Aux/IAA11 Is Required for UV-AB Tolerance and Auxin Sensing in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
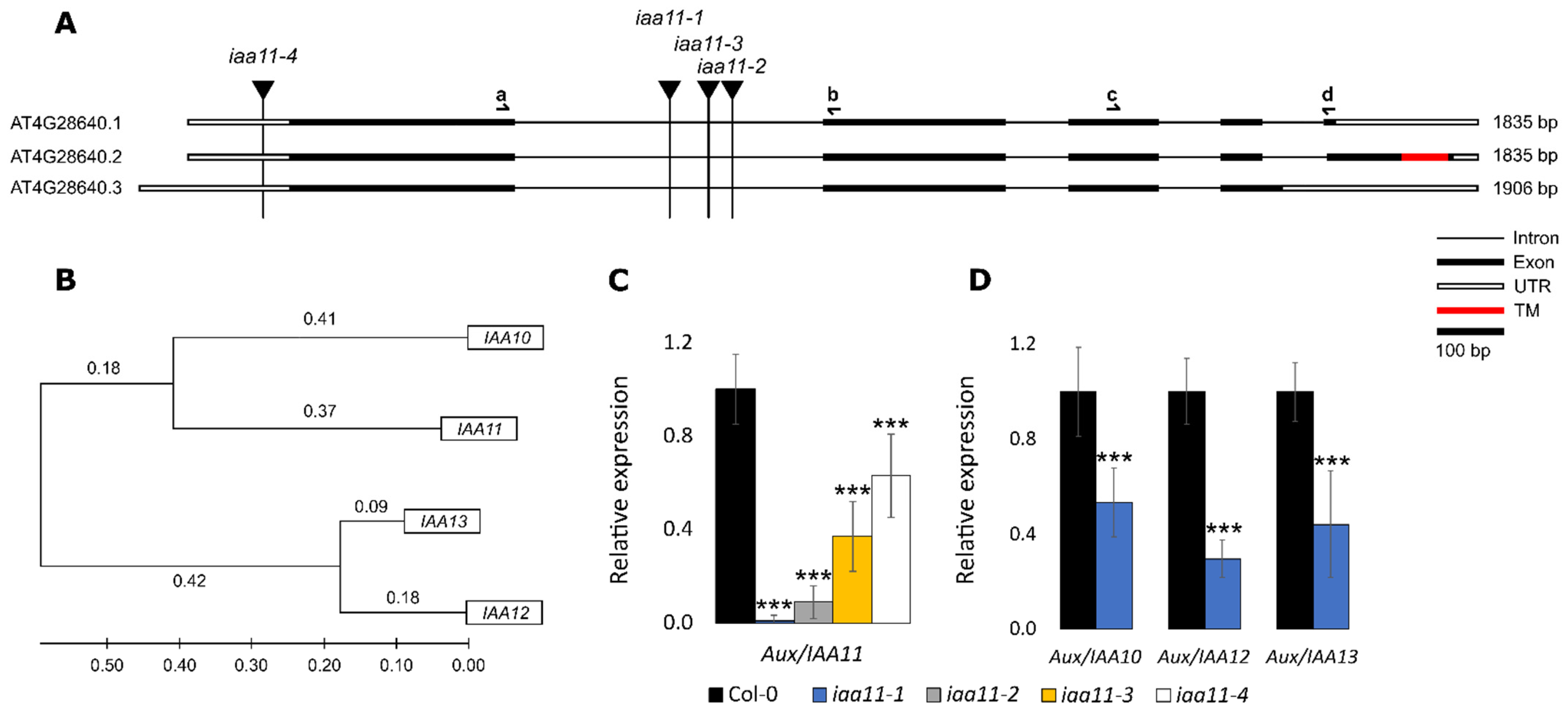
2.1. Reverse Genetic Screen for Membrane Bound Transcription Factors Involved in Retrograde Signaling
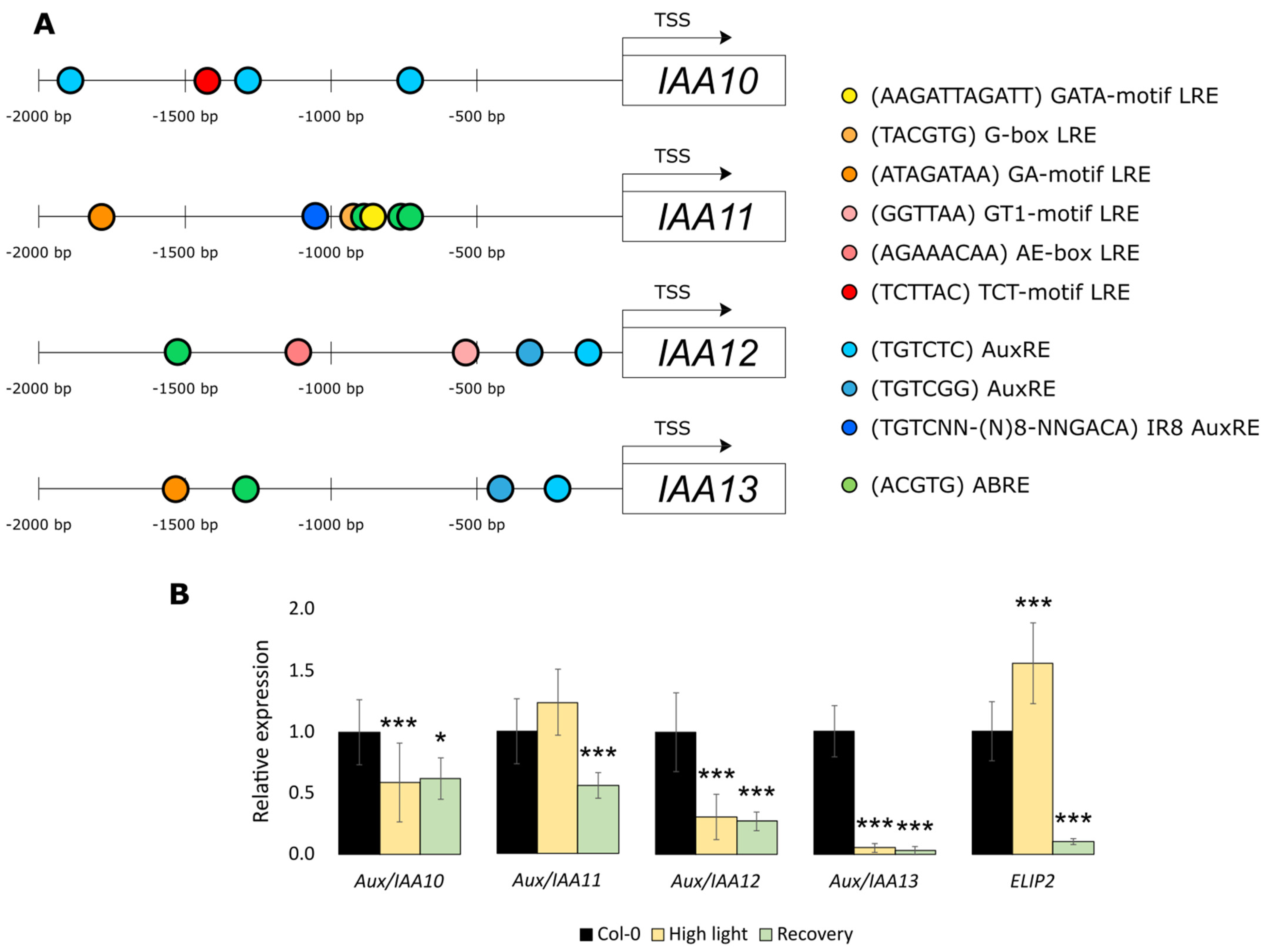
2.2. Expression of Aux/IAA11 Gene in the Tested Lines and Expression Patterns
2.3. Aux/IAA11 Is Involved in UV-AB Stress Tolerance
2.4. Aux/IAA11 Is Required for Carrying Out Photosynthesis under Inhibitory Conditions
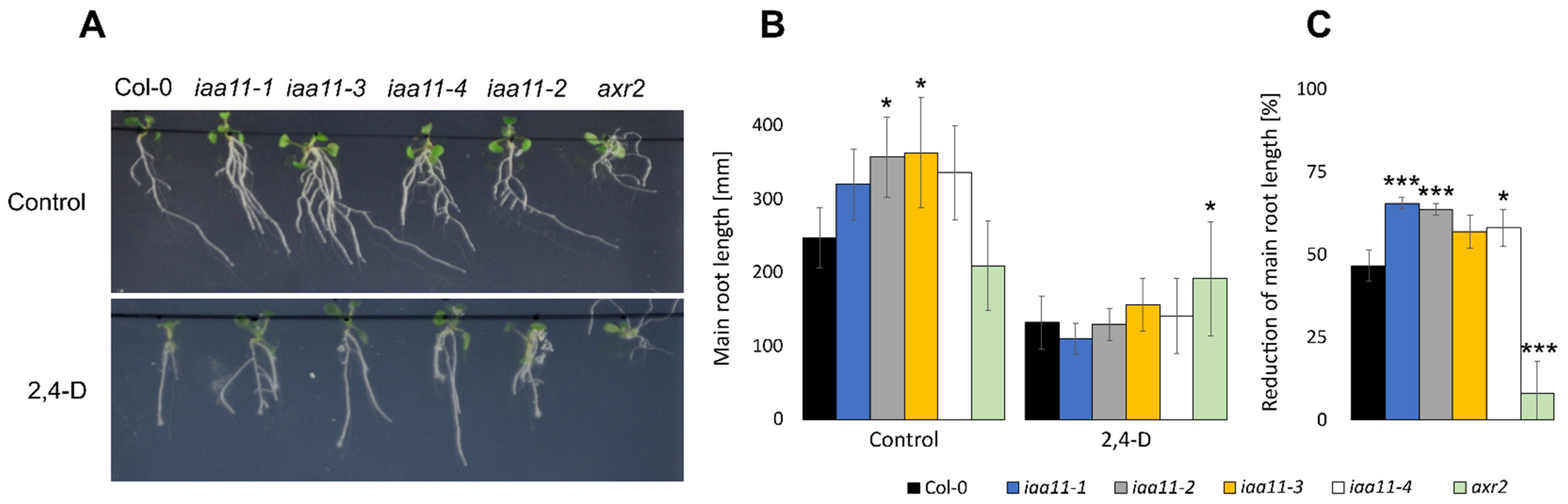
2.5. Aux/IAA11 Is Involved in Auxin Sensing
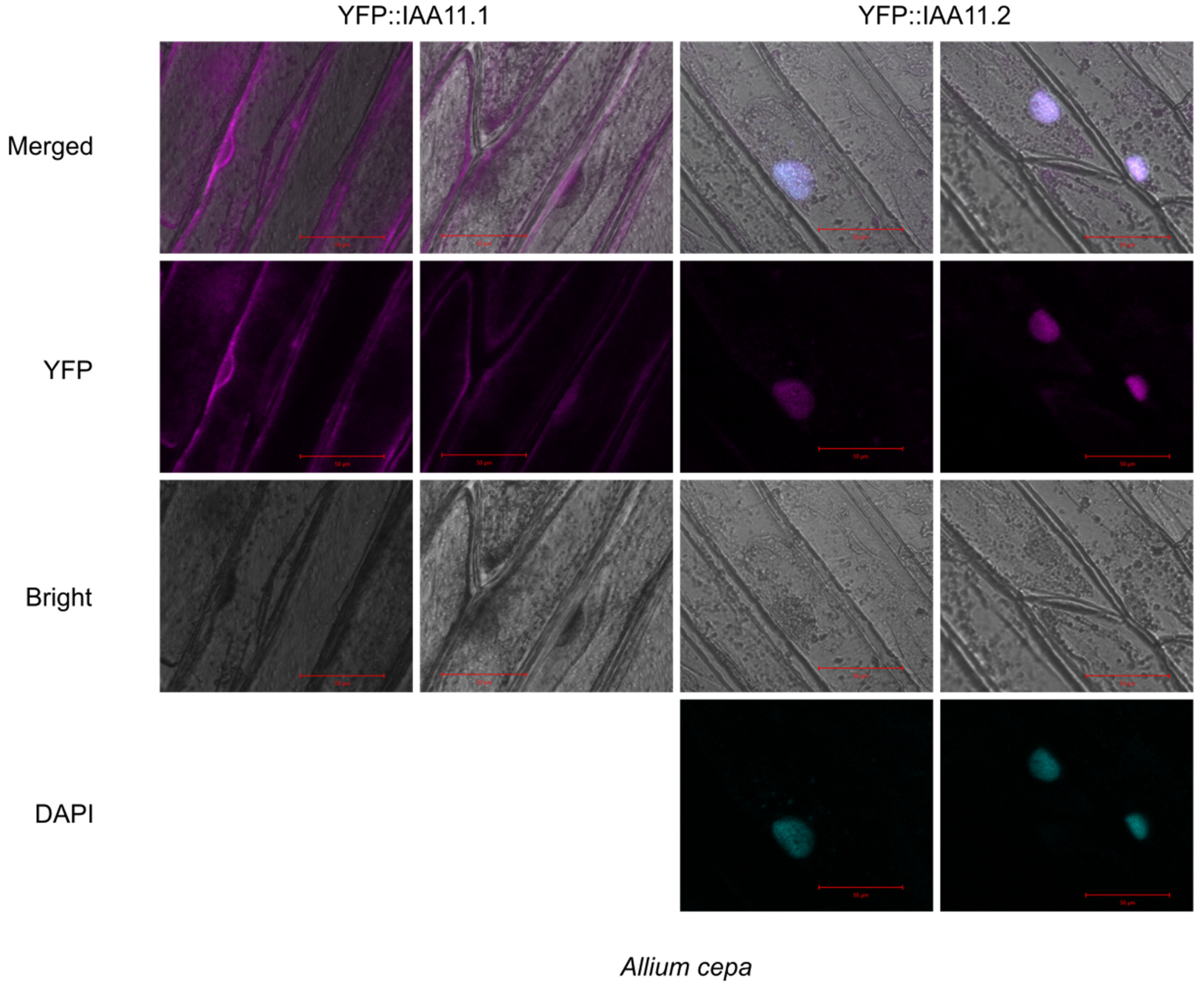
2.6. Subcellular Localization of Aux/IAA11 Protein Variants
3. Discussion
4. Materials and Methods
4.1. Preparation of Plant Material and Growth Conditions
4.2. Reverse Genetic Screen
4.3. RNA Isolation and qRT-PCR Analysis
4.4. UV-AB Treatment of Aux/IAA11 Mutants
4.5. DCMU Treatment of Aux/IAA11 Mutants
4.6. H2O2 Measurements and SOSG Staining
4.7. 2,4-Dichlorophenoxyacetic Acid Susceptibility Assay
4.8. Preparation of DNA Constructs for Tobacco and Onion Transformation
4.9. Transient Expression in Nicotiana Benthamiana and Allium Cepa Leaves
4.10. Confocal Microscopy Observations
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2,4-D | 2,4-Dichlorophenoxyacetic acid |
| ABRE | Abscisic acid responsive element |
| AFB | Auxin signaling F-BOX protein |
| ARF | Auxin responsive factor |
| AuxRE | Auxin responsive element |
| axr2 | auxin resistant 2 |
| bZIP | basic leucine zipper transcription factor |
| cDNA | Complementary DNA |
| CDS | CDS Coding site |
| DAPI | 4′,6-Diamidino-2-Phenylindole, Dihydrochloride |
| DCMU | 3-(3,4-dichlorophenyl)-1,1-dimethylurea |
| DREB2A | Dehydration-responsive element-binding protein 2A |
| GPx | Glutathione peroxidases |
| IAA | Indole-3-acetic acid |
| LOOH | Lipid hydroperoxide |
| LRE | Light responsive element |
| MV | Methyl viologen |
| NAA | 1-napthaleneacetic acid |
| NASC | Nottingham Arabidopsis Stock Centre |
| NGE | Nuclear gene expression |
| PAA | Phenylacetic acid |
| PCD | Programmed cell death |
| PIN | Pin-formed proteins |
| ROS | Reactive Oxygen Species |
| SOSG | Singlet Oxygen Sensor Green |
| TIR1 | Transport Inhibitor Response 1 |
| qPCR | Quantitative PCR |
References
- Wang, Y.; Selinski, J.; Mao, C.; Zhu, Y.; Berkowitz, O.; Whelan, J. Linking Mitochondrial and Chloroplast Retrograde Signalling in Plants. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190410. [Google Scholar] [CrossRef]
- Mielecki, J.; Gawroński, P.; Karpiński, S. Retrograde Signaling: Understanding the Communication between Organelles. Int. J. Mol. Sci. 2020, 21, 6173. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Z.; Bock, R. GUN Control in Retrograde Signaling: How GENOMES UNCOUPLED Proteins Adjust Nuclear Gene Expression to Plastid Biogenesis. Plant Cell 2021, 33, 457–474. [Google Scholar] [CrossRef]
- Jiang, J.; Dehesh, K. Plastidial Retrograde Modulation of Light and Hormonal Signaling: An Odyssey. New Phytol. 2021, 230, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Morffy, N.; Strader, L.C. Old Town Roads: Routes of Auxin Biosynthesis across Kingdoms. Curr. Opin. Plant Biol. 2020, 55, 21–27. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol 2018, 176, 465–479. [Google Scholar] [CrossRef]
- Damodaran, S.; Strader, L.C. Indole 3-Butyric Acid Metabolism and Transport in Arabidopsis Thaliana. Front. Plant Sci. 2019, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef]
- Sańko-Sawczenko, I.; Łotocka, B.; Czarnocka, W. Expression Analysis of PIN Genes in Root Tips and Nodules of Medicago Truncatula. Int. J. Mol. Sci. 2016, 17, 1197. [Google Scholar] [CrossRef]
- Zhou, J.J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Strader, L.C.; Wheeler, D.L.; Christensen, S.E.; Berens, J.C.; Cohen, J.D.; Rampey, R.A.; Bartel, B. Multiple Facets of Arabidopsis Seedling Development Require Indole-3-Butyric Acid–Derived Auxin. Plant Cell 2011, 23, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Oeller, P.W.; Theologis, A. Early Auxin-Induced Genes Encode Short-Lived Nuclear Proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of Auxin Signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Calderón Villalobos, L.I.A.; Lee, S.; de Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A Combinatorial TIR1/AFB-Aux/IAA Co-Receptor System for Differential Sensing of Auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Correa-Aragunde, N.; Colman, S.L.; Izquierdo-Álvarez, A.; Fiol, D.F.; París, R.; Sánchez-López, N.; Marina, A.; Calderón Villalobos, L.I.A.; et al. Regulation of SCFTIR1/AFBs E3 Ligase Assembly by S-Nitrosylation of Arabidopsis SKP1-Like1 Impacts on Auxin Signaling. Redox Biol. 2018, 18, 200–210. [Google Scholar] [CrossRef]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS Mediates Auxin-Dependent Transcriptional Repression during Arabidopsis Embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR Motif-Mediated Transcriptional Repression in Plants: An Underlying Mechanism for Epigenetic Regulation of Gene Expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Niemeyer, M.; Moreno Castillo, E.; Ihling, C.H.; Iacobucci, C.; Wilde, V.; Hellmuth, A.; Hoehenwarter, W.; Samodelov, S.L.; Zurbriggen, M.D.; Kastritis, P.L.; et al. Flexibility of Intrinsically Disordered Degrons in AUX/IAA Proteins Reinforces Auxin Co-Receptor Assemblies. Nat. Commun. 2020, 11, 2277. [Google Scholar] [CrossRef]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D.; et al. Variation in Auxin Sensing Guides AUX/IAA Transcriptional Repressor Ubiquitylation and Destruction. Nat. Commun. 2017, 8, 15706. [Google Scholar] [CrossRef]
- Shani, E.; Salehin, M.; Zhang, Y.; Sanchez, S.E.; Doherty, C.; Wang, R.; Mangado, C.C.; Song, L.; Tal, I.; Pisanty, O.; et al. Plant Stress Tolerance Requires Auxin-Sensitive Aux/IAA Transcriptional Repressors. Curr. Biol. 2017, 27, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-Sensitive Aux/IAA Proteins Mediate Drought Tolerance in Arabidopsis by Regulating Glucosinolate Levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Niu, H.; Xin, D.; Long, Y.; Wang, G.; Liu, Z.; Li, G.; Zhang, F.; Qi, M.; Ye, Y.; et al. OsIAA18, an Aux/IAA Transcription Factor Gene, Is Involved in Salt and Drought Tolerance in Rice. Front. Plant Sci. 2021, 12, 2571. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Sugita, K.; Qin, Y.; Rahman, A. Aux/IAA14 Regulates MicroRNA-Mediated Cold Stress Response in Arabidopsis Roots. Int. J. Mol. Sci. 2020, 21, 8441. [Google Scholar] [CrossRef]
- Shi, H.; Liu, W.; Wei, Y.; Ye, T. Integration of Auxin/Indole-3-Acetic Acid 17 and RGA-LIKE3 Confers Salt Stress Resistance through Stabilization by Nitric Oxide in Arabidopsis. J. Exp. Bot. 2016, 68, 1239–1249. [Google Scholar] [CrossRef]
- Hou, Y.; Li, H.; Zhai, L.; Xie, X.; Li, X.; Bian, S. Identification and Functional Characterization of the Aux/IAA Gene VcIAA27 in Blueberry. Plant Signal Behav. 2020, 15, 1700327. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Yamamoto, K.T. Overexpression of the Non-Canonical Aux/IAA Genes Causes Auxin-Related Aberrant Phenotypes in Arabidopsis. Physiol. Plant 2008, 133, 397–405. [Google Scholar] [CrossRef]
- Nagpal, P.; Walker, L.M.; Young, J.C.; Sonawala, A.; Timpte, C.; Estelle, M.; Reed, J.W. AXR2 Encodes a Member of the Aux/IAA Protein Family. Plant Physiol. 2000, 123, 563–573. [Google Scholar] [CrossRef]
- Yang, X.; Lee, S.; So, J.; Dharmasiri, S.; Dharmasiri, N.; Ge, L.; Jensen, C.; Hangarter, R.; Hobbie, L.; Estelle, M. The IAA1 Protein Is Encoded by AXR5 and Is a Substrate of SCFTIR1. Plant J. 2004, 40, 772–782. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, X.; Li, L.; He, Y.; Hong, G.; Li, J.; Lin, L.; Cheng, Y.; Yan, F.; Chen, J.; et al. Suppression of Auxin Signalling Promotes Rice Susceptibility to Rice Black Streaked Dwarf Virus Infection. Mol. Plant Pathol. 2019, 20, 1093–1104. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA Protein, Mediates Abiotic Stress Tolerance in Rice through an ABA Pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yuan, C.; Feng, S.; Zhong, S.; Li, H.; Zhong, J.; Shen, C.; Liu, J. Genome-Wide Analysis and Characterization of Aux/IAA Family Genes Related to Fruit Ripening in Papaya (Carica papaya L.). BMC Genom. 2017, 18, 351. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, E.; Mano, S.; Hara-Nishimura, I.; Nishimura, M.; Yamada, K. HSP90 Stabilizes Auxin Receptor TIR1 and Ensures Plasticity of Auxin Responses. Plant Signal Behav. 2017, 12, e1311439. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Yang, Y.; Yang, J.; Zhang, X.; Pan, Y.; Guo, H. IAA3-Mediated Repression of PIF Proteins Coordinates Light and Auxin Signaling in Arabidopsis. PLoS Genet. 2021, 17. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; He, S.; Zhang, J.; Mao, Z.; Wang, W.; Li, T.; Hua, J.; Du, S.; Xu, P.; Li, L.; et al. Photoactivated CRY1 and PhyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol. Plant 2018, 11, 523–541. [Google Scholar] [CrossRef]
- Jiang, J.; Xiao, Y.; Chen, H.; Hu, W.; Zeng, L.; Ke, H.; Ditengou, F.A.; Devisetty, U.; Palme, K.; Maloof, J.; et al. Retrograde Induction of PhyB Orchestrates Ethylene-Auxin Hierarchy to Regulate Growth. Plant Physiol. 2020, 183, 1268–1280. [Google Scholar] [CrossRef]
- Jiang, J.; Rodriguez-Furlan, C.; Wang, J.Z.; de Souza, A.; Ke, H.; Pasternak, T.; Lasok, H.; Ditengou, F.A.; Palme, K.; Dehesh, K. Interplay of the Two Ancient Metabolites Auxin and MEcPP Regulates Adaptive Growth. Nat Commun 2018, 9, 2262. [Google Scholar] [CrossRef]
- Phua, S.Y.; Yan, D.; Chan, K.X.; Estavillo, G.M.; Nambara, E.; Pogson, B.J. The Arabidopsis SAL1-PAP Pathway: A Case Study for Integrating Chloroplast Retrograde, Light and Hormonal Signaling in Modulating Plant Growth and Development? Front. Plant Sci. 2018, 9, 1171. [Google Scholar] [CrossRef]
- Ishiga, Y.; Watanabe, M.; Ishiga, T.; Tohge, T.; Matsuura, T.; Ikeda, Y.; Hoefgen, R.; Fernie, A.R.; Mysore, K.S. The SAL-PAP Chloroplast Retrograde Pathway Contributes to Plant Immunity by Regulating Glucosinolate Pathway and Phytohormone Signaling. Mol. Plant Microbe Interact. 2017, 30, 829–841. [Google Scholar] [CrossRef]
- Ivanova, A.; Law, S.R.; Narsai, R.; Duncan, O.; Lee, J.H.; Zhang, B.; van Aken, O.; Radomiljac, J.D.; van der Merwe, M.; Yi, K.K.; et al. A Functional Antagonistic Relationship between Auxin and Mitochondrial Retrograde Signaling Regulates Alternative Oxidase1a Expression in Arabidopsis. Plant Physiol. 2014, 165, 1233–1254. [Google Scholar] [CrossRef]
- He, C.; Liew, L.C.; Yin, L.; Lewsey, M.G.; Whelan, J.; Berkowitz, O. The Retrograde Signaling Regulator ANAC017 Recruits the MKK9–MPK3/6, Ethylene, and Auxin Signaling Pathways to Balance Mitochondrial Dysfunction with Growth. Plant Cell 2022, 34, 3460–3481. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.I.; de Clercq, I.; Denecker, J.; Mühlenbock, P.; Kumpf, R.; Nguyen, L.; Audenaert, D.; Dejonghe, W.; van Breusegem, F. Mitochondrial Perturbation Negatively Affects Auxin Signaling. Mol. Plant 2014, 7, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Millar, A.H. How Is Auxin Linked with Cellular Energy Pathways to Promote Growth? New Phytol. 2022, 233, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Gläßer, C.; Haberer, G.; Finkemeier, I.; Pfannschmidt, T.; Kleine, T.; Leister, D.; Dietz, K.J.; Häusler, R.E.; Grimm, B.; Mayer, K.F.X. Meta-Analysis of Retrograde Signaling in Arabidopsis Thaliana Reveals a Core Module of Genes Embedded in Complex Cellular Signaling Networks. Mol. Plant 2014, 7, 1167–1190. [Google Scholar] [CrossRef]
- Gawroński, P.; Burdiak, P.; Scharff, L.B.; Mielecki, J.; Górecka, M.; Zaborowska, M.; Leister, D.; Waszczak, C.; Karpiński, S. CIA2 and CIA2-LIKE Are Required for Optimal Photosynthesis and Stress Responses in Arabidopsis thaliana. Plant J. 2021, 105, 619–638. [Google Scholar] [CrossRef]
- ARAMEMNON—Plant Membrane Protein Database (Release 8.1). Available online: http://aramemnon.uni-koeln.de/index.ep (accessed on 4 July 2022).
- Schwacke, R.; Schneider, A.; van der Graaff, E.; Fischer, K.; Catoni, E.; Desimone, M.; Frommer, W.B.; Flügge, U.I.; Kunze, R. ARAMEMNON, a Novel Database for Arabidopsis Integral Membrane Proteins. Plant Physiol. 2003, 131, 16–26. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinform. 2008, 2008, 1–5. [Google Scholar] [CrossRef]
- Scholl, R.L.; May, S.T.; Ware, D.H. Seed and Molecular Resources for Arabidopsis. Plant Physiol. 2000, 124, 1477–1480. [Google Scholar] [CrossRef]
- Sun, X.; Feng, P.; Xu, X.; Guo, H.; Ma, J.; Chi, W.; Lin, R.; Lu, C.; Zhang, L. A Chloroplast Envelope-Bound PHD Transcription Factor Mediates Chloroplast Signals to the Nucleus. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.; Li, T.; Yang, Q.; Zhang, Y.; Zou, J.; Bian, Z.; Wen, X. UVA Radiation Is Beneficial for Yield and Quality of Indoor Cultivated Lettuce. Front. Plant Sci. 2019, 10, 1563. [Google Scholar] [CrossRef]
- Britt, A.B. Repair of DNA Damage Induced by Ultraviolet Radiation. Plant Physiol. 1995, 108, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, H. How Plants Protect Themselves from Ultraviolet-B Radiation Stress. Plant Physiol. 2021, 187, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Sztatelman, O.; Grzyb, J.; Gabryś, H.; Banaś, A.K. The Effect of UV-B on Arabidopsis Leaves Depends on Light Conditions after Treatment. BMC Plant Biol. 2015, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Wituszyńska, W.; Szechyńska-Hebda, M.; Sobczak, M.; Rusaczonek, A.; Kozlowska-Makulska, A.; Witoń, D.; Karpiński, S. LESION SIMULATING DISEASE 1 And ENHANCED DISEASE SUSCEPTIBILITY 1 Differentially Regulate UV-C-Induced Photooxidative Stress Signalling and Programmed Cell Death in Arabidopsis Thaliana. Plant Cell Env. 2015. [Google Scholar] [CrossRef] [PubMed]
- Flors, C.; Fryer, M.J.; Waring, J.; Reeder, B.; Bechtold, U.; Mullineaux, P.M.; Nonell, S.; Wilson, M.T.; Baker, N.R. Imaging the Production of Singlet Oxygen in Vivo Using a New Fluorescent Sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 2006, 57, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic Signaling and Acclimation in Response to Excess Excitation Energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Fufezan, C.; Rutherford, A.W.; Krieger-Liszkay, A. Singlet Oxygen Production in Herbicide-Treated Photosystem II. FEBS Lett. 2002, 532, 407–410. [Google Scholar] [CrossRef]
- Cui, F.; Brosché, M.; Shapiguzov, A.; He, X.Q.; Vainonen, J.P.; Leppälä, J.; Trotta, A.; Kangasjärvi, S.; Salojärvi, J.; Kangasjärvi, J.; et al. Interaction of Methyl Viologen-Induced Chloroplast and Mitochondrial Signalling in Arabidopsis. Free Radic. Biol. Med. 2019, 134, 555–566. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Roeland Boer, D.; et al. Architecture of DNA Elements Mediating ARF Transcription Factor Binding and Auxin-Responsive Gene Expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Li, Y.; Cai, H.; Bai, X.; Ji, W.; Ji, Z.J.; Zhu, Y.M. Arabidopsis BZIP1 Transcription Factor Binding to ABRE Cis-Element Regulates Abscisic Acid Signal Transduction. Acta Agron. Sin. 2011, 37, 612–619. [Google Scholar] [CrossRef]
- Tzvetkova-Chevolleau, T.; Franck, F.; Alawady, A.E.; Dall’Osto, L.; Carrière, F.; Bassi, R.; Grimm, B.; Nussaume, L.; Havaux, M. The Light Stress-Induced Protein ELIP2 Is a Regulator of Chlorophyll Synthesis in Arabidopsis Thaliana. Plant J. 2007, 50, 795–809. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or Foe? Reactive Oxygen Species Production, Scavenging and Signaling in Plant Response to Environmental Stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Miyamoto, S.; Ronsein, G.E.; Prado, F.M.; Uemi, M.; Corrêa, T.C.; Toma, I.N.; Bertolucci, A.; Oliveira, M.C.B.; Motta, F.D.; Medeiros, M.H.G.; et al. Biological Hydroperoxides and Singlet Molecular Oxygen Generation. IUBMB Life 2007, 59, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Mai, Y.-X.; Wang, L.; Yang, H.-Q. A Gain-of-Function Mutation in IAA7/AXR2 Confers Late Flowering under Short-Day Light in ArabidopsisF. J. Integr. Plant Biol. 2011, 53, 480–492. [Google Scholar] [CrossRef]
- Lanctot, A.; Taylor-Teeples, M.; Oki, E.A.; Nemhauser, J.L. Specificity in Auxin Responses Is Not Explained by the Promoter Preferences of Activator ARFs. Plant Physiol. 2020, 182, 1533–1536. [Google Scholar] [CrossRef]
- Kim, J.-S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE Promoter Sequence Is Involved in Osmotic Stress-Responsive Expression of the DREB2A Gene, Which Encodes a Transcription Factor Regulating Drought-Inducible Genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Geisler, M.; Kleczkowski, L.A.; Karpinski, S. A Universal Algorithm for Genome-Wide in Silicio Identification of Biologically Significant Gene Promoter Putative Cis -Regulatory-Elements; Identification of New Elements for Reactive Oxygen Species and Sucrose Signaling in Arabidopsis. Plant J. 2006, 45, 384–398. [Google Scholar] [CrossRef]
- Li, M.; Kim, C. Chloroplast ROS and Stress Signaling. Plant Commun. 2022, 3, 100264. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R.; et al. The High Light Response in Arabidopsis Involves ABA Signaling between Vascular and Bundle Sheath CellsW. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef] [PubMed]
- Pornsiriwong, W.; Estavillo, G.M.; Chan, K.X.; Tee, E.E.; Ganguly, D.; Crisp, P.A.; Phua, S.Y.; Zhao, C.; Qiu, J.; Park, J.; et al. A Chloroplast Retrograde Signal, 3’phosphoadenosine 5′-Phosphate, Acts as a Secondary Messenger in Abscisic Acid Signaling in Stomatal Closure and Germination. Elife 2017, 6, e23361. [Google Scholar] [CrossRef] [PubMed]
- Fryer, M.J.; Ball, L.; Oxborough, K.; Karpinski, S.; Mullineaux, P.M.; Baker, N.R. Control of Ascorbate Peroxidase 2 Expression by Hydrogen Peroxide and Leaf Water Status during Excess Light Stress Reveals a Functional Organisation of Arabidopsis Leaves. Plant J. 2003, 33, 691–705. [Google Scholar] [CrossRef]
- Karpinski, S.; Escobar, C.; Karpinska, B.; Creissen, G.; Mullineaux, P.M. Photosynthetic Electron Transport Regulates the Expression of Cytosolic Ascorbate Peroxidase Genes in Arabidopsis during Excess Light Stress. Plant Cell 1997, 9, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Chory, J. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019, 29, 4186–4199.e3. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, L.; Chen, P.; Liang, T.; Li, X.; Liu, H. UV-B Photoreceptor UVR8 Interacts with MYB73/MYB77 to Regulate Auxin Responses and Lateral Root Development. EMBO J. 2020, 39, e101928. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA Receptor PYL8 Promotes Lateral Root Growth by Enhancing MYB77-Dependent Transcription of Auxin-Responsive Genes. Sci. Signal 2014, 7, ra53. [Google Scholar] [CrossRef]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis Transcription Factor MYB77 Modulates Auxin Signal Transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, P.; Wang, R.; Sun, L.; Wang, W.; Zhou, H.; Xu, J. UV-B Radiation Induces Root Bending Through the Flavonoid-Mediated Auxin Pathway in Arabidopsis. Front. Plant Sci. 2018, 9, 618. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Yu, J.Q. Plant Hormones under Challenging Environmental Factors; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9789401777582. [Google Scholar]
- Wituszynska, W.; Karpinski, S. Programmed Cell Death as a Response to High Light, UV and Drought Stress in Plants. In Abiotic Stress—Plant Responses and Applications in Agriculture; InTech: Melbourne, FL, USA, 2013. [Google Scholar]
- Wituszyńska, W.; Ślesak, I.; Vanderauwera, S.; Szechyńska-Hebda, M.; Kornaś, A.; van der Kelen, K.; Mühlenbock, P.; Karpińska, B.; Maćkowski, S.; van Breusegem, F.; et al. Lesion simulating disease, enhanced disease susceptibility, and phytoalexin deficient conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 2013, 161, 1795–1805. [Google Scholar] [CrossRef]
- Biever, J.J.; Brinkman, D.; Gardner, G. UV-B Inhibition of Hypocotyl Growth in Etiolated Arabidopsis Thaliana Seedlings Is a Consequence of Cell Cycle Arrest Initiated by Photodimer Accumulation. J. Exp. Bot. 2014, 65, 2949–2961. [Google Scholar] [CrossRef] [PubMed]
- Huq, E. Direct Convergence of Light and Auxin Signaling Pathways in Arabidopsis. Mol. Plant 2018, 11, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Gören-Sağlam, N.; Harrison, E.; Breeze, E.; Öz, G.; Buchanan-Wollaston, V. Analysis of the Impact of Indole-3-Acetic Acid (IAA) on Gene Expression during Leaf Senescence in Arabidopsis Thaliana. Physiol. Mol. Biol. Plants 2020, 26, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Mou, M.; Li, P.; Huang, Y.; Zhai, X.; Ma, Y.; Liu, J.; Yu, X. Theoretical Modeling of the Aux/IAA Negative Feedback Circuit in Plants. S. Afr. J. Bot. 2015, 100, 16–19. [Google Scholar] [CrossRef]
- Lv, B.; Yu, Q.; Liu, J.; Wen, X.; Yan, Z.; Hu, K.; Li, H.; Kong, X.; Li, C.; Tian, H.; et al. Non-canonical AUX / IAA Protein IAA 33 Competes with Canonical AUX / IAA Repressor IAA 5 to Negatively Regulate Auxin Signaling. EMBO J 2020, 39. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant Development Is Regulated by a Family of Auxin Receptor F Box Proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef]
- Burdiak, P.; Rusaczonek, A.; Witoń, D.; Głów, D.; Karpiński, S. Cysteine-Rich Receptor-like Kinase CRK5 as a Regulator of Growth, Development, and Ultraviolet Radiation Responses in Arabidopsis Thaliana. J. Exp. Bot. 2015, 66, 3325–3337. [Google Scholar] [CrossRef]
- Gawroński, P.; Witoń, D.; Vashutina, K.; Bederska, M.; Betliński, B.; Rusaczonek, A.; Karpiński, S. Mitogen-Activated Protein Kinase 4 Is a Salicylic Acid-Independent Regulator of Growth but Not of Photosynthesis in Arabidopsis. Mol. Plant 2014, 7, 1151–1166. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-Free Analysis of Quantitative Real-Time Polymerase Chain Reaction (PCR) Data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of Series of Gateway Binary Vectors, PGWBs, for Realizing Efficient Construction of Fusion Genes for Plant Transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]
- Xu, K.; Huang, X.; Wu, M.; Wang, Y.; Chang, Y.; Liu, K.; Zhang, J.; Zhang, Y.; Zhang, F.; Yi, L.; et al. A Rapid, Highly Efficient and Economical Method of Agrobacterium-Mediated In Planta Transient Transformation in Living Onion Epidermis. PLoS ONE 2014, 9, e83556. [Google Scholar] [CrossRef] [PubMed]
- Klinkenberg, J. Extraction of Chloroplast Proteins from Transiently Transformed Nicotiana Benthamiana Leaves. Bio. Protoc. 2014, 4, e1238. [Google Scholar] [CrossRef]
- Statistica—StatSoft Poland. Available online: https://www.statsoft.pl/Pelna-lista-programow-Statistica/ (accessed on 4 July 2022).
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mielecki, J.; Gawroński, P.; Karpiński, S. Aux/IAA11 Is Required for UV-AB Tolerance and Auxin Sensing in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13386. https://doi.org/10.3390/ijms232113386
Mielecki J, Gawroński P, Karpiński S. Aux/IAA11 Is Required for UV-AB Tolerance and Auxin Sensing in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(21):13386. https://doi.org/10.3390/ijms232113386
Chicago/Turabian StyleMielecki, Jakub, Piotr Gawroński, and Stanisław Karpiński. 2022. "Aux/IAA11 Is Required for UV-AB Tolerance and Auxin Sensing in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 21: 13386. https://doi.org/10.3390/ijms232113386
APA StyleMielecki, J., Gawroński, P., & Karpiński, S. (2022). Aux/IAA11 Is Required for UV-AB Tolerance and Auxin Sensing in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(21), 13386. https://doi.org/10.3390/ijms232113386








