Generation of Functional Immortalized Human Corneal Stromal Stem Cells
Abstract
1. Introduction
2. Results
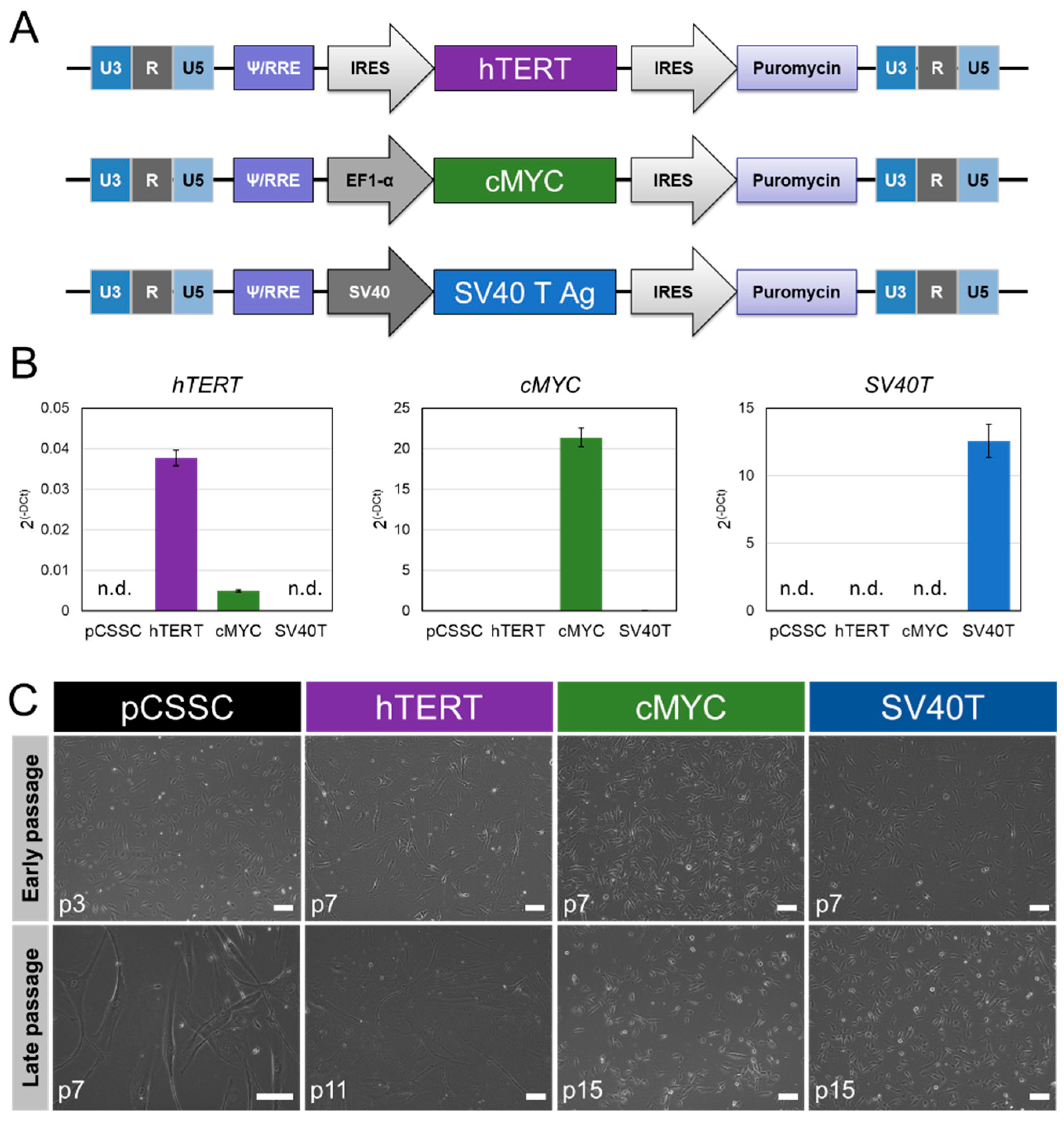
2.1. Generation of Immortalized CSSC
2.2. Phenotypic Characterization of Immortalized CSSC
2.3. Keratocyte Lineage Commitment
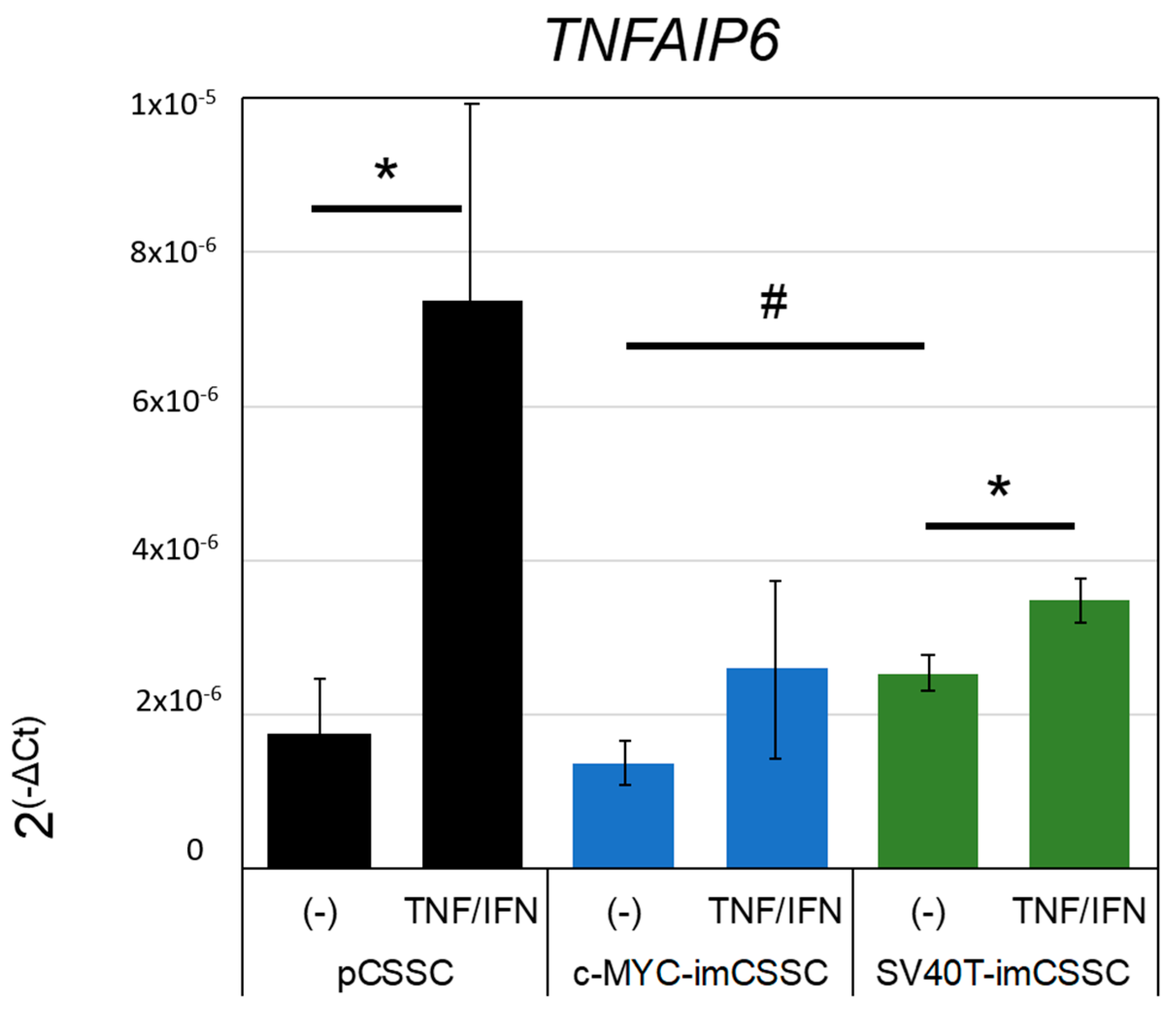
2.4. Modulation of Inflammation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of Immortalized CSSC Lines
4.3. Multilineage Differentiation
4.4. Immunofluorescence
4.5. TNFAIP6 Expression
4.6. Flow Cytometry
4.7. RNA Extraction, cDNA Transcription, and Real-Time PCR
4.8. Image Acquisition
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Basu, S.; Hertsenberg, A.J.; Funderburgh, M.L.; Burrow, M.K.; Mann, M.M.; Du, Y.; Lathrop, K.L.; Syed-Picard, F.N.; Adams, S.M.; Birk, D.E.; et al. Human limbal biopsy-derived stromal stem cells prevent corneal scarring. Sci. Transl. Med. 2014, 6, 266ra172. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.; Balayan, A.; Funderburgh, M.L.; Ngo, J.; Funderburgh, J.L.; Deng, S.X. Differentiation Capacity of Human Mesenchymal Stem Cells into Keratocyte Lineage. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3013–3023. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Funderburgh, M.L.; Mann, M.M.; SundarRaj, N.; Funderburgh, J.L. Multipotent stem cells in human corneal stroma. Stem Cells 2005, 23, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Hertsenberg, A.J.; Shojaati, G.; Funderburgh, M.L.; Mann, M.M.; Du, Y.; Funderburgh, J.L. Corneal stromal stem cells reduce corneal scarring by mediating neutrophil infiltration after wounding. PLoS ONE 2017, 12, e0171712. [Google Scholar] [CrossRef]
- Eslani, M.; Putra, I.; Shen, X.; Hamouie, J.; Afsharkhamseh, N.; Besharat, S.; Rosenblatt, M.I.; Dana, R.; Hematti, P.; Djalilian, A.R. Corneal Mesenchymal Stromal Cells Are Directly Antiangiogenic via PEDF and sFLT-1. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5507–5517. [Google Scholar] [CrossRef]
- Eslani, M.; Putra, I.; Shen, X.; Hamouie, J.; Tadepalli, A.; Anwar, K.N.; Kink, J.A.; Ghassemi, S.; Agnihotri, G.; Reshetylo, S.; et al. Cornea-Derived Mesenchymal Stromal Cells Therapeutically Modulate Macrophage Immunophenotype and Angiogenic Function. Stem Cells 2018, 36, 775–784. [Google Scholar] [CrossRef]
- Du, Y.; Carlson, E.C.; Funderburgh, M.L.; Birk, D.E.; Pearlman, E.; Guo, N.; Kao, W.W.; Funderburgh, J.L. Stem cell therapy restores transparency to defective murine corneas. Stem Cells 2009, 27, 1635–1642. [Google Scholar] [CrossRef]
- Wang, H.S.; Hwang, L.L.; Sue, H.F.; Lee, K.M.; Chen, C.T. A simple quantitative method for evaluation of angiogenesis activity. Assay Drug Dev. Technol. 2004, 2, 31–38. [Google Scholar] [CrossRef]
- Glinos, G.D.; Verne, S.H.; Aldahan, A.S.; Liang, L.; Nouri, K.; Elliot, S.; Glassberg, M.; Cabrera DeBuc, D.; Koru-Sengul, T.; Tomic-Canic, M.; et al. Optical coherence tomography for assessment of epithelialization in a human ex vivo wound model. Wound Repair Regen. 2017, 25, 1017–1026. [Google Scholar] [CrossRef]
- Krutzke, L.; Allmendinger, E.; Hirt, K.; Kochanek, S. Chorioallantoic membrane (CAM) tumor model for evaluating oncolytic viruses. Hum. Gene Ther. 2020, 31, 1100–1113. [Google Scholar] [CrossRef]
- Dou, L.; Wu, Y.; Yan, Q.; Wang, J.; Zhang, Y.; Ji, P. Secretome profiles of immortalized dental follicle cells using iTRAQ-based proteomic analysis. Sci. Rep. 2017, 7, 7300. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Yang, C.; Ji, X.; Zhang, L.; Bi, Y.; Yang, K.; Gong, M.; Liu, X.; Guo, Q.; Su, Y.; et al. Reversibly immortalized human umbilical cord-derived mesenchymal stem cells (UC-MSCs) are responsive to BMP9-induced osteogenic and adipogenic differentiation. J. Cell Biochem. 2018, 119, 8872–8886. [Google Scholar] [CrossRef] [PubMed]
- Wagner, W.; Horn, P.; Castoldi, M.; Diehlmann, A.; Bork, S.; Saffrich, R.; Benes, V.; Blake, J.; Pfister, S.; Eckstein, V.; et al. Replicative senescence of mesenchymal stem cells: A continuous and organized process. PLoS ONE 2008, 3, e2213. [Google Scholar] [CrossRef] [PubMed]
- Ferreira Lopes, S.; Vacher, G.; Ciarlo, E.; Savova-Bianchi, D.; Roger, T.; Niculita-Hirzel, H. Primary and Immortalized Human Respiratory Cells Display Different Patterns of Cytotoxicity and Cytokine Release upon Exposure to Deoxynivalenol, Nivalenol and Fusarenon-X. Toxins 2017, 9, 337. [Google Scholar] [CrossRef] [PubMed]
- Qadhi, R.; Alsaleh, N.; Samokhvalov, V.; El-Sikhry, H.; Bellenger, J.; Seubert, J.M. Differential responses to docosahexaenoic acid in primary and immortalized cardiac cells. Toxicol. Lett. 2013, 219, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Dobson-Belaire, W.N.; Cochrane, A.; Ostrowski, M.A.; Gray-Owen, S.D. Differential response of primary and immortalized CD4+ T cells to Neisseria gonorrhoeae-induced cytokines determines the effect on HIV-1 replication. PLoS ONE 2011, 6, e18133. [Google Scholar] [CrossRef]
- Knight, R.; Board-Davies, E.; Brown, H.; Clayton, A.; Davis, T.; Karatas, B.; Burston, J.; Tabi, Z.; Falcon-Perez, J.M.; Paisey, S.; et al. Oral Progenitor Cell Line-Derived Small Extracellular Vesicles as a Treatment for Preferential Wound Healing Outcome. Stem Cells Transl. Med. 2022, 11, 861–875. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Peshdary, V.; Calzadilla, G.; Landry, A.; Sorisky, A.; Atlas, E. Dechlorane Plus increases adipogenesis in 3T3-L1 and human primary preadipocytes independent of peroxisome proliferator-activated receptor γ transcriptional activity. Int. J. Obes. 2019, 43, 545–555. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.-Y.; Wang, Y.-P.; Wu, Z.-H.; Yu, B. Dynamic Expression Profiles of Marker Genes in Osteogenic Differentiation of Human Bone Marrow-derived Mesenchymal Stem Cells. Chin. Med. Sci. J. 2015, 30, 108–113. [Google Scholar] [CrossRef]
- Nuttelman, C.R.; Tripodi, M.C.; Anseth, K.S. In vitro osteogenic differentiation of human mesenchymal stem cells photoencapsulated in PEG hydrogels. J. Biomed. Mater. Res. Part A 2004, 68A, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Rim, Y.A.; Jung, S.M.; Ju, J.H. Cord blood cell-derived iPSCs as a new candidate for chondrogenic differentiation and cartilage regeneration. Stem Cell Res. Ther. 2017, 8, 16. [Google Scholar] [CrossRef]
- Wu, J.; Du, Y.; Mann, M.M.; Yang, E.; Funderburgh, J.L.; Wagner, W.R. Bioengineering organized, multilamellar human corneal stromal tissue by growth factor supplementation on highly aligned synthetic substrates. Tissue Eng. Part A 2013, 19, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Lakshman, N.; Kim, A.; Petroll, W.M. Characterization of corneal keratocyte morphology and mechanical activity within 3-D collagen matrices. Exp. Eye Res. 2010, 90, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Roh, D.S.; Funderburgh, M.L.; Mann, M.M.; Marra, K.G.; Rubin, J.P.; Li, X.; Funderburgh, J.L. Adipose-derived stem cells differentiate to keratocytes in vitro. Mol. Vis. 2010, 16, 2680–2689. [Google Scholar]
- Funderburgh, J.L.; Mann, M.M.; Funderburgh, M.L. Keratocyte Phenotype Mediates Proteoglycan Structure: A Role for Fibroblasts in Corneal Fibrosis. J. Biol. Chem. 2011, 278, 45629–45637. [Google Scholar] [CrossRef]
- Chakravarti, S.; Wu, F.; Vij, N.; Roberts, L.; Joyce, S. Microarray studies reveal macrophage-like function of stromal keratocytes in the cornea. Invest. Ophthalmol. Vis. Sci. 2004, 45, 3475–3484. [Google Scholar] [CrossRef][Green Version]
- Funderburgh, M.L.; Du, Y.; Mann, M.M.; SundarRaj, N.; Funderburgh, J.L. PAX6 expression identifies progenitor cells for corneal keratocytes. Faseb. J. 2005, 19, 1371–1373. [Google Scholar] [CrossRef]
- Lee, R.H.; Yu, J.M.; Foskett, A.M.; Peltier, G.; Reneau, J.C.; Bazhanov, N.; Oh, J.Y.; Prockop, D.J. TSG-6 as a biomarker to predict efficacy of human mesenchymal stem/progenitor cells (hMSCs) in modulating sterile inflammation in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 16766–16771. [Google Scholar] [CrossRef]
- Lee, K.M.; Choi, K.H.; Ouellette, M.M. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology 2004, 45, 33–38. [Google Scholar] [CrossRef]
- Mihara, K.; Imai, C.; Coustan-Smith, E.; Dome, J.S.; Dominici, M.; Vanin, E.; Campana, D. Development and functional characterization of human bone marrow mesenchymal cells immortalized by enforced expression of telomerase. Br. J. Haematol. 2003, 120, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Bentzon, J.F.; Stenderup, K.; Hansen, F.D.; Schroder, H.D.; Abdallah, B.M.; Jensen, T.G.; Kassem, M. Tissue distribution and engraftment of human mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene. Biochem. Biophys. Res. Commun. 2005, 330, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Siska, E.K.; Weisman, I.; Romano, J.; Ivics, Z.; Izsvák, Z.; Barkai, U.; Petrakis, S.; Koliakos, G. Generation of an immortalized mesenchymal stem cell line producing a secreted biosensor protein for glucose monitoring. PLoS ONE 2017, 12, e0185498. [Google Scholar] [CrossRef] [PubMed]
- Böcker, W.; Yin, Z.; Drosse, I.; Haasters, F.; Rossmann, O.; Wierer, M.; Popov, C.; Locher, M.; Mutschler, W.; Docheva, D.; et al. Introducing a single-cell-derived human mesenchymal stem cell line expressing hTERT after lentiviral gene transfer. J. Cell Mol. Med. 2008, 12, 1347–1359. [Google Scholar] [CrossRef]
- Ramboer, E.; De Craene, B.; De Kock, J.; Vanhaecke, T.; Berx, G.; Rogiers, V.; Vinken, M. Strategies for immortalization of primary hepatocytes. J. Hepatol. 2014, 61, 925–943. [Google Scholar] [CrossRef]
- Piñeiro-Ramil, M.; Sanjurjo-Rodríguez, C.; Castro-Viñuelas, R.; Rodríguez-Fernández, S.; Fuentes-Boquete, I.M.; Blanco, F.J.; Díaz-Prado, S.M. Usefulness of Mesenchymal Cell Lines for Bone and Cartilage Regeneration Research. Int. J. Mol. Sci. 2019, 20, 6286. [Google Scholar] [CrossRef]
- Fujita, S.; Toguchida, J.; Morita, Y.; Iwata, H. Clonal analysis of hematopoiesis-supporting activity of human mesenchymal stem cells in association with Jagged1 expression and osteogenic potential. Cell Transplant. 2008, 17, 1169–1179. [Google Scholar] [CrossRef]
- Liu, T.M.; Ng, W.M.; Tan, H.S.; Vinitha, D.; Yang, Z.; Fan, J.B.; Zou, Y.; Hui, J.H.; Lee, E.H.; Lim, B. Molecular basis of immortalization of human mesenchymal stem cells by combination of p53 knockdown and human telomerase reverse transcriptase overexpression. Stem Cells Dev. 2013, 22, 268–278. [Google Scholar] [CrossRef]
- Mori, T.; Kiyono, T.; Imabayashi, H.; Takeda, Y.; Tsuchiya, K.; Miyoshi, S.; Makino, H.; Matsumoto, K.; Saito, H.; Ogawa, S.; et al. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol. Cell Biol. 2005, 25, 5183–5195. [Google Scholar] [CrossRef]
- Okamoto, T.; Aoyama, T.; Nakayama, T.; Nakamata, T.; Hosaka, T.; Nishijo, K.; Nakamura, T.; Kiyono, T.; Toguchida, J. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2002, 295, 354–361. [Google Scholar] [CrossRef]
- Takeuchi, M.; Takeuchi, K.; Kohara, A.; Satoh, M.; Shioda, S.; Ozawa, Y.; Ohtani, A.; Morita, K.; Hirano, T.; Terai, M.; et al. Chromosomal instability in human mesenchymal stem cells immortalized with human papilloma virus E6, E7, and hTERT genes. In Vitro Cell Dev. Biol. Anim. 2007, 43, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Thalmeier, K.; Meissner, P.; Reisbach, G.; Falk, M.; Brechtel, A.; Dörmer, P. Establishment of two permanent human bone marrow stromal cell lines with long-term post irradiation feeder capacity. Blood 1994, 83, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Glowacki, J.; Hahne, J.; Xie, L.; LeBoff, M.S.; Zhou, S. Dehydroepiandrosterone Stimulation of Osteoblastogenesis in Human MSCs Requires IGF-I Signaling. J. Cell Biochem. 2016, 117, 1769–1774. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, K.; Handa, H. Generation of functional clonal cell lines from human bone marrow stroma. Proc. Natl. Acad. Sci. USA 1985, 82, 3477–3480. [Google Scholar] [CrossRef]
- Song, D.; Zhang, F.; Reid, R.R.; Ye, J.; Wei, Q.; Liao, J.; Zou, Y.; Fan, J.; Ma, C.; Hu, X. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J. Cell Mol. Med. 2017, 21, 2782–2795. [Google Scholar] [CrossRef]
- Alexander, D.; Biller, R.; Rieger, M.; Ardjomandi, N.; Reinert, S. Phenotypic characterization of a human immortalized cranial periosteal cell line. Cell Physiol. Biochem. 2015, 35, 2244–2254. [Google Scholar] [CrossRef]
- Wu, Y.; Feng, G.; Song, J.; Zhang, Y.; Yu, Y.; Huang, L.; Zheng, L.; Deng, F. TrAmplification of Human Dental Follicle Cells by piggyBac Transposon—Mediated Reversible Immortalization System. PLoS ONE 2015, 10, e0130937. [Google Scholar] [CrossRef][Green Version]
- Jayasuriya, C.T.; Hu, N.; Li, J.; Lemme, N.; Terek, R.; Ehrlich, M.G.; Chen, Q. Molecular characterization of mesenchymal stem cells in human osteoarthritis cartilage reveals contribution to the OA phenotype. Sci. Rep. 2018, 8, 7044. [Google Scholar] [CrossRef]
- Hu, N.; Gao, Y.; Jayasuriya, C.T.; Liu, W.; Du, H.; Ding, J.; Feng, M.; Chen, Q. Chondrogenic induction of human osteoarthritic cartilage-derived mesenchymal stem cells activates mineralization and hypertrophic and osteogenic gene expression through a mechanomiR. Arthritis Res. Ther. 2019, 21, 167. [Google Scholar] [CrossRef]
- An, P.; Sáenz Robles, M.T.; Pipas, J.M. Large T antigens of polyomaviruses: Amazing molecular machines. Annu. Rev. Microbiol. 2012, 66, 213–236. [Google Scholar] [CrossRef]
- Ahuja, D.; Sáenz-Robles, M.T.; Pipas, J.M. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005, 24, 7729–7745. [Google Scholar] [CrossRef] [PubMed]
- Melnik, S.; Werth, N.; Boeuf, S.; Hahn, E.M.; Gotterbarm, T.; Anton, M.; Richter, W. Impact of c-MYC expression on proliferation, differentiation, and risk of neoplastic transformation of human mesenchymal stromal cells. Stem Cell Res. Ther. 2019, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Kami, D.; Kitani, T.; Kawasaki, T.; Gojo, S. Cardiac mesenchymal progenitors differentiate into adipocytes via Klf4 and c-Myc. Cell Death Dis. 2016, 7, e2190. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.S.; Arslan, F.; Yin, Y.; Tan, S.S.; Lai, R.C.; Choo, A.B.H.; Padmanabhan, J.; Lee, C.N.; de Kleijn, D.P.V.; Lim, S.K. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. J. Transl. Med. 2011, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.F.; Gagnon, J. Neuronal cell Thy-1 glycoprotein: Homology with immunoglobulin. Science 1982, 216, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Raff, M.C. Surface antigenic markers for distinguishing T and B lymphocytes in mice. Transplant. Rev. 1971, 6, 52–80. [Google Scholar] [CrossRef]
- Sibov, T.T.; Severino, P.; Marti, L.C.; Pavon, L.F.; Oliveira, D.M.; Tobo, P.R.; Campos, A.H.; Paes, A.T.; Amaro, E., Jr.; Gamarra, L.; et al. Mesenchymal stem cells from umbilical cord blood: Parameters for isolation, characterization and adipogenic differentiation. Cytotechnology 2012, 64, 511–521. [Google Scholar] [CrossRef]
- Wiesmann, A.; Bühring, H.J.; Mentrup, C.; Wiesmann, H.P. Decreased CD90 expression in human mesenchymal stem cells by applying mechanical stimulation. Head Face Med. 2006, 2, 8. [Google Scholar] [CrossRef]
- Chen, X.D.; Qian, H.Y.; Neff, L.; Satomura, K.; Horowitz, M.C. Thy-1 antigen expression by cells in the osteoblast lineage. J. Bone Miner Res. 1999, 14, 362–375. [Google Scholar] [CrossRef]
- Saldanha-Araujo, F.; Ferreira, F.I.; Palma, P.V.; Araujo, A.G.; Queiroz, R.H.; Covas, D.T.; Zago, M.A.; Panepucci, R.A. Mesenchymal stromal cells up-regulate CD39 and increase adenosine production to suppress activated T-lymphocytes. Stem Cell Res. 2011, 7, 66–74. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wu, P.K.; Chen, P.C.; Lee, C.W.; Chen, W.M.; Hung, S.C. Generation of Osteosarcomas from a Combination of Rb Silencing and c-Myc Overexpression in Human Mesenchymal Stem Cells. Stem Cells Transl. Med. 2017, 6, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Murphy, M.J.; Oskarsson, T.; Kaloulis, K.; Bettess, M.D.; Oser, G.M.; Pasche, A.C.; Knabenhans, C.; Macdonald, H.R.; Trumpp, A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004, 18, 2747–2763. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.; McKee, C.; Bakshi, S.; Walker, K.; Hakman, E.; Halassy, S.; Svinarich, D.; Dodds, R.; Govind, C.K.; Chaudhry, G.R. Mesenchymal stem cells: Cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 2019, 13, 1738–1755. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dos Santos, A.; Lyu, N.; Balayan, A.; Knight, R.; Zhuo, K.S.; Sun, Y.; Xu, J.; Funderburgh, M.L.; Funderburgh, J.L.; Deng, S.X. Generation of Functional Immortalized Human Corneal Stromal Stem Cells. Int. J. Mol. Sci. 2022, 23, 13399. https://doi.org/10.3390/ijms232113399
Dos Santos A, Lyu N, Balayan A, Knight R, Zhuo KS, Sun Y, Xu J, Funderburgh ML, Funderburgh JL, Deng SX. Generation of Functional Immortalized Human Corneal Stromal Stem Cells. International Journal of Molecular Sciences. 2022; 23(21):13399. https://doi.org/10.3390/ijms232113399
Chicago/Turabian StyleDos Santos, Aurelie, Ning Lyu, Alis Balayan, Rob Knight, Katherine Sun Zhuo, Yuzhao Sun, Jianjiang Xu, Martha L. Funderburgh, James L. Funderburgh, and Sophie X. Deng. 2022. "Generation of Functional Immortalized Human Corneal Stromal Stem Cells" International Journal of Molecular Sciences 23, no. 21: 13399. https://doi.org/10.3390/ijms232113399
APA StyleDos Santos, A., Lyu, N., Balayan, A., Knight, R., Zhuo, K. S., Sun, Y., Xu, J., Funderburgh, M. L., Funderburgh, J. L., & Deng, S. X. (2022). Generation of Functional Immortalized Human Corneal Stromal Stem Cells. International Journal of Molecular Sciences, 23(21), 13399. https://doi.org/10.3390/ijms232113399





