What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review
Abstract
1. Introduction

2. Methods
2.1. Search Strategy
2.2. Data Extraction
2.3. Statistical Analysis
3. Results
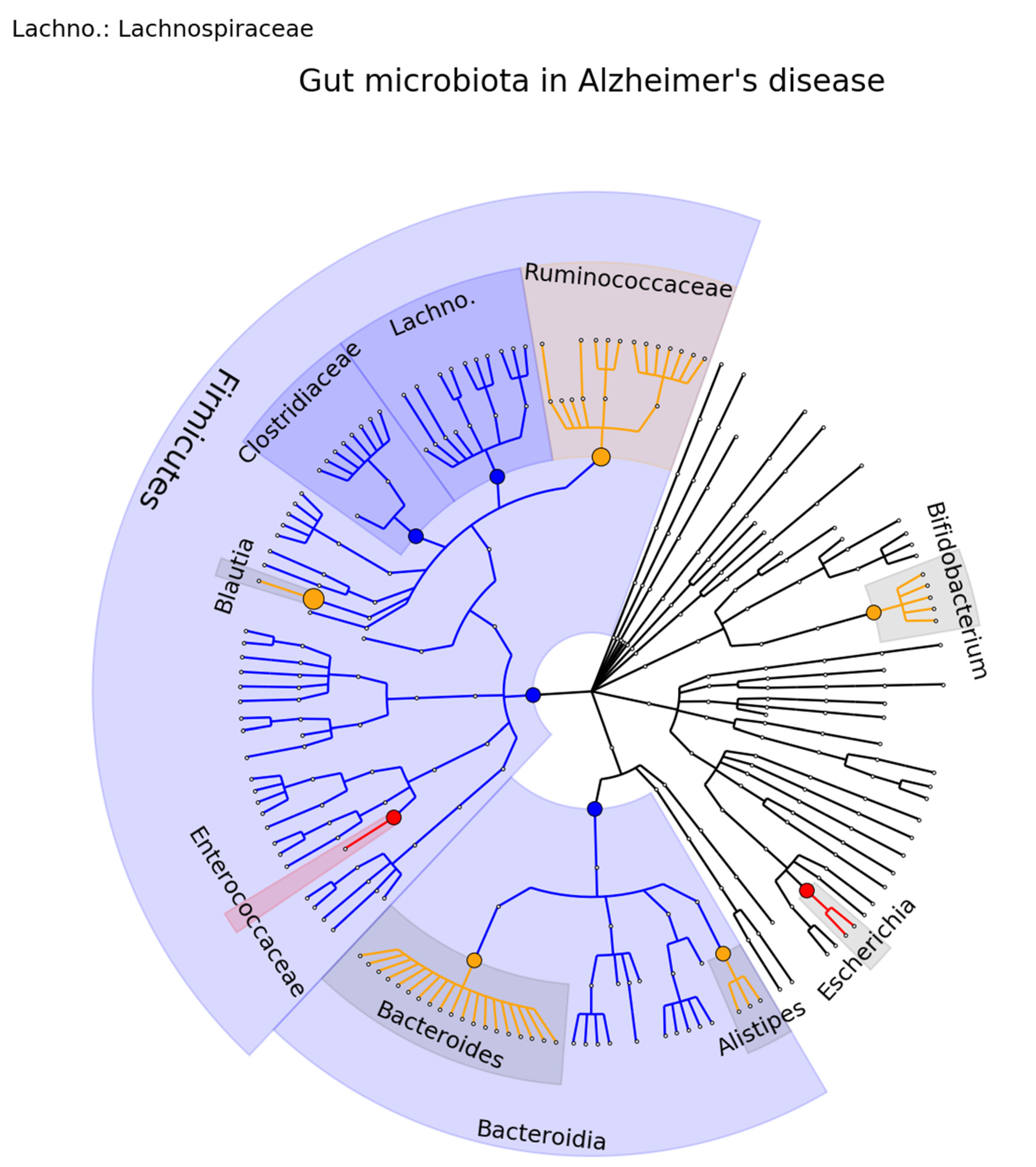
3.1. Review of the Gut Microbiome in PD, AD, MS, Stroke, and ALS
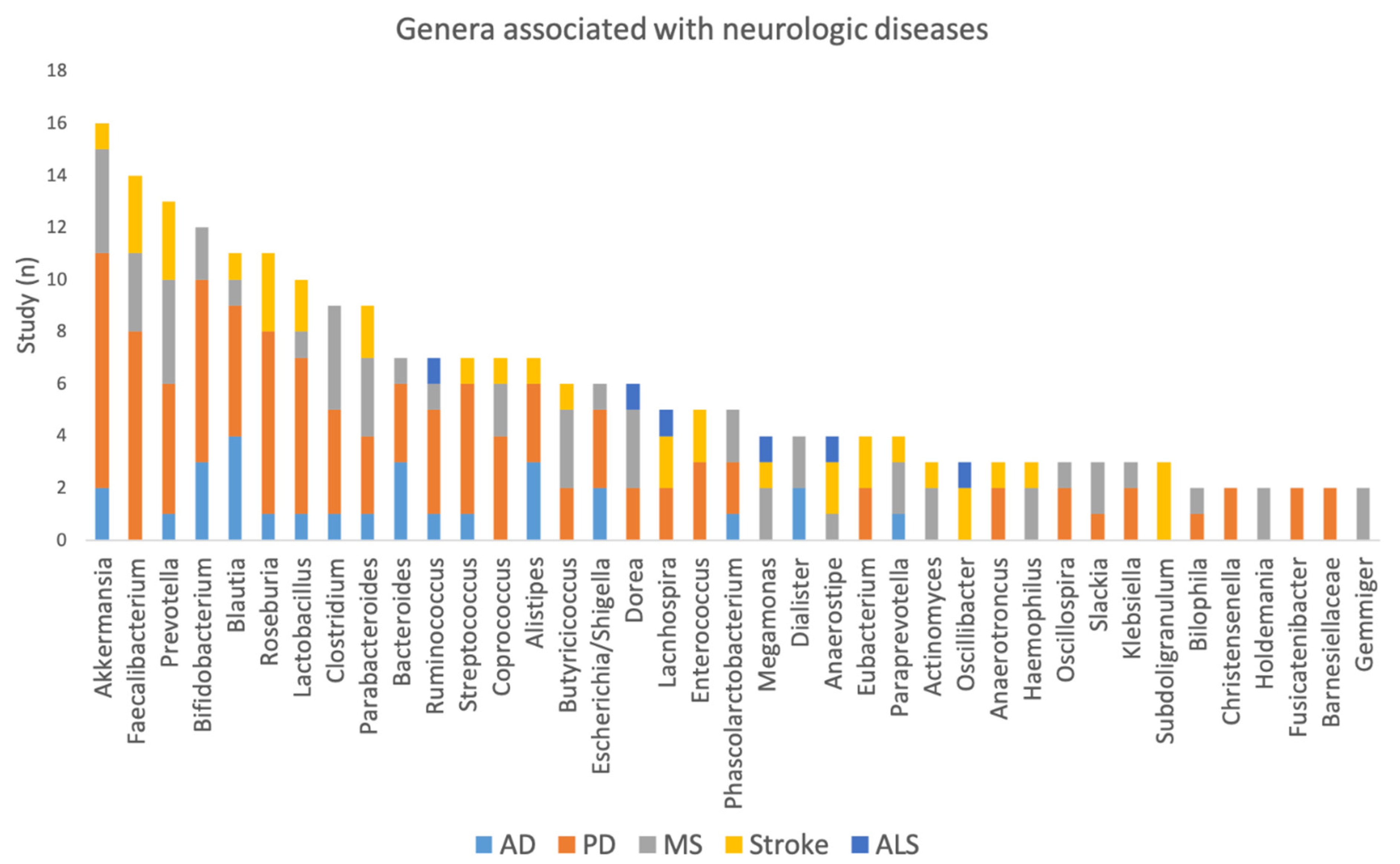
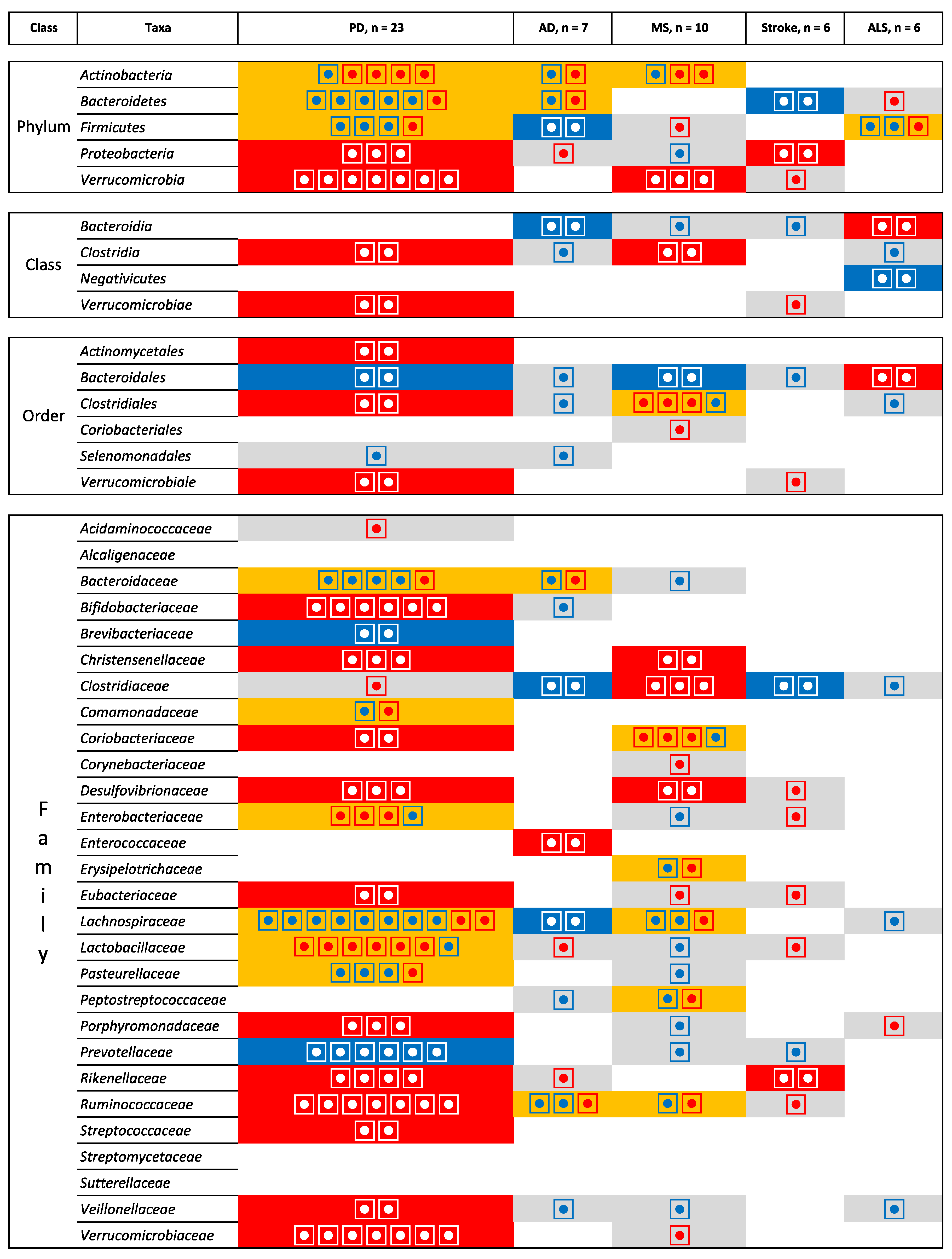
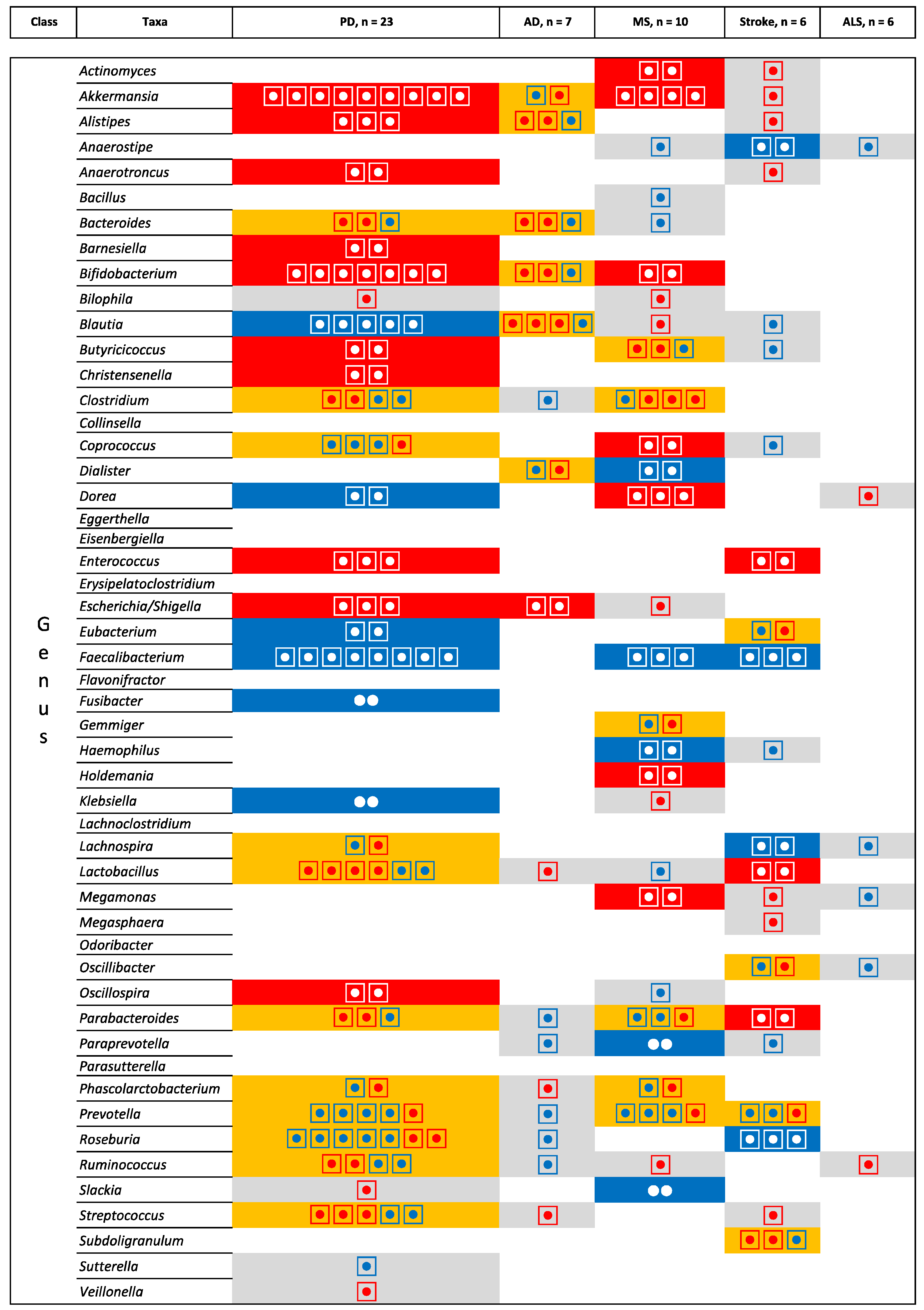
3.2. Integrating Data across Neurological Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial Genes, Brain & Behaviour—Epigenetic Regulation of the Gut-Brain Axis. Genes Brain Behav. 2014, 13, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L.H. The Role of Microbiome in Central Nervous System Disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Seguella, L.; Capuano, R.; Sarnelli, G.; Esposito, G. Play in Advance against Neurodegeneration: Exploring Enteric Glial Cells in Gut-Brain Axis during Neurodegenerative Diseases. Expert Rev. Clin. Pharmacol. 2019, 12, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Sasmita, A.O. Modification of the Gut Microbiome to Combat Neurodegeneration. Rev. Neurosci. 2019, 30, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Heiss, C.N.; Olofsson, L.E. The Role of the Gut Microbiota in Development, Function and Disorders of the Central Nervous System and the Enteric Nervous System. J. Neuroendocrinol. 2019, 31, e12684. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Human Microbiome Project Consortium Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, Probiotics and Neurodegenerative Diseases: Deciphering the Gut Brain Axis. Cell. Mol. Life Sci. 2017, 74, 3769–3787. [Google Scholar] [CrossRef]
- Abdel-Haq, R.; Schlachetzki, J.C.M.; Glass, C.K.; Mazmanian, S.K. Microbiome–Microglia Connections via the Gut–Brain Axis. J. Exp. Med. 2019, 216, 41–59. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Socała, K.; Doboszewska, U.; Szopa, A.; Serefko, A.; Włodarczyk, M.; Zielińska, A.; Poleszak, E.; Fichna, J.; Wlaź, P. The Role of Microbiota-Gut-Brain Axis in Neuropsychiatric and Neurological Disorders. Pharmacol. Res. 2021, 172, 105840. [Google Scholar] [CrossRef]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef]
- Angelucci, F.; Cechova, K.; Amlerova, J.; Hort, J. Antibiotics, Gut Microbiota, and Alzheimer’s Disease. J. Neuroinflamm. 2019, 16, 108. [Google Scholar] [CrossRef]
- Bostanciklioğlu, M. The Role of Gut Microbiota in Pathogenesis of Alzheimer’s Disease. J. Appl. Microbiol. 2019, 127, 954–967. [Google Scholar] [CrossRef]
- Sochocka, M.; Donskow-Łysoniewska, K.; Diniz, B.S.; Kurpas, D.; Brzozowska, E.; Leszek, J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease-a Critical Review. Mol. Neurobiol. 2019, 56, 1841–1851. [Google Scholar] [CrossRef]
- Doifode, T.; Giridharan, V.V.; Generoso, J.S.; Bhatti, G.; Collodel, A.; Schulz, P.E.; Forlenza, O.V.; Barichello, T. The Impact of the Microbiota-Gut-Brain Axis on Alzheimer’s Disease Pathophysiology. Pharmacol. Res. 2021, 164, 105314. [Google Scholar] [CrossRef]
- Liu, S.; Gao, J.; Zhu, M.; Liu, K.; Zhang, H.-L. Gut Microbiota and Dysbiosis in Alzheimer’s Disease: Implications for Pathogenesis and Treatment. Mol. Neurobiol. 2020, 57, 5026–5043. [Google Scholar] [CrossRef]
- Chapelet, G.; Leclair-Visonneau, L.; Clairembault, T.; Neunlist, M.; Derkinderen, P. Can the Gut Be the Missing Piece in Uncovering PD Pathogenesis? Parkinsonism Relat. Disord. 2019, 59, 26–31. [Google Scholar] [CrossRef]
- Santos, S.F.; de Oliveira, H.L.; Yamada, E.S.; Neves, B.C.; Pereira, A. The Gut and Parkinson’s Disease-A Bidirectional Pathway. Front. Neurol. 2019, 10, 574. [Google Scholar] [CrossRef]
- Chiang, H.-L.; Lin, C.-H. Altered Gut Microbiome and Intestinal Pathology in Parkinson’s Disease. J. Mov. Disord. 2019, 12, 67–83. [Google Scholar] [CrossRef]
- Sharma, S.; Awasthi, A.; Singh, S. Altered Gut Microbiota and Intestinal Permeability in Parkinson’s Disease: Pathological Highlight to Management. Neurosci. Lett. 2019, 712, 134516. [Google Scholar] [CrossRef]
- Miraglia, F.; Colla, E. Microbiome, Parkinson’s Disease and Molecular Mimicry. Cells 2019, 8, 222. [Google Scholar] [CrossRef]
- Nowak, J.M.; Kopczyński, M.; Friedman, A.; Koziorowski, D.; Figura, M. Microbiota Dysbiosis in Parkinson Disease-In Search of a Biomarker. Biomedicines 2022, 10, 2057. [Google Scholar] [CrossRef] [PubMed]
- Grant, H.; Anderton, R.; Gasson, N.; Lawrence, B.J. The Gut Microbiome and Cognition in Parkinson’s Disease: A Systematic Review. Nutr. Neurosci. 2022, 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sato, W.; Yamamura, T. Multiple Sclerosis: Possibility of a Gut Environment-Induced Disease. Neurochem. Int. 2019, 130, 104475. [Google Scholar] [CrossRef] [PubMed]
- Buscarinu, M.C.; Fornasiero, A.; Romano, S.; Ferraldeschi, M.; Mechelli, R.; Reniè, R.; Morena, E.; Romano, C.; Pellicciari, G.; Landi, A.C.; et al. The Contribution of Gut Barrier Changes to Multiple Sclerosis Pathophysiology. Front. Immunol. 2019, 10, 1916. [Google Scholar] [CrossRef]
- Ghadiri, F.; Ebadi, Z.; Asadollahzadeh, E.; Naser Moghadasi, A. Gut Microbiome in Multiple Sclerosis-Related Cognitive Impairment. Mult. Scler. Relat. Disord. 2022, 67, 104165. [Google Scholar] [CrossRef]
- Correale, J.; Hohlfeld, R.; Baranzini, S.E. The Role of the Gut Microbiota in Multiple Sclerosis. Nat. Rev. Neurol. 2022, 18, 544–558. [Google Scholar] [CrossRef]
- Bao, Z.; Zhang, Z.; Zhou, G.; Zhang, A.; Shao, A.; Zhou, F. Novel Mechanisms and Therapeutic Targets for Ischemic Stroke: A Focus on Gut Microbiota. Front. Cell. Neurosci. 2022, 16, 871720. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; He, J.; Xiong, X. The Role of the Gut Microbiota in the Development of Ischemic Stroke. Front. Immunol. 2022, 13, 845243. [Google Scholar] [CrossRef]
- Boddy, S.L.; Giovannelli, I.; Sassani, M.; Cooper-Knock, J.; Snyder, M.P.; Segal, E.; Elinav, E.; Barker, L.A.; Shaw, P.J.; McDermott, C.J. The Gut Microbiome: A Key Player in the Complexity of Amyotrophic Lateral Sclerosis (ALS). BMC Med. 2021, 19, 13. [Google Scholar] [CrossRef]
- Martin, S.; Battistini, C.; Sun, J. A Gut Feeling in Amyotrophic Lateral Sclerosis: Microbiome of Mice and Men. Front. Cell. Infect. Microbiol. 2022, 12, 839526. [Google Scholar] [CrossRef]
- Mancuso, C.; Santangelo, R. Alzheimer’s Disease and Gut Microbiota Modifications: The Long Way between Preclinical Studies and Clinical Evidence. Pharmacol. Res. 2018, 129, 329–336. [Google Scholar] [CrossRef]
- Sun, M.-F.; Shen, Y.-Q. Dysbiosis of Gut Microbiota and Microbial Metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef]
- Mirza, A.; Mao-Draayer, Y. The Gut Microbiome and Microbial Translocation in Multiple Sclerosis. Clin. Immunol. 2017, 183, 213–224. [Google Scholar] [CrossRef]
- Zhang, Y.-G.; Wu, S.; Yi, J.; Xia, Y.; Jin, D.; Zhou, J.; Sun, J. Target Intestinal Microbiota to Alleviate Disease Progression in Amyotrophic Lateral Sclerosis. Clin. Ther. 2017, 39, 322–336. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The Gut Microbiota-Brain Axis in Behaviour and Brain Disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Suganya, K.; Koo, B.-S. Gut–Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 7551. [Google Scholar] [CrossRef]
- Collins, S.M.; Surette, M.; Bercik, P. The Interplay between the Intestinal Microbiota and the Brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef]
- Xin, Y.; Diling, C.; Jian, Y.; Ting, L.; Guoyan, H.; Hualun, L.; Xiaocui, T.; Guoxiao, L.; Ou, S.; Chaoqun, Z.; et al. Effects of Oligosaccharides from Morinda Officinalis on Gut Microbiota and Metabolome of APP/PS1 Transgenic Mice. Front. Neurol. 2018, 9, 412. [Google Scholar] [CrossRef]
- Peng, W.; Yi, P.; Yang, J.; Xu, P.; Wang, Y.; Zhang, Z.; Huang, S.; Wang, Z.; Zhang, C. Association of Gut Microbiota Composition and Function with a Senescence-Accelerated Mouse Model of Alzheimer’s Disease Using 16S RRNA Gene and Metagenomic Sequencing Analysis. Aging 2018, 10, 4054–4065. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.K.; Verma, S.; Jain, V.; Surapaneni, B.K.; Vinayek, R.; Phillips, L.; Nair, P.P. Parkinson’s Disease: The Emerging Role of Gut Dysbiosis, Antibiotics, Probiotics, and Fecal Microbiota Transplantation. J. Neurogastroenterol. Motil. 2019, 25, 363–376. [Google Scholar] [CrossRef] [PubMed]
- De Lara-Sánchez, S.S.; Sánchez-Pérez, A.M. Probiotics Treatment Can Improve Cognition in Patients with Mild Cognitive Impairment: A Systematic Review. J. Alzheimers Dis. 2022, 89, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-M.; Letchumanan, V.; Tan, L.T.-H.; Hong, K.-W.; Wong, S.-H.; Ab Mutalib, N.-S.; Lee, L.-H.; Law, J.W.-F. Ketogenic Diet: A Dietary Intervention via Gut Microbiome Modulation for the Treatment of Neurological and Nutritional Disorders (a Narrative Review). Nutrients 2022, 14, 3566. [Google Scholar] [CrossRef] [PubMed]
- Vamanu, E.; Rai, S.N. The Link between Obesity, Microbiota Dysbiosis, and Neurodegenerative Pathogenesis. Diseases 2021, 9, 45. [Google Scholar] [CrossRef]
- Feng, W.; Liu, J.; Ao, H.; Yue, S.; Peng, C. Targeting Gut Microbiota for Precision Medicine: Focusing on the Efficacy and Toxicity of Drugs. Theranostics 2020, 10, 11278–11301. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.B.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut Microbiota Are Related to Parkinson’s Disease and Clinical Phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal Dysbiosis and Lowered Serum Lipopolysaccharide-Binding Protein in Parkinson’s Disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic Bacterial Composition in Parkinson’s Disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.-H. Short Chain Fatty Acids and Gut Microbiota Differ between Patients with Parkinson’s Disease and Age-Matched Controls. Parkinsonism Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Li, W.; Wu, X.; Hu, X.; Wang, T.; Liang, S.; Duan, Y.; Jin, F.; Qin, B. Structural Changes of Gut Microbiota in Parkinson’s Disease and Its Correlation with Clinical Features. Sci. China Life Sci. 2017, 60, 1223–1233. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s Disease and Parkinson’s Disease Medications Have Distinct Signatures of the Gut Microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional Implications of Microbial and Viral Gut Metagenome Changes in Early Stage L-DOPA-Naïve Parkinson’s Disease Patients. Genome Med. 2017, 9, 39. [Google Scholar] [CrossRef]
- Hopfner, F.; Künstner, A.; Müller, S.H.; Künzel, S.; Zeuner, K.E.; Margraf, N.G.; Deuschl, G.; Baines, J.F.; Kuhlenbäumer, G. Gut Microbiota in Parkinson Disease in a Northern German Cohort. Brain Res. 2017, 1667, 41–45. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Döring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The Nasal and Gut Microbiome in Parkinson’s Disease and Idiopathic Rapid Eye Movement Sleep Behavior Disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Wu, C.; Song, Y.; Qin, N.; Chen, S.-D.; Xiao, Q. Alteration of the Fecal Microbiota in Chinese Patients with Parkinson’s Disease. Brain Behav. Immun. 2018, 70, 194–202. [Google Scholar] [CrossRef]
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H.; et al. Gut Microbiota in Patients with Parkinson’s Disease in Southern China. Parkinsonism Relat. Disord. 2018, 53, 82–88. [Google Scholar] [CrossRef]
- Barichella, M.; Severgnini, M.; Cilia, R.; Cassani, E.; Bolliri, C.; Caronni, S.; Ferri, V.; Cancello, R.; Ceccarani, C.; Faierman, S.; et al. Unraveling Gut Microbiota in Parkinson’s Disease and Atypical Parkinsonism. Mov. Disord. 2019, 34, 396–405. [Google Scholar] [CrossRef]
- Li, C.; Cui, L.; Yang, Y.; Miao, J.; Zhao, X.; Zhang, J.; Cui, G.; Zhang, Y. Gut Microbiota Differs Between Parkinson’s Disease Patients and Healthy Controls in Northeast China. Front. Mol. Neurosci. 2019, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, P.; Chen, Z.; Sui, X.; Xie, X.; Zhang, J. Alteration of the Fecal Microbiota in North-Eastern Han Chinese Population with Sporadic Parkinson’s Disease. Neurosci. Lett. 2019, 707, 134297. [Google Scholar] [CrossRef] [PubMed]
- Aho, V.T.E.; Pereira, P.A.B.; Voutilainen, S.; Paulin, L.; Pekkonen, E.; Auvinen, P.; Scheperjans, F. Gut Microbiota in Parkinson’s Disease: Temporal Stability and Relations to Disease Progression. EBioMedicine 2019, 44, 691–707. [Google Scholar] [CrossRef]
- Weis, S.; Schwiertz, A.; Unger, M.M.; Becker, A.; Faßbender, K.; Ratering, S.; Kohl, M.; Schnell, S.; Schäfer, K.-H.; Egert, M. Effect of Parkinson’s Disease and Related Medications on the Composition of the Fecal Bacterial Microbiota. npj Parkinson’s Dis. 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Pietrucci, D.; Cerroni, R.; Unida, V.; Farcomeni, A.; Pierantozzi, M.; Mercuri, N.B.; Biocca, S.; Stefani, A.; Desideri, A. Dysbiosis of Gut Microbiota in a Selected Population of Parkinson’s Patients. Parkinsonism Relat. Disord. 2019, 65, 124–130. [Google Scholar] [CrossRef]
- Ren, T.; Gao, Y.; Qiu, Y.; Jiang, S.; Zhang, Q.; Zhang, J.; Wang, L.; Zhang, Y.; Wang, L.; Nie, K. Gut Microbiota Altered in Mild Cognitive Impairment Compared with Normal Cognition in Sporadic Parkinson’s Disease. Front. Neurol. 2020, 11, 137. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut Microbiota and Metabolome Alterations Associated with Parkinson’s Disease. mSystems 2020, 5, e00561. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Ito, M.; Ishida, T.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Meta-Analysis of Gut Dysbiosis in Parkinson’s Disease. Mov. Disord. 2020, 35, 1626–1635. [Google Scholar] [CrossRef]
- Cirstea, M.S.; Yu, A.C.; Golz, E.; Sundvick, K.; Kliger, D.; Radisavljevic, N.; Foulger, L.H.; Mackenzie, M.; Huan, T.; Finlay, B.B.; et al. Microbiota Composition and Metabolism Are Associated with Gut Function in Parkinson’s Disease. Mov. Disord. 2020, 35, 1208–1217. [Google Scholar] [CrossRef]
- Cosma-Grigorov, A.; Meixner, H.; Mrochen, A.; Wirtz, S.; Winkler, J.; Marxreiter, F. Changes in Gastrointestinal Microbiome Composition in PD: A Pivotal Role of Covariates. Front. Neurol. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut Microbiome Alterations in Alzheimer’s Disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C.; et al. Association of Brain Amyloidosis with Pro-Inflammatory Gut Bacterial Taxa and Peripheral Inflammation Markers in Cognitively Impaired Elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.-L.; et al. Gut Microbiota Is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef]
- Haran, J.P.; Bhattarai, S.K.; Foley, S.E.; Dutta, P.; Ward, D.V.; Bucci, V.; McCormick, B.A. Alzheimer’s Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway. MBio 2019, 10, e00632-19. [Google Scholar] [CrossRef]
- Lopizzo, N.; Provasi, S.; Marizzoni, M.; Borruso, L.; Andryszak, P.; Frisoni, G.B.; Cattaneo, A. Identification of Gut Microbiota Signature in Alzheimer’s Disease: Possible Role in Influencing Peripheral Inflammation. Eur. Neuropsychopharmacol. 2019, 29, S167–S168. [Google Scholar] [CrossRef]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K.; et al. Altered Microbiomes Distinguish Alzheimer’s Disease from Amnestic Mild Cognitive Impairment and Health in a Chinese Cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild Cognitive Impairment Has Similar Alterations as Alzheimer’s Disease in Gut Microbiota. Alzheimer’s Dement. 2019, 15, 1357–1366. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut Microbiota in Multiple Sclerosis: Possible Influence of Immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple Sclerosis Patients Have a Distinct Gut Microbiota Compared to Healthy Controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E. US Network of Pediatric MS Centers Gut Microbiota in Early Pediatric Multiple Sclerosis: A Case-Control Study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the Human Gut Microbiome in Multiple Sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut Bacteria from Multiple Sclerosis Patients Modulate Human T Cells and Exacerbate Symptoms in Mouse Models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Chen, C.; Knox, N.C.; Marrie, R.-A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A Comparative Study of the Gut Microbiota in Immune-Mediated Inflammatory Diseases—Does a Common Dysbiosis Exist? Microbiome 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Kozhieva, M.; Naumova, N.; Alikina, T.; Boyko, A.; Vlassov, V.; Kabilov, M.R. Primary Progressive Multiple Sclerosis in a Russian Cohort: Relationship with Gut Bacterial Diversity. BMC Microbiol. 2019, 19, 309. [Google Scholar] [CrossRef]
- Ventura, R.E.; Iizumi, T.; Battaglia, T.; Liu, M.; Perez-Perez, G.I.; Herbert, J.; Blaser, M.J. Gut Microbiome of Treatment-Naïve MS Patients of Different Ethnicities Early in Disease Course. Sci. Rep. 2019, 9, 16396. [Google Scholar] [CrossRef]
- Oezguen, N.; Yalcinkaya, N.; Kücükali, C.I.; Dahdouli, M.; Hollister, E.B.; Luna, R.A.; Türkoglu, R.; Kürtüncü, M.; Eraksoy, M.; Savidge, T.C.; et al. Microbiota Stratification Identifies Disease-Specific Alterations in Neuro-Behçet’s Disease and Multiple Sclerosis. Clin. Exp. Rheumatol. 2019, 37 (Suppl. 121), 58–66. [Google Scholar]
- Yin, J.; Liao, S.-X.; He, Y.; Wang, S.; Xia, G.-H.; Liu, F.-T.; Zhu, J.-J.; You, C.; Chen, Q.; Zhou, L.; et al. Dysbiosis of Gut Microbiota with Reduced Trimethylamine-N-Oxide Level in Patients With Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 2015, 4, e002699. [Google Scholar] [CrossRef]
- Yamashiro, K.; Tanaka, R.; Urabe, T.; Ueno, Y.; Yamashiro, Y.; Nomoto, K.; Takahashi, T.; Tsuji, H.; Asahara, T.; Hattori, N. Gut Dysbiosis Is Associated with Metabolism and Systemic Inflammation in Patients with Ischemic Stroke. PLoS ONE 2017, 12, e0171521. [Google Scholar] [CrossRef]
- Xia, G.-H.; You, C.; Gao, X.-X.; Zeng, X.-L.; Zhu, J.-J.; Xu, K.-Y.; Tan, C.-H.; Xu, R.-T.; Wu, Q.-H.; Zhou, H.-W.; et al. Stroke Dysbiosis Index (SDI) in Gut Microbiome Are Associated with Brain Injury and Prognosis of Stroke. Front. Neurol. 2019, 10, 397. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.; Sun, C.; Wu, X.; Lu, M.; Si, Y.; Ye, X.; Wang, T.; Yu, X.; Zhao, X.; et al. Change of Intestinal Microbiota in Cerebral Ischemic Stroke Patients. BMC Microbiol. 2019, 19, 191. [Google Scholar] [CrossRef]
- Haak, B.W.; Westendorp, W.F.; van Engelen, T.S.R.; Brands, X.; Brouwer, M.C.; Vermeij, J.-D.; Hugenholtz, F.; Verhoeven, A.; Derks, R.J.; Giera, M.; et al. Disruptions of Anaerobic Gut Bacteria Are Associated with Stroke and Post-Stroke Infection: A Prospective Case-Control Study. Transl. Stroke Res. 2020, 12, 581–592. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.; Pan, D.; Liu, Y.; Yan, X.; Tang, Y.; Tao, M.; Gong, L.; Zhang, T.; Woods, C.R.; et al. Dysbiosis Characteristics of Gut Microbiota in Cerebral Infarction Patients. Transl. Neurosci. 2020, 11, 124–133. [Google Scholar] [CrossRef]
- Fang, X.; Wang, X.; Yang, S.; Meng, F.; Wang, X.; Wei, H.; Chen, T. Evaluation of the Microbial Diversity in Amyotrophic Lateral Sclerosis Using High-Throughput Sequencing. Front. Microbiol. 2016, 7, 1479. [Google Scholar] [CrossRef]
- Brenner, D.; Hiergeist, A.; Adis, C.; Mayer, B.; Gessner, A.; Ludolph, A.C.; Weishaupt, J.H. The Fecal Microbiome of ALS Patients. Neurobiol. Aging 2018, 61, 132–137. [Google Scholar] [CrossRef]
- Mazzini, L.; Mogna, L.; De Marchi, F.; Amoruso, A.; Pane, M.; Aloisio, I.; Cionci, N.B.; Gaggìa, F.; Lucenti, A.; Bersano, E.; et al. Potential Role of Gut Microbiota in ALS Pathogenesis and Possible Novel Therapeutic Strategies. J. Clin. Gastroenterol. 2018, 52, S68–S70. [Google Scholar] [CrossRef]
- Zhai, C.-D.; Zheng, J.-J.; An, B.-C.; Huang, H.-F.; Tan, Z.-C. Intestinal Microbiota Composition in Patients with Amyotrophic Lateral Sclerosis: Establishment of Bacterial and Archaeal Communities Analyses. Chin. Med. J. 2019, 132, 1815–1822. [Google Scholar] [CrossRef]
- Di Gioia, D.; Bozzi Cionci, N.; Baffoni, L.; Amoruso, A.; Pane, M.; Mogna, L.; Gaggìa, F.; Lucenti, M.A.; Bersano, E.; Cantello, R.; et al. A Prospective Longitudinal Study on the Microbiota Composition in Amyotrophic Lateral Sclerosis. BMC Med. 2020, 18, 153. [Google Scholar] [CrossRef]
- Zeng, Q.; Shen, J.; Chen, K.; Zhou, J.; Liao, Q.; Lu, K.; Yuan, J.; Bi, F. The Alteration of Gut Microbiome and Metabolism in Amyotrophic Lateral Sclerosis Patients. Sci. Rep. 2020, 10, 12998. [Google Scholar] [CrossRef]
- Rowin, J.; Xia, Y.; Jung, B.; Sun, J. Gut Inflammation and Dysbiosis in Human Motor Neuron Disease. Physiol. Rep. 2017, 5, e13443. [Google Scholar] [CrossRef]
- Xing, J.; Li, X.; Sun, Y.; Zhao, J.; Miao, S.; Xiong, Q.; Zhang, Y.; Zhang, G. Comparative Genomic and Functional Analysis of Akkermansia Muciniphila and Closely Related Species. Genes Genom. 2019, 41, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia Muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Rezende, R.M.; Moreira, T.G.; Tankou, S.K.; Cox, L.M.; Wu, M.; Song, A.; Dhang, F.H.; Wei, Z.; Costamagna, G.; et al. Oral Administration of MiR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia Muciniphila. Cell Host Microbe 2019, 26, 779–794.e8. [Google Scholar] [CrossRef]
- Nandwana, V.; Nandwana, N.K.; Das, Y.; Saito, M.; Panda, T.; Das, S.; Almaguel, F.; Hosmane, N.S.; Das, B.C. The Role of Microbiome in Brain Development and Neurodegenerative Diseases. Molecules 2022, 27, 3402. [Google Scholar] [CrossRef]
- Hirsch, E.C.; Hunot, S. Neuroinflammation in Parkinson’s Disease: A Target for Neuroprotection? Lancet Neurol. 2009, 8, 382–397. [Google Scholar] [CrossRef]
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson Disease Epidemiology, Pathology, Genetics, and Pathophysiology. Clin. Geriatr. Med. 2020, 36, 1–12. [Google Scholar] [CrossRef]
- De Virgilio, A.; Greco, A.; Fabbrini, G.; Inghilleri, M.; Rizzo, M.I.; Gallo, A.; Conte, M.; Rosato, C.; Ciniglio Appiani, M.; de Vincentiis, M. Parkinson’s Disease: Autoimmunity and Neuroinflammation. Autoimmun. Rev. 2016, 15, 1005–1011. [Google Scholar] [CrossRef]
- EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA); Turck, D.; Bohn, T.; Castenmiller, J.; De Henauw, S.; Hirsch-Ernst, K.I.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Safety of Pasteurised Akkermansia Muciniphila as a Novel Food Pursuant to Regulation (EU) 2015/2283. EFS2 2021, 19, e06780. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-Analysis of the Parkinson’s Disease Gut Microbiome Suggests Alterations Linked to Intestinal Inflammation. npj Parkinsons Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Szilagyi, A.; Shrier, I.; Heilpern, D.; Je, J.; Park, S.; Chong, G.; Lalonde, C.; Cote, L.-F.; Lee, B. Differential Impact of Lactose/Lactase Phenotype on Colonic Microflora. Can. J. Gastroenterol. 2010, 24, 373–379. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Liu, G.; Shen, X.; Tang, Y. Association between Cigarette Smoking and Parkinson’s Disease: A Meta-Analysis. Arch. Gerontol. Geriatr. 2015, 61, 510–516. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, F.; Liu, Y.; Ma, T.; Jin, H.; Quan, K.; Leng, B.; Zhao, J.; Yuan, X.; Li, Z.; et al. Probiotics Synergized with Conventional Regimen in Managing Parkinson’s Disease. npj Parkinsons Dis. 2022, 8, 62. [Google Scholar] [CrossRef]
- Messaoudi, M.; Lalonde, R.; Violle, N.; Javelot, H.; Desor, D.; Nejdi, A.; Bisson, J.-F.; Rougeot, C.; Pichelin, M.; Cazaubiel, M.; et al. Assessment of Psychotropic-like Properties of a Probiotic Formulation (Lactobacillus Helveticus R0052 and Bifidobacterium Longum R0175) in Rats and Human Subjects. Br. J. Nutr. 2011, 105, 755–764. [Google Scholar] [CrossRef]
- Sarkar, A.; Lehto, S.M.; Harty, S.; Dinan, T.G.; Cryan, J.F.; Burnet, P.W.J. Psychobiotics and the Manipulation of Bacteria-Gut-Brain Signals. Trends Neurosci. 2016, 39, 763–781. [Google Scholar] [CrossRef]
- Savignac, H.M.; Couch, Y.; Stratford, M.; Bannerman, D.M.; Tzortzis, G.; Anthony, D.C.; Burnet, P.W.J. Prebiotic Administration Normalizes Lipopolysaccharide (LPS)-Induced Anxiety and Cortical 5-HT2A Receptor and IL1-β Levels in Male Mice. Brain Behav. Immun. 2016, 52, 120–131. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Biagi, E.; Heilig, H.G.H.J.; Kajander, K.; Kekkonen, R.A.; Tims, S.; de Vos, W.M. Global and Deep Molecular Analysis of Microbiota Signatures in Fecal Samples from Patients with Irritable Bowel Syndrome. Gastroenterology 2011, 141, 1792–1801. [Google Scholar] [CrossRef]
- Maini Rekdal, V.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and Inhibition of an Interspecies Gut Bacterial Pathway for Levodopa Metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef]
- Van Kessel, S.P.; Frye, A.K.; El-Gendy, A.O.; Castejon, M.; Keshavarzian, A.; van Dijk, G.; El Aidy, S. Gut Bacterial Tyrosine Decarboxylases Restrict Levels of Levodopa in the Treatment of Parkinson’s Disease. Nat. Commun. 2019, 10, 310. [Google Scholar] [CrossRef] [PubMed]
- Connil, N.; Le Breton, Y.; Dousset, X.; Auffray, Y.; Rincé, A.; Prévost, H. Identification of the Enterococcus Faecalis Tyrosine Decarboxylase Operon Involved in Tyramine Production. Appl. Environ. Microbiol. 2002, 68, 3537–3544. [Google Scholar] [CrossRef] [PubMed]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.J.; Hsiao, E.Y. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018, 173, 1728–1741.e13. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Seo, S.-U.; Chen, G.Y.; Núñez, G. Role of the Gut Microbiota in Immunity and Inflammatory Disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.; Cristiano, C.; Avagliano, C.; De Caro, C.; La Rana, G.; Raso, G.M.; Canani, R.B.; Meli, R.; Calignano, A. Gut-Brain Axis: Role of Lipids in the Regulation of Inflammation, Pain and CNS Diseases. Curr. Med. Chem. 2018, 25, 3930–3952. [Google Scholar] [CrossRef]
- Tsunoda, I. Lymphatic System and Gut Microbiota Affect Immunopathology of Neuroinflammatory Diseases, Including Multiple Sclerosis, Neuromyelitis Optica and Alzheimer’s Disease. Clin. Exp. Neuroimmunol. 2017, 8, 177–179. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Pindo, M.; Renzi, D.; et al. Altered Gut Microbiota in Rett Syndrome. Microbiome 2016, 4, 41. [Google Scholar] [CrossRef]
- Xu, R.; Tan, C.; Zhu, J.; Zeng, X.; Gao, X.; Wu, Q.; Chen, Q.; Wang, H.; Zhou, H.; He, Y.; et al. Dysbiosis of the Intestinal Microbiota in Neurocritically Ill Patients and the Risk for Death. Crit. Care 2019, 23, 195. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut Microbiota Dysbiosis Contributes to the Development of Hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef]
- Yan, Q.; Gu, Y.; Li, X.; Yang, W.; Jia, L.; Chen, C.; Han, X.; Huang, Y.; Zhao, L.; Li, P.; et al. Alterations of the Gut Microbiome in Hypertension. Front. Cell. Infect. Microbiol. 2017, 7, 381. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Hylemon, P.B.; Ridlon, J.M.; Heuman, D.M.; Daita, K.; White, M.B.; Monteith, P.; Noble, N.A.; Sikaroodi, M.; Gillevet, P.M. Colonic Mucosal Microbiome Differs from Stool Microbiome in Cirrhosis and Hepatic Encephalopathy and Is Linked to Cognition and Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G675–G685. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered Profile of Human Gut Microbiome Is Associated with Cirrhosis and Its Complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef]
- Ni, J.; Huang, R.; Zhou, H.; Xu, X.; Li, Y.; Cao, P.; Zhong, K.; Ge, M.; Chen, X.; Hou, B.; et al. Analysis of the Relationship Between the Degree of Dysbiosis in Gut Microbiota and Prognosis at Different Stages of Primary Hepatocellular Carcinoma. Front. Microbiol. 2019, 10, 1458. [Google Scholar] [CrossRef]
- Chen, P.-C.; Chien, Y.-W.; Yang, S.-C. The Alteration of Gut Microbiota in Newly Diagnosed Type 2 Diabetic Patients. Nutrition 2019, 63, 51–56. [Google Scholar] [CrossRef]
- Chen, B.; Sun, L.; Zhang, X. Integration of Microbiome and Epigenome to Decipher the Pathogenesis of Autoimmune Diseases. J. Autoimmun. 2017, 83, 31–42. [Google Scholar] [CrossRef]
- Luo, X.M.; Edwards, M.R.; Mu, Q.; Yu, Y.; Vieson, M.D.; Reilly, C.M.; Ahmed, S.A.; Bankole, A.A. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 2018, 84, e02288-17. [Google Scholar] [CrossRef]
- Ipci, K.; Altıntoprak, N.; Muluk, N.B.; Senturk, M.; Cingi, C. The Possible Mechanisms of the Human Microbiome in Allergic Diseases. Eur. Arch. Otorhinolaryngol. 2017, 274, 617–626. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, N.; Wu, C.; Zhang, X.; Wang, Q.; Huang, X.; Du, L.; Cao, Q.; Tang, J.; Zhou, C.; et al. A Metagenomic Study of the Gut Microbiome in Behcet’s Disease. Microbiome 2018, 6, 135. [Google Scholar] [CrossRef]
- Patrone, V.; Puglisi, E.; Cardinali, M.; Schnitzler, T.S.; Svegliati, S.; Festa, A.; Gabrielli, A.; Morelli, L. Gut Microbiota Profile in Systemic Sclerosis Patients with and without Clinical Evidence of Gastrointestinal Involvement. Sci. Rep. 2017, 7, 14874. [Google Scholar] [CrossRef]
- Liu, X.; Zou, Q.; Zeng, B.; Fang, Y.; Wei, H. Analysis of Fecal Lactobacillus Community Structure in Patients with Early Rheumatoid Arthritis. Curr. Microbiol. 2013, 67, 170–176. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Xu, Q.; Hu, C.; Gao, Y.; Chen, M.; Hu, R.; Liu, Y.; Chi, H.; Yin, Q.; et al. Dysbiotic Gut Microbes May Contribute to Hypertension by Limiting Vitamin D Production. Clin. Cardiol. 2019, 42, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-N.; Yun, Y.; Ryu, S.; Chang, Y.; Kwon, M.-J.; Cho, J.; Shin, H.; Kim, H.-L. Correlation between Gut Microbiota and Personality in Adults: A Cross-Sectional Study. Brain Behav. Immun. 2018, 69, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Libbey, J.E.; Sanchez, J.M.; Doty, D.J.; Sim, J.T.; Cusick, M.F.; Cox, J.E.; Fischer, K.F.; Round, J.L.; Fujinami, R.S. Variations in Diet Cause Alterations in Microbiota and Metabolites That Follow Changes in Disease Severity in a Multiple Sclerosis Model. Benef. Microbes 2018, 9, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.W.; Pandya, J.D.; Shear, D.A. Gut Microbiota as a Therapeutic Target to Ameliorate the Biochemical, Neuroanatomical, and Behavioral Effects of Traumatic Brain Injuries. Front. Neurol. 2019, 10, 875. [Google Scholar] [CrossRef] [PubMed]
- Cignarella, F.; Cantoni, C.; Ghezzi, L.; Salter, A.; Dorsett, Y.; Chen, L.; Phillips, D.; Weinstock, G.M.; Fontana, L.; Cross, A.H.; et al. Intermittent Fasting Confers Protection in CNS Autoimmunity by Altering the Gut Microbiota. Cell Metab. 2018, 27, 1222–1235.e6. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.; Zhou, Y.; Yu, L.; Zhang, L.; Wang, Y. Altered Gut Microbiome Composition in Children with Refractory Epilepsy after Ketogenic Diet. Epilepsy Res. 2018, 145, 163–168. [Google Scholar] [CrossRef]
- Lindefeldt, M.; Eng, A.; Darban, H.; Bjerkner, A.; Zetterström, C.K.; Allander, T.; Andersson, B.; Borenstein, E.; Dahlin, M.; Prast-Nielsen, S. The Ketogenic Diet Influences Taxonomic and Functional Composition of the Gut Microbiota in Children with Severe Epilepsy. npj Biofilms Microbiomes 2019, 5, 5. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, H.; Liu, X.; Zhang, J.; Liu, G. Crosstalk between the Ketogenic Diet and Epilepsy: From the Perspective of Gut Microbiota. Mediat. Inflamm. 2019, 2019, 8373060. [Google Scholar] [CrossRef]
- Spinelli, E.; Blackford, R. Gut Microbiota, the Ketogenic Diet and Epilepsy. Pediatr. Neurol. Briefs 2018, 32, 10. [Google Scholar] [CrossRef]
- Cui, B.; Li, P.; Xu, L.; Peng, Z.; Xiang, J.; He, Z.; Zhang, T.; Ji, G.; Nie, Y.; Wu, K.; et al. Step-up Fecal Microbiota Transplantation (FMT) Strategy. Gut Microbes 2016, 7, 323–328. [Google Scholar] [CrossRef]
- Kao, D.; Roach, B.; Silva, M.; Beck, P.; Rioux, K.; Kaplan, G.G.; Chang, H.-J.; Coward, S.; Goodman, K.J.; Xu, H.; et al. Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium Difficile Infection: A Randomized Clinical Trial. JAMA 2017, 318, 1985–1993. [Google Scholar] [CrossRef]
- Sun, M.-F.; Zhu, Y.-L.; Zhou, Z.-L.; Jia, X.-B.; Xu, Y.-D.; Yang, Q.; Cui, C.; Shen, Y.-Q. Neuroprotective Effects of Fecal Microbiota Transplantation on MPTP-Induced Parkinson’s Disease Mice: Gut Microbiota, Glial Reaction and TLR4/TNF-α Signaling Pathway. Brain Behav. Immun. 2018, 70, 48–60. [Google Scholar] [CrossRef]
- Evrensel, A.; Ceylan, M.E. Fecal Microbiota Transplantation and Its Usage in Neuropsychiatric Disorders. Clin. Psychopharmacol. Neurosci. 2016, 14, 231–237. [Google Scholar] [CrossRef]
- Zhao, Z.; Ning, J.; Bao, X.-Q.; Shang, M.; Ma, J.; Li, G.; Zhang, D. Fecal Microbiota Transplantation Protects Rotenone-Induced Parkinson’s Disease Mice via Suppressing Inflammation Mediated by the Lipopolysaccharide-TLR4 Signaling Pathway through the Microbiota-Gut-Brain Axis. Microbiome 2021, 9, 226. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Gazerani, P. Probiotics for Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 74121. [Google Scholar] [CrossRef]
- Kesika, P.; Suganthy, N.; Sivamaruthi, B.S.; Chaiyasut, C. Role of Gut-Brain Axis, Gut Microbial Composition, and Probiotic Intervention in Alzheimer’s Disease. Life Sci. 2021, 264, 118627. [Google Scholar] [CrossRef]
- Ghezzi, L.; Cantoni, C.; Pinget, G.V.; Zhou, Y.; Piccio, L. Targeting the Gut to Treat Multiple Sclerosis. J. Clin. Investig. 2021, 131, 143774. [Google Scholar] [CrossRef]
- Zhong, D.-Y.; Li, L.; Ma, R.-M.; Deng, Y.-H. The Effect of Probiotics in Stroke Treatment. Evid. Based Complement. Altern. Med. 2021, 2021, 4877311. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef]
- Shen, L.; Liu, L.; Ji, H.-F. Alzheimer’s Disease Histological and Behavioral Manifestations in Transgenic Mice Correlate with Specific Gut Microbiome State. J. Alzheimers Dis. 2017, 56, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Harach, T.; Marungruang, N.; Duthilleul, N.; Cheatham, V.; Mc Coy, K.D.; Frisoni, G.; Neher, J.J.; Fåk, F.; Jucker, M.; Lasser, T.; et al. Reduction of Abeta Amyloid Pathology in APPPS1 Transgenic Mice in the Absence of Gut Microbiota. Sci. Rep. 2017, 7, 41802. [Google Scholar] [CrossRef] [PubMed]
- Bonfili, L.; Cecarini, V.; Berardi, S.; Scarpona, S.; Suchodolski, J.S.; Nasuti, C.; Fiorini, D.; Boarelli, M.C.; Rossi, G.; Eleuteri, A.M. Microbiota Modulation Counteracts Alzheimer’s Disease Progression Influencing Neuronal Proteolysis and Gut Hormones Plasma Levels. Sci. Rep. 2017, 7, 2426. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Ling, Y.; Wang, F.; Gong, T.; Yang, C.; Ye, S.; Ye, K.; Wei, D.; Song, Z.; et al. Fecal Microbiota Transplantation Alleviated Alzheimer’s Disease-like Pathogenesis in APP/PS1 Transgenic Mice. Transl. Psychiatry 2019, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Amakye, W.K.; Guo, L.; Gong, C.; Zhao, Y.; Yao, M.; Ren, J. Walnut-Derived Peptide PW5 Ameliorates Cognitive Impairments and Alters Gut Microbiota in APP/PS1 Transgenic Mice. Mol. Nutr. Food Res. 2019, 63, 1900326. [Google Scholar] [CrossRef]
- Tran, T.T.T.; Corsini, S.; Kellingray, L.; Hegarty, C.; Le Gall, G.; Narbad, A.; Müller, M.; Tejera, N.; O’Toole, P.W.; Minihane, A.-M.; et al. APOE Genotype Influences the Gut Microbiome Structure and Function in Humans and Mice: Relevance for Alzheimer’s Disease Pathophysiology. FASEB J. 2019, 33, 8221–8231. [Google Scholar] [CrossRef]
- Zhan, J.; Liang, Y.; Liu, D.; Ma, X.; Li, P.; Liu, C.; Liu, X.; Wang, P.; Zhou, Z. Antibiotics May Increase Triazine Herbicide Exposure Risk via Disturbing Gut Microbiota. Microbiome 2018, 6, 224. [Google Scholar] [CrossRef]
- Brandscheid, C.; Schuck, F.; Reinhardt, S.; Schäfer, K.-H.; Pietrzik, C.U.; Grimm, M.; Hartmann, T.; Schwiertz, A.; Endres, K. Altered Gut Microbiome Composition and Tryptic Activity of the 5xFAD Alzheimer’s Mouse Model. J. Alzheimers Dis. 2017, 56, 775–788. [Google Scholar] [CrossRef]
- Maldonado Weng, J.; Parikh, I.; Naqib, A.; York, J.; Green, S.J.; Estus, S.; LaDu, M.J. Synergistic Effects of APOE and Sex on the Gut Microbiome of Young EFAD Transgenic Mice. Mol. Neurodegener. 2019, 14, 47. [Google Scholar] [CrossRef]
- Hoffman, J.D.; Yanckello, L.M.; Chlipala, G.; Hammond, T.C.; McCulloch, S.D.; Parikh, I.; Sun, S.; Morganti, J.M.; Green, S.J.; Lin, A.-L. Dietary Inulin Alters the Gut Microbiome, Enhances Systemic Metabolism and Reduces Neuroinflammation in an APOE4 Mouse Model. PLoS ONE 2019, 14, e0221828. [Google Scholar] [CrossRef]
- Lee, H.-J.; Lee, K.-E.; Kim, J.-K.; Kim, D.-H. Suppression of Gut Dysbiosis by Bifidobacterium Longum Alleviates Cognitive Decline in 5XFAD Transgenic and Aged Mice. Sci. Rep. 2019, 9, 11814. [Google Scholar] [CrossRef]
- Shen, L.; Liu, L.; Li, X.-Y.; Ji, H.-F. Regulation of Gut Microbiota in Alzheimer’s Disease Mice by Silibinin and Silymarin and Their Pharmacological Implications. Appl. Microbiol. Biotechnol. 2019, 103, 7141–7149. [Google Scholar] [CrossRef]
- Jackson, A.; Engen, P.A.; Forsyth, C.B.; Shaikh, M.; Naqib, A.; Wilber, S.; Frausto, D.M.; Raeisi, S.; Green, S.J.; Bradaric, B.D.; et al. Intestinal Barrier Dysfunction in the Absence of Systemic Inflammation Fails to Exacerbate Motor Dysfunction and Brain Pathology in a Mouse Model of Parkinson’s Disease. Front. Neurol. 2022, 13, 882628. [Google Scholar] [CrossRef]
- Lai, F.; Jiang, R.; Xie, W.; Liu, X.; Tang, Y.; Xiao, H.; Gao, J.; Jia, Y.; Bai, Q. Intestinal Pathology and Gut Microbiota Alterations in a Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine (MPTP) Mouse Model of Parkinson’s Disease. Neurochem. Res. 2018, 43, 1986–1999. [Google Scholar] [CrossRef]
- Zhou, X.; Lu, J.; Wei, K.; Wei, J.; Tian, P.; Yue, M.; Wang, Y.; Hong, D.; Li, F.; Wang, B.; et al. Neuroprotective Effect of Ceftriaxone on MPTP-Induced Parkinson’s Disease Mouse Model by Regulating Inflammation and Intestinal Microbiota. Oxid. Med. Cell. Longev. 2021, 2021, 9424582. [Google Scholar] [CrossRef]
- Cui, C.; Han, Y.; Li, H.; Yu, H.; Zhang, B.; Li, G. Curcumin-Driven Reprogramming of the Gut Microbiota and Metabolome Ameliorates Motor Deficits and Neuroinflammation in a Mouse Model of Parkinson’s Disease. Front. Cell. Infect. Microbiol. 2022, 12, 887407. [Google Scholar] [CrossRef]
- Liu, M.-M.; Zhou, N.; Jiang, N.; Lu, K.-M.; Wu, C.-F.; Bao, J.-K. Neuroprotective Effects of Oligosaccharides from Periplaneta Americana on Parkinson’s Disease Models In Vitro and In Vivo. Front. Pharmacol. 2022, 13, 936818. [Google Scholar] [CrossRef]
- Wang, N.; Feng, B.-N.; Hu, B.; Cheng, Y.-L.; Guo, Y.-H.; Qian, H. Neuroprotection of Chicoric Acid in a Mouse Model of Parkinson’s Disease Involves Gut Microbiota and TLR4 Signaling Pathway. Food Funct. 2022, 13, 2019–2032. [Google Scholar] [CrossRef]
- Zahra, W.; Birla, H.; Singh, S.S.; Rathore, A.S.; Dilnashin, H.; Singh, R.; Keshri, P.K.; Gautam, P.; Singh, S.P. Neuroprotection by Mucuna Pruriens in Neurodegenerative Diseases. Neurochem. Res. 2022, 47, 1816–1829. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing Dysbiosis of Gut Microbiome in PD: Evidence for Overabundance of Opportunistic Pathogens. npj Parkinsons Dis. 2020, 6, 11. [Google Scholar] [CrossRef]
- Simões, C.D.; Maukonen, J.; Kaprio, J.; Rissanen, A.; Pietiläinen, K.H.; Saarela, M. Habitual Dietary Intake Is Associated with Stool Microbiota Composition in Monozygotic Twins. J. Nutr. 2013, 143, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Yahagi, K.; Mori, H.; Matsumoto, H.; Hara, T.; Tajima, S.; Ogawa, E.; Kodama, H.; Yamamoto, K.; Yamada, T.; et al. A Key Genetic Factor for Fucosyllactose Utilization Affects Infant Gut Microbiota Development. Nat. Commun. 2016, 7, 11939. [Google Scholar] [CrossRef] [PubMed]
- Bowyer, R.C.E.; Jackson, M.A.; Le Roy, C.I.; Ni Lochlainn, M.; Spector, T.D.; Dowd, J.B.; Steves, C.J. Socioeconomic Status and the Gut Microbiome: A TwinsUK Cohort Study. Microorganisms 2019, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Bibbò, S.; Ianiro, G.; Giorgio, V.; Scaldaferri, F.; Masucci, L.; Gasbarrini, A.; Cammarota, G. The Role of Diet on Gut Microbiota Composition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4742–4749. [Google Scholar] [PubMed]
- Chassaing, B.; Vijay-Kumar, M.; Gewirtz, A.T. How Diet Can Impact Gut Microbiota to Promote or Endanger Health. Curr. Opin. Gastroenterol. 2017, 33, 417–421. [Google Scholar] [CrossRef]
- Proctor, C.; Thiennimitr, P.; Chattipakorn, N.; Chattipakorn, S.C. Diet, Gut Microbiota and Cognition. Metab. Brain Dis. 2017, 32, 1–17. [Google Scholar] [CrossRef]
- Sandhu, K.V.; Sherwin, E.; Schellekens, H.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Feeding the Microbiota-Gut-Brain Axis: Diet, Microbiome, and Neuropsychiatry. Transl. Res. 2017, 179, 223–244. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Temko, J.E.; Bouhlal, S.; Farokhnia, M.; Lee, M.R.; Cryan, J.F.; Leggio, L. The Microbiota, the Gut and the Brain in Eating and Alcohol Use Disorders: A “Ménage à Trois”? Alcohol Alcohol. 2017, 52, 403–413. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, X.; Huang, L.; Shen, R.; Qin, H. Gut Microbiota Alteration After Long-Term Consumption of Probiotics in the Elderly. Probiotics Antimicrob. Proteins 2019, 11, 655–666. [Google Scholar] [CrossRef]
- Kim, J.E.; Park, J.J.; Lee, M.R.; Choi, J.Y.; Song, B.R.; Park, J.W.; Kang, M.J.; Son, H.J.; Hong, J.T.; Hwang, D.Y. Constipation in Tg2576 Mice Model for Alzheimer’s Disease Associated with Dysregulation of Mechanism Involving the MAChR Signaling Pathway and ER Stress Response. PLoS ONE 2019, 14, e0215205. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, Gut Microbiota, and Their Influence on Host Health and Disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef]
- Roe, K. An Alternative Explanation for Alzheimer’s Disease and Parkinson’s Disease Initiation from Specific Antibiotics, Gut Microbiota Dysbiosis and Neurotoxins. Neurochem. Res. 2022, 47, 517–530. [Google Scholar] [CrossRef]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline Human Gut Microbiota Profile in Healthy People and Standard Reporting Template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef]
- Xiang, L.; Lou, Y.; Liu, L.; Liu, Y.; Zhang, W.; Deng, J.; Guan, Y.; She, M.; You, X.; Liu, M.; et al. Gut Microbiotic Features Aiding the Diagnosis of Acute Ischemic Stroke. Front. Cell. Infect. Microbiol. 2020, 10, 587284. [Google Scholar] [CrossRef]
- Ling, Y.; Gong, T.; Zhang, J.; Gu, Q.; Gao, X.; Weng, X.; Liu, J.; Sun, J. Gut Microbiome Signatures Are Biomarkers for Cognitive Impairment in Patients with Ischemic Stroke. Front. Aging Neurosci. 2020, 12, 511562. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonnechère, B.; Amin, N.; van Duijn, C. What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review. Int. J. Mol. Sci. 2022, 23, 13665. https://doi.org/10.3390/ijms232213665
Bonnechère B, Amin N, van Duijn C. What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review. International Journal of Molecular Sciences. 2022; 23(22):13665. https://doi.org/10.3390/ijms232213665
Chicago/Turabian StyleBonnechère, Bruno, Najaf Amin, and Cornelia van Duijn. 2022. "What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review" International Journal of Molecular Sciences 23, no. 22: 13665. https://doi.org/10.3390/ijms232213665
APA StyleBonnechère, B., Amin, N., & van Duijn, C. (2022). What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review. International Journal of Molecular Sciences, 23(22), 13665. https://doi.org/10.3390/ijms232213665







