Transcriptome Analysis Reveals a Comprehensive Virus Resistance Response Mechanism in Pecan Infected by a Novel Badnavirus Pecan Virus
Abstract
:1. Introduction
2. Results
2.1. Virus Discovery by Deep Sequencing and Assembly of Virus-Derived Small Silencing RNAs
2.2. Leaf Cross-Section Microstructure Analysis
2.3. Characterization of the PMV-Infected Pecan Transcriptome
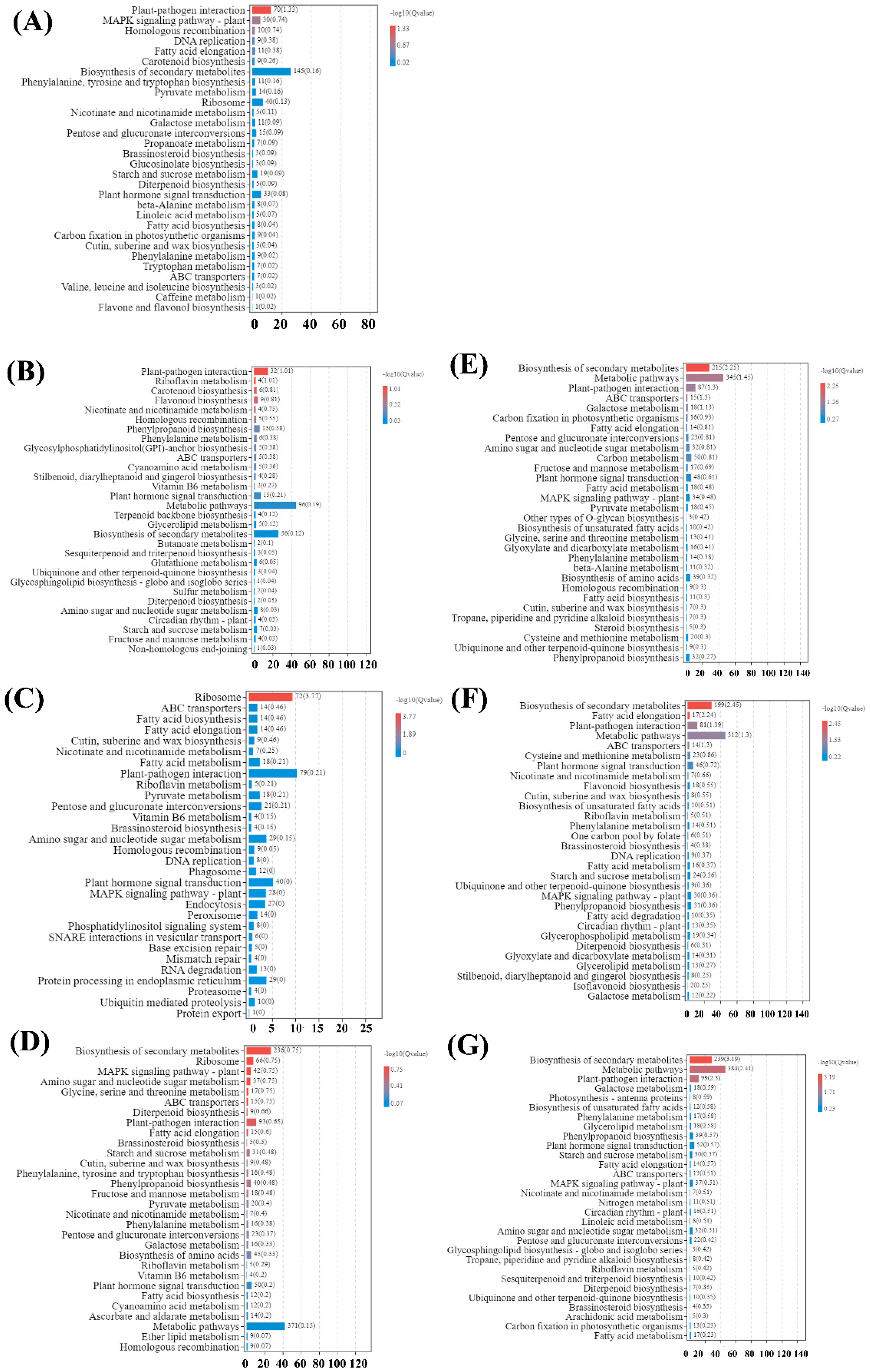
2.4. Global Transcriptome Changes in Pecan Infected with PMV
2.5. Inferred Pecan-PMV Interaction Pathway Suggests PTI and ETI
2.6. Identified Differentially Expressed Plant Disease Resistance (R) Protein Genes in Pecan in Response to PMV
2.7. PMV Infection Downregulated the Genes Related to the Photosynthesis
2.8. DEGs Analysis of Plant Hormone Biosynthesis and Signal Transduction Pathways
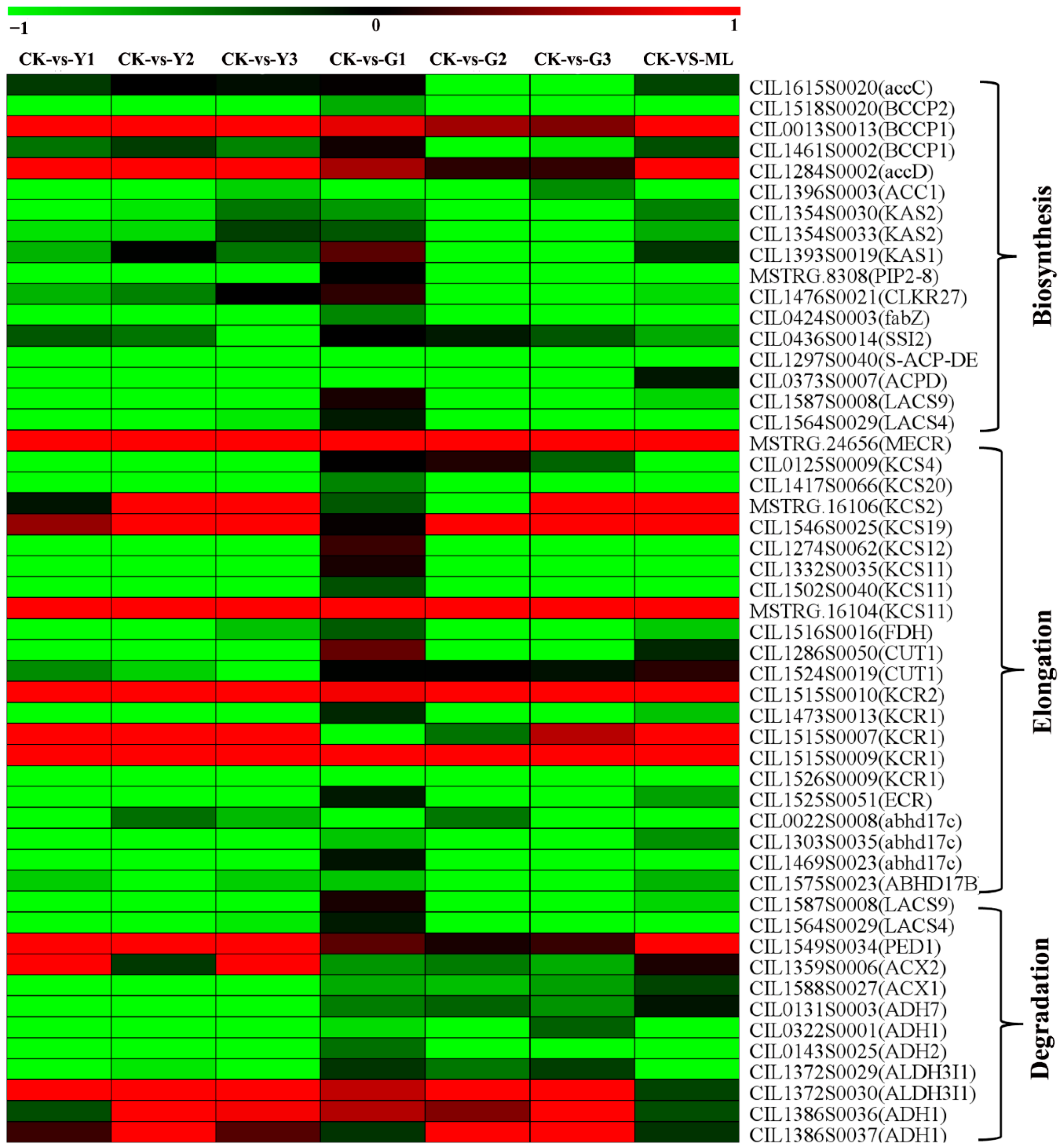
2.9. DEGs of Fatty Acid Biosynthesis, Elongation, and Degradation Pathways
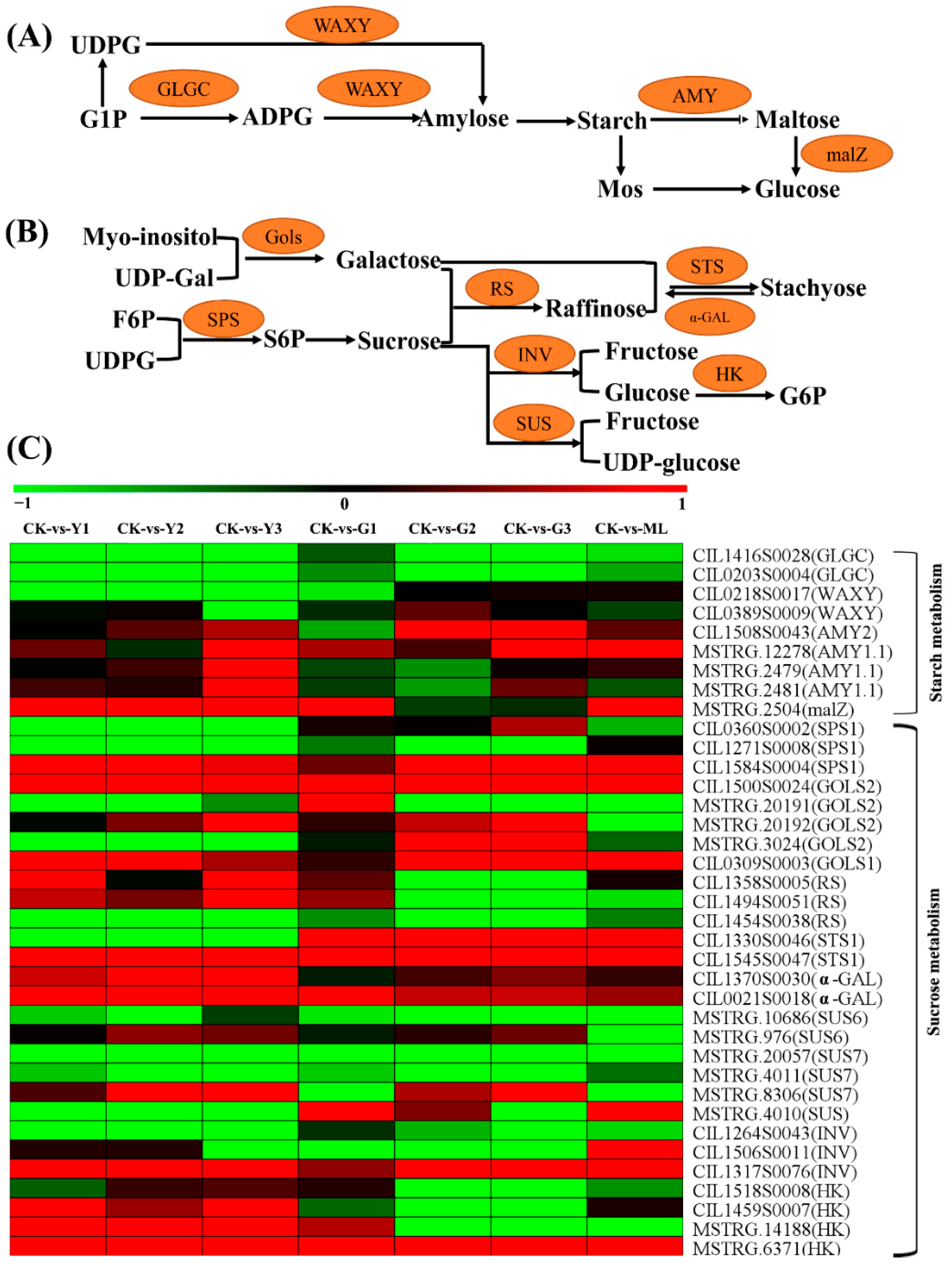
2.10. DEGs Involved in Starch and Sucrose Metabolism Pathway
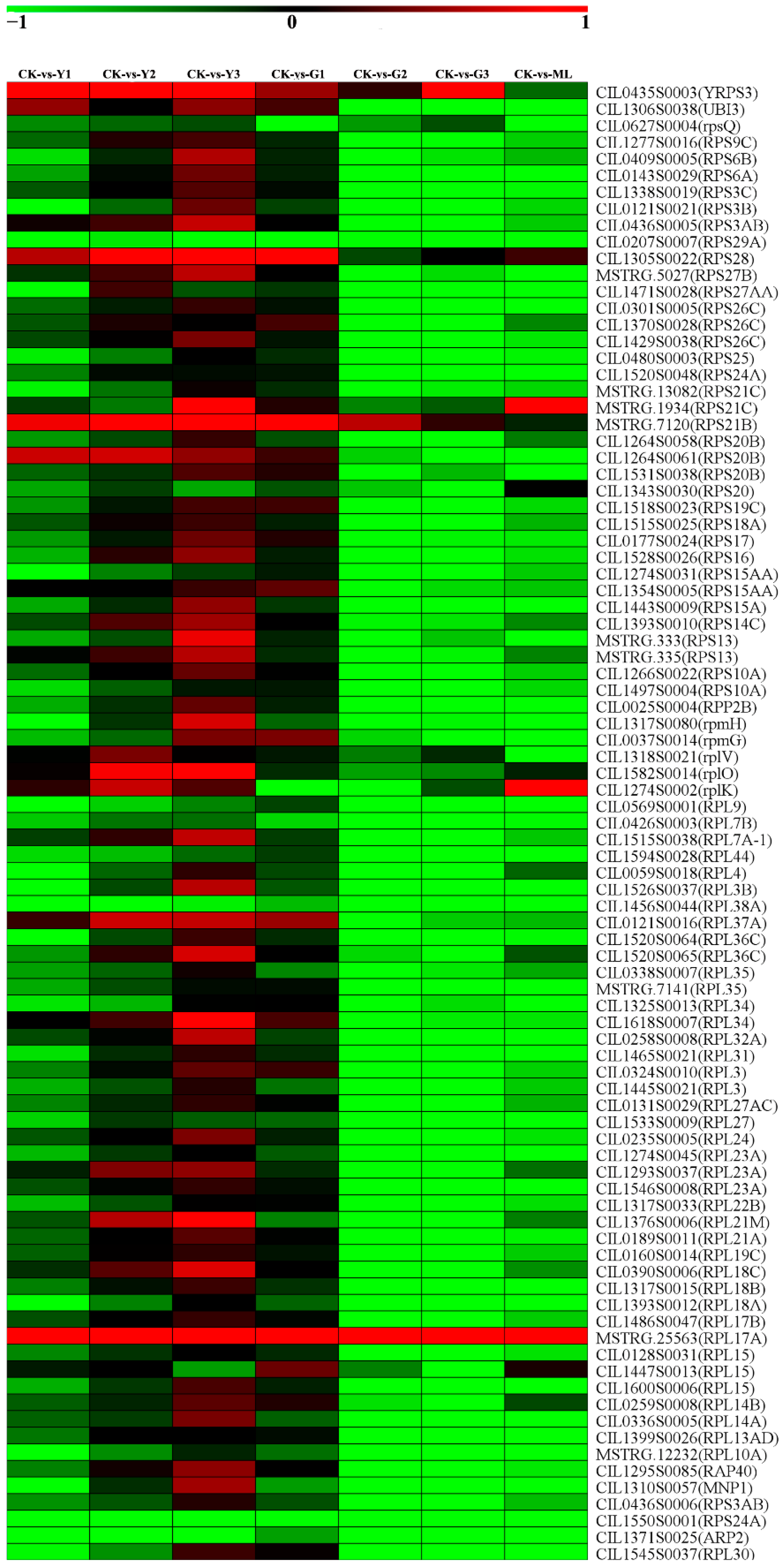
2.11. DEGs of Ribosome and Ribosome Biogenesis in Eukaryotes Pathways
3. Discussion
3.1. PMV-Infected Pecan Trigged PTI and ETI
3.2. ETI or PTI Might Induce Activation of MAPK and Inhibited Photosynthesis, Leading to Leaf Chlorosis
3.3. PMV Impairs Chloroplasts Clustering to Facilitate Viral Infection
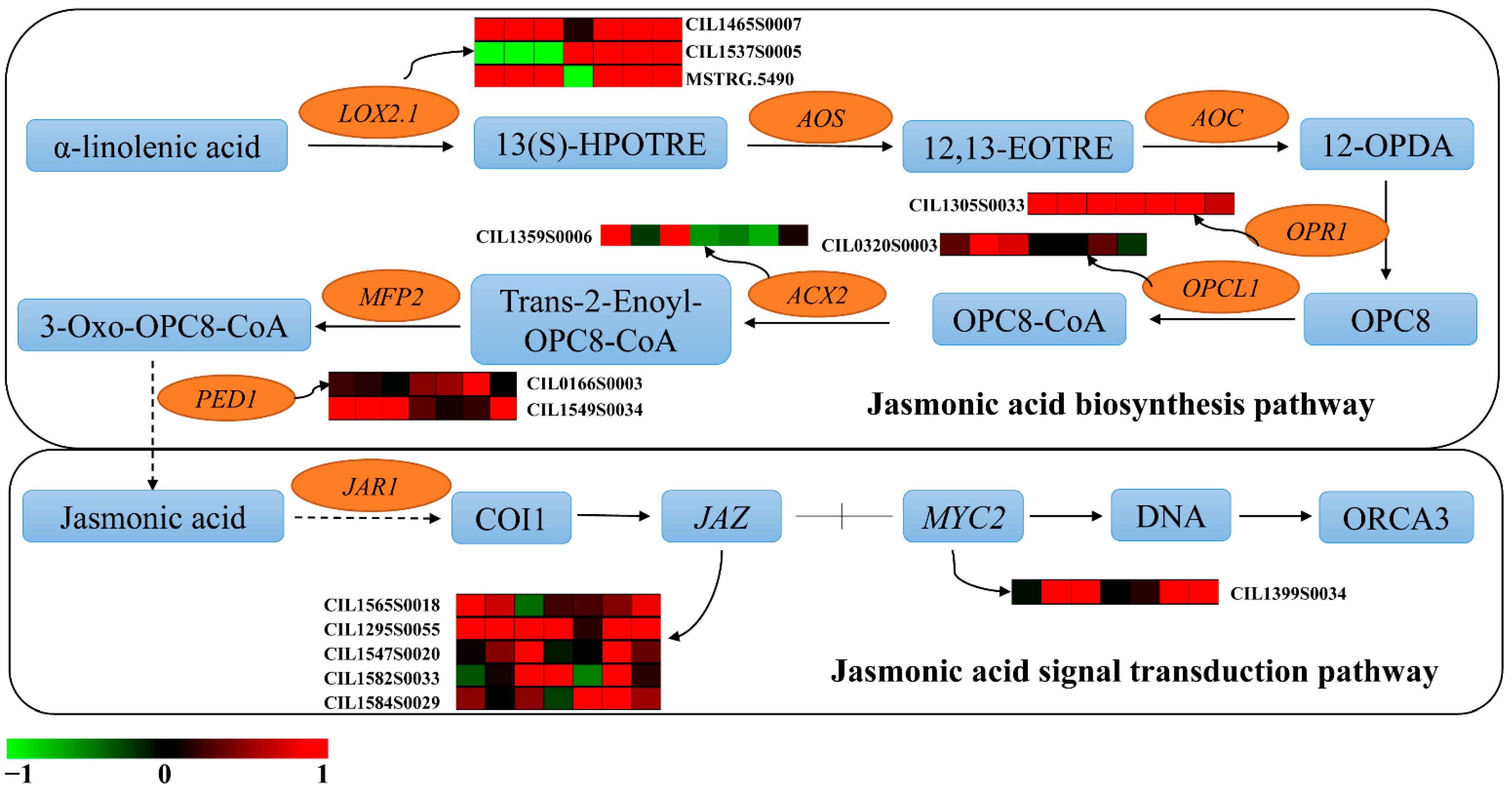
3.4. The Involvement of the JA Pathway in Auxin-Mediated Defense against PMV Infection
3.5. Cytokinins Signaling Difference Was One of the Key Reasons for Leaves Color in PMV Infection Pecan Plants
3.6. Carbohydrate Partitioning Was Dramatic Changed in PMV Infection Pecan Plants
3.7. Disturbance of the Ribosome Biogenesis Might Enhance the Resistance to PMV Infection and Lead to Leaves Staying Green in Pecans
3.8. Fatty Acids-Derived Signaling Was One of the Important Defense Pathways in Resistance to PMV Infection Pecan
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. Paraffin Section
4.3. Sequencing, Assembly, and Analysis of Small RNA Libraries
4.4. RNA Isolation and cDNA Library Preparation and Sequencing
4.5. Analysis of Differential Gene Expression
4.6. Illumina RNA-Seq Results Validations by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Zhu, L.; Hull, J.J.; Liang, S.; Daniell, H.; Jin, S.; Zhang, X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly). Plant Biotechnol. J. 2016, 14, 1956–1975. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.P.; Reddy, M.G. Chapter 38—Overview of yield losses due to plant viruses. In Applied Plant Virology; Awasthi, L.P., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 531–562. [Google Scholar]
- Bhattacharyya, D.; Chakraborty, S. Chloroplast: The Trojan horse in plant-virus interaction. Mol. Plant Pathol. 2018, 19, 504–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Huang, T.S.; McNeil, J.; Laliberte, J.F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Caplan, J.L.; Kumar, A.S.; Park, E.; Padmanabhan, M.S.; Hoban, K.; Modla, S.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplast stromules function during innate immunity. Dev. Cell 2015, 34, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Fu, F.-Q.; Zhang, D.-W.; Deng, X.-G.; Li, J.-Y.; Peng, X.-J.; Tang, H.; Lin, H.-H. Role of plastid signals in modulating Arabidopsis responses to Cucumber mosaic virus. Plant Growth Regul. 2015, 75, 761–769. [Google Scholar] [CrossRef]
- Angra-Sharma, R.; Sharma, D.K. Cytokinins in pathogenesis and disease resistance of Pyrenophora teres-barley and Dreschslera maydis-maize interactions during early stages of infection. Mycopathologia 2000, 148, 87–95. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Epple, P.; To, J.P.; Hutchison, C.E.; Schaller, G.E.; Dangl, J.L.; Kieber, J.J. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012, 8, e1002448. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, X.; Li, L.; He, Y.; Hong, G.; Li, J.; Lin, L.; Cheng, Y.; Yan, F.; Chen, J.; et al. Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol. Plant Pathol. 2019, 20, 1093–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, S.; Zhang, Y.; Wang, X.; Yang, J.; Sun, Z.; Zhang, D.; Chen, B.; Wang, G.; Ke, H.; Liu, Z.; et al. Cotton GhSSI2 isoforms from the stearoyl acyl carrier protein fatty acid desaturase family regulate Verticillium wilt resistance. Mol. Plant Pathol. 2021, 22, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, S.; Padmanabhan, C.; Li, R.; Galvez, M.; Gutierrez, D.; Fuentes, S.; Ling, K.S.; Kreuze, J.; Fei, Z. VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 2017, 500, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, L.; Fu, S.; Wu, J.; Zhou, X. Population diversity of rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus. PLoS ONE 2012, 7, e46238. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Kalischuk, M.L.; Fusaro, A.F.; Waterhouse, P.M.; Pappu, H.R.; Kawchuk, L.M. Complete genomic sequence of a Rubus yellow net virus isolate and detection of genome-wide pararetrovirus-derived small RNAs. Virus Res. 2013, 178, 306–313. [Google Scholar] [CrossRef]
- Xu, D.; Mock, R.; Kinard, G.; Li, R. Molecular analysis of the complete genomic sequences of four isolates of Gooseberry vein banding associated virus. Virus Genes 2011, 43, 130–137. [Google Scholar] [CrossRef]
- Kazmi, S.A.; Yang, Z.; Hong, N.; Wang, G.; Wang, Y. Characterization by Small RNA Sequencing of Taro Bacilliform CH Virus (TaBCHV), a Novel Badnavirus. PLoS ONE 2015, 10, e0134147. [Google Scholar]
- Wang, Y.; Cheng, X.; Wu, X.; Wang, A.; Wu, X. Characterization of complete genome and small RNA profile of pagoda yellow mosaic associated virus, a novel badnavirus in China. Virus Res. 2014, 188, 103–108. [Google Scholar] [CrossRef]
- Calle Garcia, J.; Guadagno, A.; Paytuvi-Gallart, A.; Saera-Vila, A.; Amoroso, C.G.; D’Esposito, D.; Andolfo, G.; Aiese Cigliano, R.; Sanseverino, W.; Ercolano, M.R. PRGdb 4.0: An updated database dedicated to genes involved in plant disease resistance process. Nucleic Acids Res. 2022, 50, D1483–D1490. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X.; Dai, L.; Wang, G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genom. 2007, 34, 765–776. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Yin, L.; Lu, J. Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 2017, 254, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Zang, P.; Xu, Y. Transcriptome analysis reveals underlying immune response mechanism of fungal (Penicillium oxalicum) disease in Gastrodia elata Bl. f. glauca S. chow (Orchidaceae). BMC Plant Biol. 2020, 20, 445. [Google Scholar] [CrossRef] [PubMed]
- Irieda, H.; Inoue, Y.; Mori, M.; Yamada, K.; Oshikawa, Y.; Saitoh, H.; Uemura, A.; Terauchi, R.; Kitakura, S.; Kosaka, A.; et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 496–505. [Google Scholar] [CrossRef] [Green Version]
- Bian, X.; Zhao, Y.; Xiao, S.; Yang, H.; Han, Y.; Zhang, L. Metabolome and transcriptome analysis reveals the molecular profiles underlying the ginseng response to rusty root symptoms. BMC Plant Biol. 2021, 21, 215. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F., III; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tian, W.; Tao, F.; Wang, J.; Shang, H.; Chen, X.; Xu, X.; Hu, X. TaRPM1 positively regulates wheat high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2019, 10, 1679. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Su, J.; Zhang, M.; Zhang, L.; Sun, T.; Liu, Y.; Lukowitz, W.; Xu, J.; Zhang, S. Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 2017, 29, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.; Clough, S.J.; Ort, D.R.; DeLucia, E.H. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 2018, 16, e2004122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, Q.; Zhang, H.; Jia, Q.; Hong, Y.; Liu, Y. The rubisco small subunit is involved in tobamovirus movement and Tm-2(2)-mediated extreme resistance. Plant Physiol. 2013, 161, 374–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, D.; Gnanasekaran, P.; Kumar, R.K.; Kushwaha, N.K.; Sharma, V.K.; Yusuf, M.A.; Chakraborty, S. A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J. Exp. Bot. 2015, 66, 5881–5895. [Google Scholar] [CrossRef] [Green Version]
- Prod’homme, D.; Jakubiec, A.; Tournier, V.; Drugeon, G.; Jupin, I. Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J. Virol. 2003, 77, 9124–9135. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.M.; Ishikawa, M.; Ahlquist, P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 2001, 75, 2097–2106. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yan, Z.; Xia, Z.; Cheng, Y.; Jiao, Z.; Sun, B.; Zhou, T.; Fan, Z. A violaxanthin deepoxidase interacts with a viral suppressor of RNA silencing to inhibit virus amplification. Plant Physiol. 2017, 175, 1774–1794. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Yuan, Q.; Qiu, S.; Li, S.; Li, M.; Zheng, H.; Wu, G.; Lu, Y.; Peng, J.; Rao, S.; et al. Turnip mosaic virus impairs perinuclear chloroplast clustering to facilitate viral infection. Plant Cell Environ. 2021, 44, 3681–3699. [Google Scholar] [CrossRef]
- Yang, X.; Lu, Y.; Wang, F.; Chen, Y.; Tian, Y.; Jiang, L.; Peng, J.; Zheng, H.; Lin, L.; Yan, C.; et al. Involvement of the chloroplast gene ferredoxin 1 in multiple responses of Nicotiana benthamiana to Potato virus X infection. J. Exp. Bot. 2020, 71, 2142–2156. [Google Scholar] [CrossRef]
- Bhat, S.; Folimonova, S.Y.; Cole, A.B.; Ballard, K.D.; Lei, Z.; Watson, B.S.; Sumner, L.W.; Nelson, R.S. Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 2013, 161, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babosha, A.V. Regulation of resistance and susceptibility in wheat-powdery mildew pathosystem with exogenous cytokinins. J. Plant Physiol. 2009, 166, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Grosskinsky, D.K.; Naseem, M.; Abdelmohsen, U.R.; Plickert, N.; Engelke, T.; Griebel, T.; Zeier, J.; Novak, O.; Strnad, M.; Pfeifhofer, H.; et al. Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol. 2011, 157, 815–830. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [Green Version]
- Pogány, M.; Koehl, J.; Heiser, I.; Elstner, E.F.; Barna, B. Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to Tobacco necrosis virus and confers tolerance to oxidative stress. Physiol. Mol. Plant Pathol. 2004, 65, 39–47. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Tecsi, L.I.; Smith, A.M.; Maule, A.J.; Leegood, R.C. A spatial analysis of physiological changes associated with infection of cotyledons of marrow plants with Cucumber mosaic virus. Plant Physiol. 1996, 111, 975–985. [Google Scholar] [CrossRef] [Green Version]
- Shalitin, D.; Wolf, S. Cucumber mosaic virus infection affects sugar transport in Melon plants. Plant Physiol. 2000, 123, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.L.; Lehrer, A.T.; Hajirezaei, M.R.; Springer, A.; Komor, E. Modulation of carbohydrate metabolism and chloroplast structure in sugarcane leaves which were infected by Sugarcane Yellow Leaf Virus (SCYLV). Physiol. Mol. Plant Pathol. 2008, 73, 78–87. [Google Scholar] [CrossRef]
- Berger, S.; Papadopoulos, M.; Schreiber, U.; Kaiser, W.; Roitsch, T. Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant. 2004, 122, 419–428. [Google Scholar] [CrossRef]
- Gamm, M.; Héloir, M.C.; Bligny, R.; Vaillant-Gaveau, N.; Trouvelot, S.; Alcaraz, G.; Frettinger, P.; Clément, C.; Pugin, A.; Wendehenne, D.; et al. Changes in carbohydrate metabolism in Plasmopara viticola-infected grapevine leaves. Mol. Plant-Microbe Interact. MPMI 2011, 24, 1061–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Arae, T.; Green, P.J.; Yamaguchi, J.; Chiba, Y. AtCCR4a and AtCCR4b are involved in determining the Poly(A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 863–874. [Google Scholar] [CrossRef] [Green Version]
- Asatsuma, S.; Sawada, C.; Itoh, K.; Okito, M.; Kitajima, A.; Mitsui, T. Involvement of alpha-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005, 46, 858–869. [Google Scholar] [CrossRef]
- Berger, S.; Sinha, A.K.; Roitsch, T. Plant Physiol.ogy meets phytopathology: Plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 2007, 58, 4019–4026. [Google Scholar] [CrossRef]
- Ribeiro, C.; Stitt, M.; Hotta, C.T. How stress affects your budget-stress impacts on starch metabolism. Front. Plant Sci. 2022, 13, 774060. [Google Scholar] [CrossRef]
- Barbosa, P.M.G.; de Morais, T.P.; de Andrade Silva, C.A.; da Silva Santos, F.R.; Garcia, N.F.L.; Fonseca, G.G.; Leite, R.S.R.; da Paz, M.F. Biochemical characterization and evaluation of invertases produced from Saccharomyces cerevisiae CAT-1 and Rhodotorula mucilaginosa for the production of fructooligosaccharides. Prep. Biochem. Biotechnol. 2018, 48, 506–513. [Google Scholar] [CrossRef]
- Chou, H.-M.; Bundock, N.; Rolfe, S.A.; Scholes, J.D. Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Mol. Plant Pathol. 2000, 1, 99–113. [Google Scholar] [CrossRef]
- Bianco, C.; Mohr, I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. eLife 2019, 8, e49551. [Google Scholar] [CrossRef]
- Petelski, A.A.; Slavov, N. Analyzing ribosome remodeling in health and disease. Proteomics 2020, 20, e2000039. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; de Loubresse, N.G.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armache, J.P.; Jarasch, A.; Anger, A.M.; Villa, E.; Becker, T.; Bhushan, S.; Jossinet, F.; Habeck, M.; Dindar, G.; Franckenberg, S.; et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 19748–19753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Lai, S.; Jia, R.; Xu, A.; Zhang, L.; Lu, J.; Ye, K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 2011, 469, 559–563. [Google Scholar] [CrossRef]
- Woolford, J.L., Jr.; Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Gao, L.; Teng, L.; Ge, J.; Oo, Z.M.; Kumar, A.R.; Gilliland, D.G.; Mason, P.J.; Tan, K.; Speck, N.A. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 2015, 17, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Prava, J.; Pan, A. In silico analysis of Leishmania proteomes and protein-protein interaction network: Prioritizing therapeutic targets and drugs for repurposing to treat leishmaniasis. Acta Trop. 2022, 229, 106337. [Google Scholar] [CrossRef]
- Hang, R.; Wang, Z.; Deng, X.; Liu, C.; Yan, B.; Yang, C.; Song, X.; Mo, B.; Cao, X. Ribosomal RNA biogenesis and its response to chilling stress in Oryza sativa. Plant Physiol. 2018, 177, 381–397. [Google Scholar] [CrossRef] [Green Version]
- Greseth, M.D.; Traktman, P. De novo fatty acid biosynthesis contributes significantly to establishment of a bioenergetically favorable environment for vaccinia virus infection. PLoS Pathog. 2014, 10, e1004021. [Google Scholar] [CrossRef] [Green Version]
- Hardie, D.G.; Pan, D.A. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem. Soc. Trans. 2002, 30 Pt 6, 1064–1070. [Google Scholar] [CrossRef]
- Davies, S.P.; Sim, A.T.R.; Hardie, D.G. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur. J. Biochem. 1990, 187, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Moser, T.S.; Schieffer, D.; Cherry, S. AMP-activated kinase restricts Rift Valley fever virus infection by inhibiting fatty acid synthesis. PLoS Pathog. 2012, 8, e1002661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2007, 63, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef] [Green Version]
- Chandra-Shekara, A.C.; Venugopal, S.C.; Barman, S.R.; Kachroo, A.; Kachroo, P. Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 7277–7282. [Google Scholar] [CrossRef] [Green Version]
- Kachroo, A.; Fu, D.Q.; Havens, W.; Navarre, D.; Kachroo, P.; Ghabrial, S.A. An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant-Microbe Interact. MPMI 2008, 21, 564–575. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.J.; Shimono, M.; Maeda, S.; Inoue, H.; Mori, M.; Hasegawa, M.; Sugano, S.; Takatsuji, H. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant-Microbe Interact. MPMI 2009, 22, 820–829. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.W.; Kliebenstein, D.J.; Bostock, R.M.; Dehesh, K. Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 2013, 16, 520–526. [Google Scholar] [CrossRef] [Green Version]
- Batsale, M.; Bahammou, D.; Fouillen, L.; Mongrand, S.; Joubes, J.; Domergue, F. Biosynthesis and functions of very-long-chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells 2021, 10, 1284. [Google Scholar] [CrossRef]
- Millar, A.A.; Clemens, S.; Zachgo, S.; Giblin, E.M.; Taylor, D.C.; Kunst, L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 1999, 11, 825–838. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, D.; Liu, Y. Ectopic expression of CsKCS6 from navel orange promotes the production of very-long-chain fatty acids (VLCFAs) and increases the abiotic stress tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef] [PubMed]
- Weidenbach, D.; Jansen, M.; Franke, R.B.; Hensel, G.; Weissgerber, W.; Ulferts, S.; Jansen, I.; Schreiber, L.; Korzun, V.; Pontzen, R.; et al. Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 2014, 166, 1621–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Haslam, T.M.; Kruger, A.; Schneider, L.M.; Mishina, K.; Samuels, L.; Yang, H.; Kunst, L.; Schaffrath, U.; Nawrath, C.; et al. The beta-Ketoacyl-CoA synthase HvKCS1, encoded by Cer-zh, plays a key role in synthesis of barley leaf wax and germination of barley powdery mildew. Plant Cell Physiol. 2018, 59, 806–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.G.; Seo, P.J. MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis. Nat. Commun. 2019, 10, 1713. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, X.; Chen, Z.; Zhang, J.; Si, K.; Xu, R.; He, Y.; Zhu, F.; Cheng, Y. Function and transcriptional regulation of CsKCS20 in the elongation of very-long-chain fatty acids and wax biosynthesis in Citrus sinensis flavedo. Hortic. Res. 2022, 9, uhab027. [Google Scholar] [CrossRef]
- Oshima, Y.; Mitsuda, N. The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signal. Behav. 2013, 8, e26826. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 2015, 56, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 2013, 25, 1609–1624. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, N.; Chen, L. Paraffin section observation of flower bud differentiation of Chimonanthus praecox in Kunming and comparison of the differentiation processes in different regions, China. Hortic. Plant J. 2022, 8, 221–229. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, T.; Zhang, F.; Liu, Y.; Wang, G. Comparative analysis of the transcriptomes of persisting and abscised fruitlets: Insights into plant hormone and carbohydrate metabolism regulated self-thinning of pecan fruitlets during the early stage. Curr. Issues Mol. Biol. 2021, 44, 176–193. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.-L.; Wang, G.; Wang, T.; Jia, Z.-H.; Guo, Z.-R.; Zhang, J.-Y. AdRAP2.3, a novel ethylene response factor VII from Actinidia deliciosa, enhances waterlogging resistance in transgenic tobacco through improving expression levels of PDC and ADH genes. Int. J. Mol. Sci. 2019, 20, 1189. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Feng, G.; Su, W.; Liu, Z.; Peng, F.J.G. Transcriptomic analysis provides insights into grafting union development in pecan (Carya illinoinensis). Genes 2018, 9, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kachroo, A.; Lapchyk, L.; Fukushige, H.; Hildebrand, D.; Klessig, D.; Kachroo, P. Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 2003, 15, 2952–2965. [Google Scholar] [CrossRef] [Green Version]
- Kachroo, P.; Kachroo, A.; Lapchyk, L.; Hildebrand, D.; Klessig, D.F. Restoration of defective cross talk in ssi2 mutants: Role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant-Microbe Interact. MPMI 2003, 16, 1022–1029. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, T.; Jia, Z.; Jia, X.; Liu, Y.; Xuan, J.; Wang, G.; Zhang, F. Transcriptome Analysis Reveals a Comprehensive Virus Resistance Response Mechanism in Pecan Infected by a Novel Badnavirus Pecan Virus. Int. J. Mol. Sci. 2022, 23, 13576. https://doi.org/10.3390/ijms232113576
Zhang J, Wang T, Jia Z, Jia X, Liu Y, Xuan J, Wang G, Zhang F. Transcriptome Analysis Reveals a Comprehensive Virus Resistance Response Mechanism in Pecan Infected by a Novel Badnavirus Pecan Virus. International Journal of Molecular Sciences. 2022; 23(21):13576. https://doi.org/10.3390/ijms232113576
Chicago/Turabian StyleZhang, Jiyu, Tao Wang, Zhanhui Jia, Xiaodong Jia, Yongzhi Liu, Jiping Xuan, Gang Wang, and Fan Zhang. 2022. "Transcriptome Analysis Reveals a Comprehensive Virus Resistance Response Mechanism in Pecan Infected by a Novel Badnavirus Pecan Virus" International Journal of Molecular Sciences 23, no. 21: 13576. https://doi.org/10.3390/ijms232113576





