-induced obesity model in mice. Int. J. Parasitol. 2021, 51, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Khudhair, Z.; Alhallaf, R.; Eichenberger, R.M.; Whan, J.; Kupz, A.; Field, M.; Krause, L.; Wilson, D.T.; Daly, N.L.; Giacomin, P.; et al. Gastrointestinal Helminth Infection Improves Insulin Sensitivity, Decreases Systemic Inflammation, and Alters the Composition of Gut Microbiota in Distinct Mouse Models of Type 2 Diabetes. Front. Endocrinol. 2021, 11, 606530. [Google Scholar] [CrossRef] [PubMed]
- Pace, F.; Carvalho, B.M.; Zanotto, T.M.; Santos, A.; Guadagnini, D.; Silva, K.L.C.; Mendes, M.C.S.; Rocha, G.Z.; Alegretti, S.M.; Santos, G.A.; et al. Helminth infection in mice improves insulin sensitivity via modulation of gut microbiota and fatty acid metabolism. Pharmacol. Res. 2018, 132, 33–46. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, A.J.; McSorley, H.J.; Davidson, D.J.; Fitch, P.M.; Errington, C.; Mackenzie, K.J.; Gollwitzer, E.S.; Johnston, C.J.C.; MacDonald, A.S.; Edwards, M.R.; et al. Enteric helminth-induced type I interferon signaling protects against pulmonary virus infection through interaction with the microbiota. J. Allergy Clin. Immunol. 2017, 140, 1068–1078.e1066. [Google Scholar] [CrossRef] [Green Version]
- Croese, J.; Giacomin, P.; Navarro, S.; Clouston, A.; McCann, L.; Dougall, A.; Ferreira, I.; Susianto, A.; O’Rourke, P.; Howlett, M.; et al. Experimental hookworm infection and gluten microchallenge promote tolerance in celiac disease. J. Allergy Clin. Immunol. 2015, 135, 508–516.e505. [Google Scholar] [CrossRef]
- Cantacessi, C.; Giacomin, P.; Croese, J.; Zakrzewski, M.; Sotillo, J.; McCann, L.; Nolan, M.J.; Mitreva, M.; Krause, L.; Loukas, A. Impact of experimental hookworm infection on the human gut microbiota. J. Infect. Dis. 2014, 210, 1431–1434. [Google Scholar] [CrossRef]
- Giacomin, P.; Zakrzewski, M.; Croese, J.; Su, X.; Sotillo, J.; McCann, L.; Navarro, S.; Mitreva, M.; Krause, L.; Loukas, A.; et al. Experimental hookworm infection and escalating gluten challenges are associated with increased microbial richness in celiac subjects. Sci. Rep. 2015, 5, 13797. [Google Scholar] [CrossRef]
- Giacomin, P.; Zakrzewski, M.; Jenkins, T.P.; Su, X.; Al-Hallaf, R.; Croese, J.; De Vries, S.; Grant, A.; Mitreva, M.; Loukas, A.; et al. Changes in duodenal tissue-associated microbiota following hookworm infection and consecutive gluten challenges in humans with coeliac disease. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Bisgaard, H.; Li, N.; Bonnelykke, K.; Chawes, B.L.K.; Skov, T.; Paludan-Müller, G.; Stokholm, J.; Smith, B.; Krogfelt, K.A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011, 128, 646–652.e645. [Google Scholar] [CrossRef]
- Sjögren, Y.M.; Jenmalm, M.C.; Böttcher, M.F.; Björkstén, B.; Sverremark-Ekström, E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin. Exp. Allergy 2009, 39, 518–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012, 129, 434–440.e432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin Study Indicates Loss of Interaction Between Microbiota and Mucosa of Patients with Ulcerative Colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [Green Version]
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Stewart, A.W.; Mitchell, E.A.; Pearce, N.; Strachan, D.P.; Weiland, S.K.; Asthma, I.S.C.I.S.f.; Allergy in, C. The relationship of per capita gross national product to the prevalence of symptoms of asthma and other atopic diseases in children (ISAAC). Int. J. Epidemiol. 2001, 30, 173–179. [Google Scholar] [CrossRef]
- Mallol, J.; Crane, J.; von Mutius, E.; Odhiambo, J.; Keil, U.; Stewart, A.; Group, I.P.T.S. The International Study of Asthma and Allergies in Childhood (ISAAC) Phase Three: A global synthesis. Allergol. Immunopathol. 2013, 41, 73–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.-O.; Kim, H.-J.; Kim, Y.-J.; Kang, M.-J.; Kwon, J.-W.; Seo, J.-H.; Kim, H.Y.; Kim, B.-J.; Yu, J.; Hong, S.-J. Asthma Prevention by Lactobacillus Rhamnosus in a Mouse Model is Associated with CD4 + CD25 + Foxp3 + T Cells. Allergy Asthma Immunol. Res. 2012, 4, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, K.; Inman, M.D.; Bienenstock, J.; Forsythe, P. Lactobacillus reuteri –induced Regulatory T cells Protect against an Allergic Airway Response in Mice. Am. J. Respir. Crit. Care Med. 2009, 179, 186–193. [Google Scholar] [CrossRef]
- Kim, H.-J.; Kim, Y.-J.; Kang, M.-J.; Seo, J.-H.; Kim, H.-Y.; Jeong, S.K.; Lee, S.-H.; Kim, J.-M.; Hong, S.-J. A novel mouse model of atopic dermatitis with epicutaneous allergen sensitization and the effect of Lactobacillus rhamnosus. Exp. Dermatol. 2012, 21, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.W.; Myers, L.E.S.; Ray, L.; Song, S.-C.; Nasr, T.R.; Berardinelli, A.J.; Kundu, K.; Murthy, N.; Hansen, J.M.; Neish, A.S. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic. Biol. Med. 2009, 47, 1205–1211. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, S.; Hara, T.; Nagaoka, M.; Mike, A.; Mitsuyama, K.; Sako, T.; Yamamoto, M.; Kado, S.; Takada, T. A component of polysaccharide peptidoglycan complex on Lactobacillus induced an improvement of murine model of inflammatory bowel disease and colitis-associated cancer. Immunology 2009, 128, e170–e180. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [Green Version]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic T reg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.-W.; Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.-S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–254. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. The microbiota regulates type 2 immunity through ROR t+ T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, Y.; Uchiyama, K.; Takagi, T. A next-generation beneficial microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 2018, 63, 33–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tielens, A.G.M.; van Grinsven, K.W.A.; Henze, K.; van Hellemond, J.J.; Martin, W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 2010, 40, 387–397. [Google Scholar] [CrossRef]
- Acevedo, N.; Alhamwe, B.A.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and early-life nutrition, epigenetics, and allergy. Nutrients 2021, 13, 724. [Google Scholar] [CrossRef]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.Y.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef] [Green Version]
- Zakzuk, J.; Acevedo, N.; Harb, H.; Eick, L.; Renz, H.; Potaczek, D.P.; Caraballo, L. IgE Levels to Ascaris and House Dust Mite Allergens Are Associated with Increased Histone Acetylation at Key Type-2 Immune Genes. Front. Immunol. 2020, 11, 756. [Google Scholar] [CrossRef]
- Midha, A.; Janek, K.; Niewienda, A.; Henklein, P.; Guenther, S.; Serra, D.O.; Schlosser, J.; Hengge, R.; Hartmann, S. The Intestinal Roundworm Ascaris suum Releases Antimicrobial Factors Which Interfere with Bacterial Growth and Biofilm Formation. Front. Cell. Infect. Microbiol. 2018, 8, 271. [Google Scholar] [CrossRef] [Green Version]
- Midha, A.; Goyette-Desjardins, G.; Goerdeler, F.; Moscovitz, O.; Seeberger, P.H.; Tedin, K.; Bertzbach, L.D.; Lepenies, B.; Hartmann, S. Lectin-Mediated Bacterial Modulation by the Intestinal Nematode Ascaris suum. Int. J. Mol. Sci. 2021, 22, 8739. [Google Scholar] [CrossRef]
- Hansen, E.P.; Fromm, B.; Andersen, S.D.; Marcilla, A.; Andersen, K.L.; Borup, A.; Williams, A.R.; Jex, A.R.; Gasser, R.B.; Young, N.D.; et al. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite-host cross talk. J. Extracell. Vesicles 2019, 8, 1578116. [Google Scholar] [CrossRef] [Green Version]
- Rooney, J.; Northcote, H.M.; Williams, T.L.; Cortes, A.; Cantacessi, C.; Morphew, R.M. Parasitic helminths and the host microbiome-a missing ’extracellular vesicle-sized’ link? Trends Parasitol. 2022, 38, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, C.; Sifuentes-Dominguez, L.; Zarek, C.M.; Propheter, D.C.; Kuang, Z.; Wang, Y.; Pendse, M.; Ruhn, K.A.; Hassell, B.; et al. Small proline-rich protein 2A is a gut bactericidal protein deployed during helminth infection. Science 2021, 374, eabe6723. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Song, Q.; Liu, J.; Chen, F.; Zhang, Y.; Wu, Z.; Sun, X.; Wu, X. Potential Gut Microbiota Features for Non-Invasive Detection of Schistosomiasis. Front. Immunol. 2022, 13, 941530. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llinás-Caballero, K.; Caraballo, L. Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”. Int. J. Mol. Sci. 2022, 23, 13358. https://doi.org/10.3390/ijms232113358
Llinás-Caballero K, Caraballo L. Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”. International Journal of Molecular Sciences. 2022; 23(21):13358. https://doi.org/10.3390/ijms232113358
Chicago/Turabian StyleLlinás-Caballero, Kevin, and Luis Caraballo. 2022. "Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”" International Journal of Molecular Sciences 23, no. 21: 13358. https://doi.org/10.3390/ijms232113358
APA StyleLlinás-Caballero, K., & Caraballo, L. (2022). Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”. International Journal of Molecular Sciences, 23(21), 13358. https://doi.org/10.3390/ijms232113358