In Silico Discovery of Anticancer Peptides from Sanghuang
Abstract
:1. Introduction
2. Results
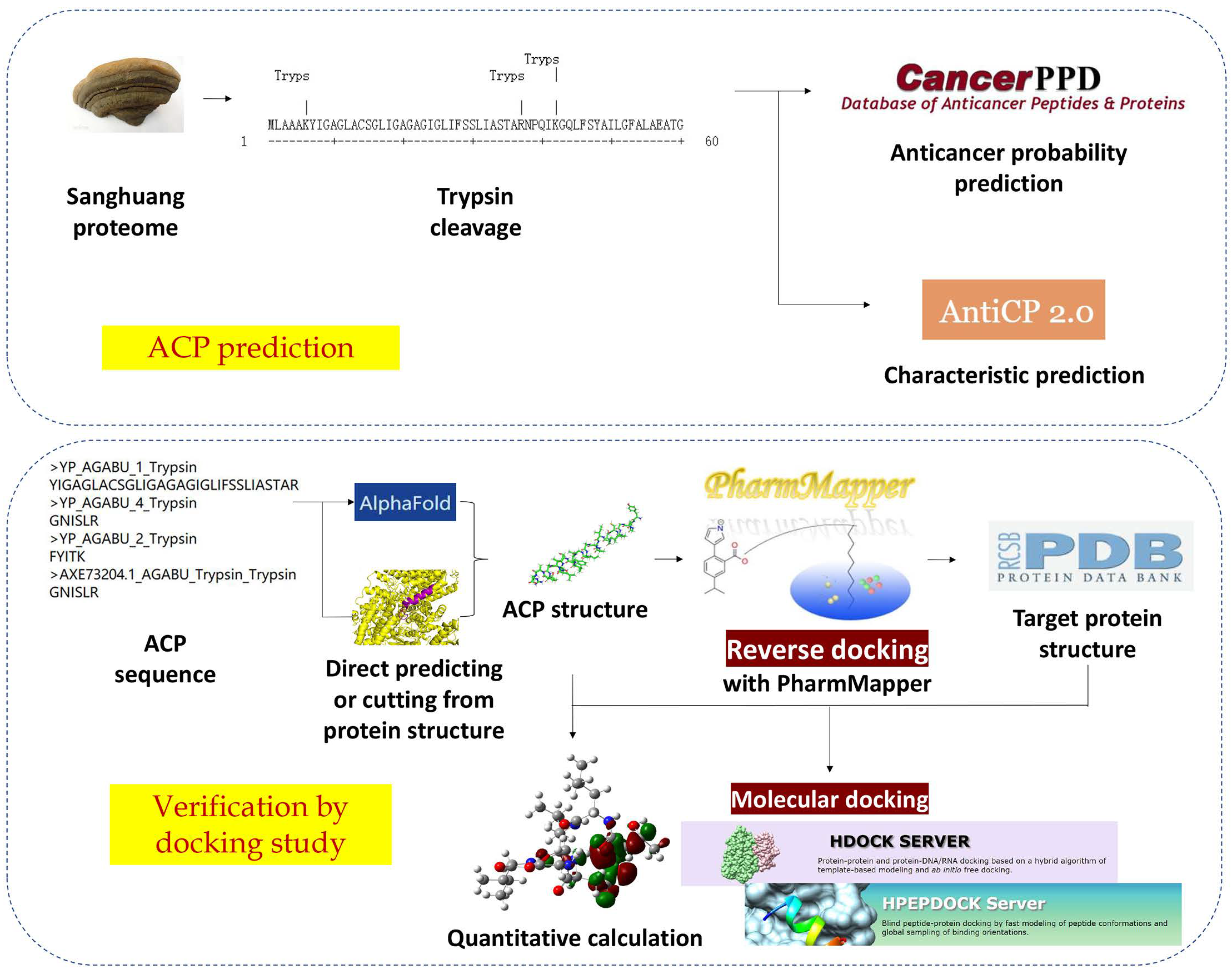
2.1. ACP Screening
2.1.1. Prediction and Screening of Anticancer Peptides of Sanghuang Proteins
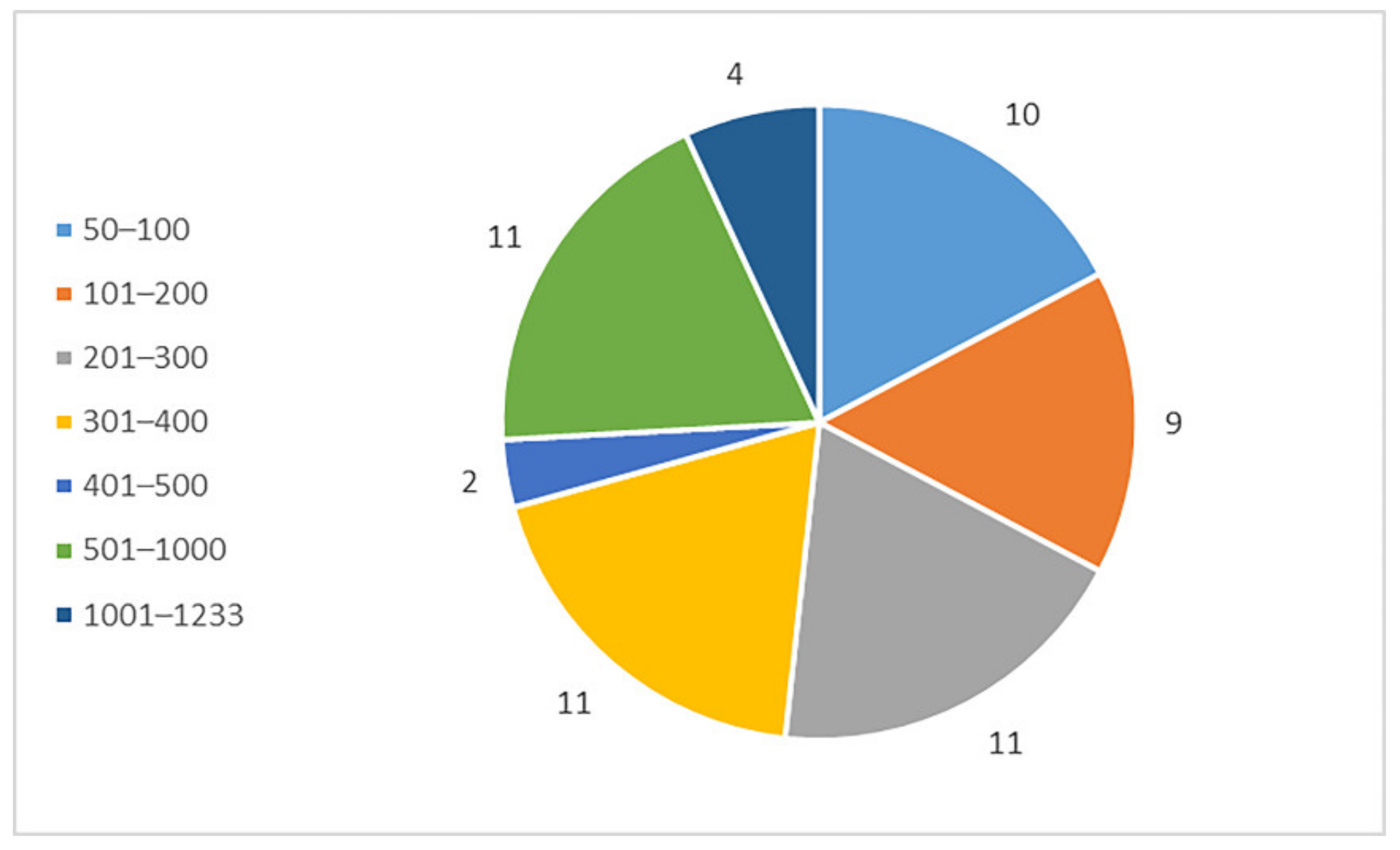
2.1.2. In Silico Digestion and Feature Prediction of Cleaved Fragments
2.2. Docking Study
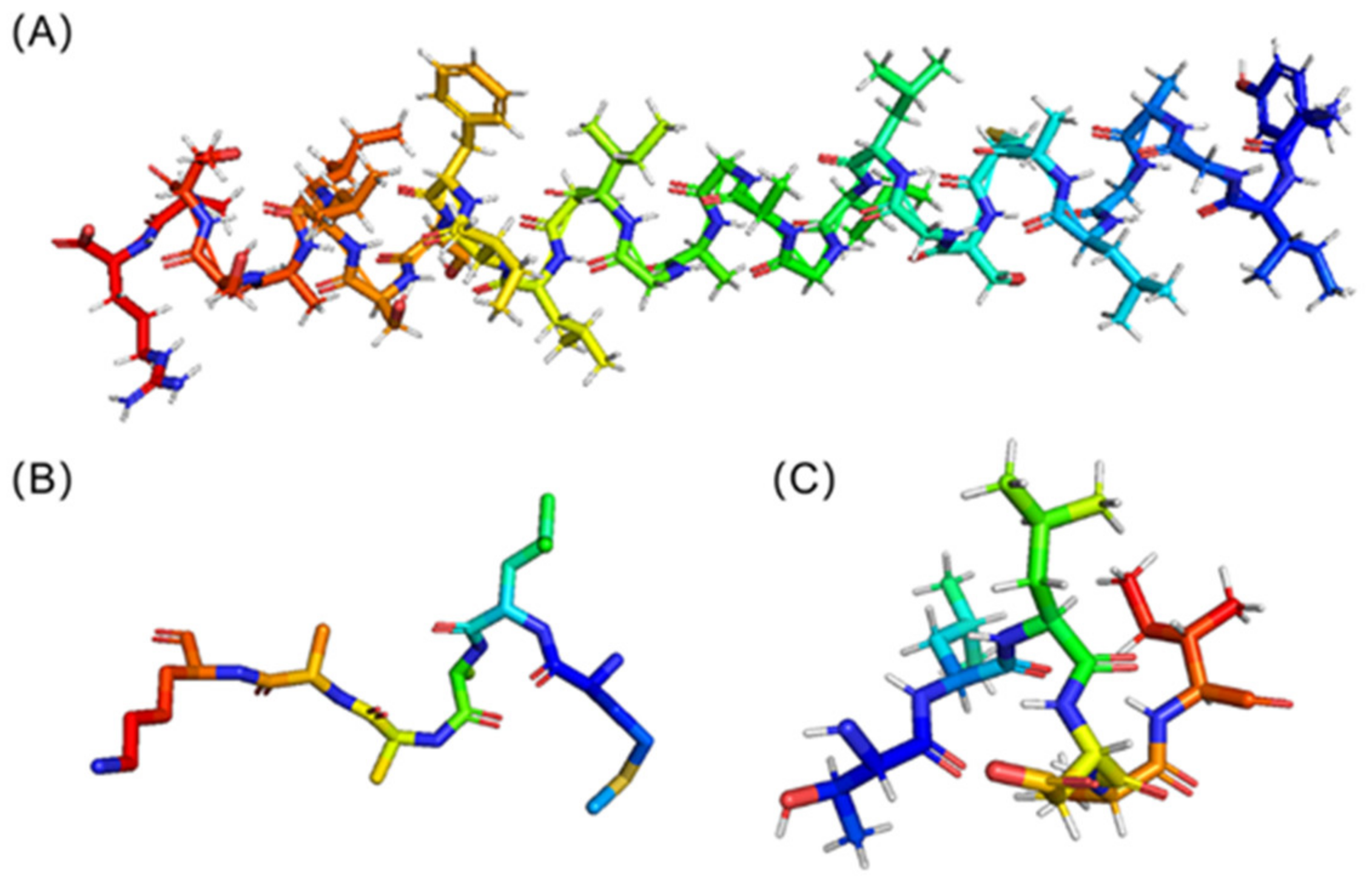
2.2.1. ACP Structure Prediction and Receptor Structure Obtained Based on Reverse Docking
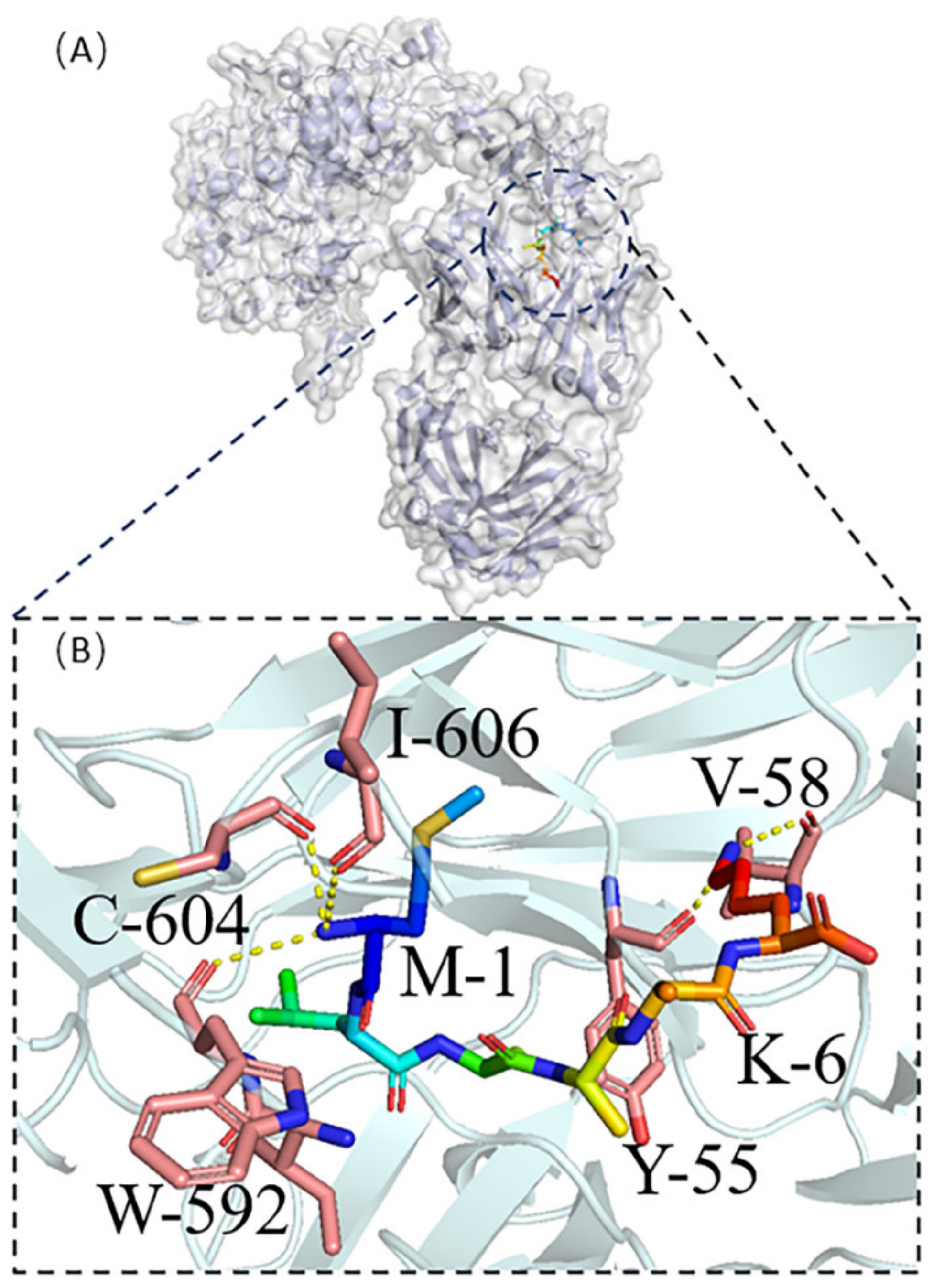
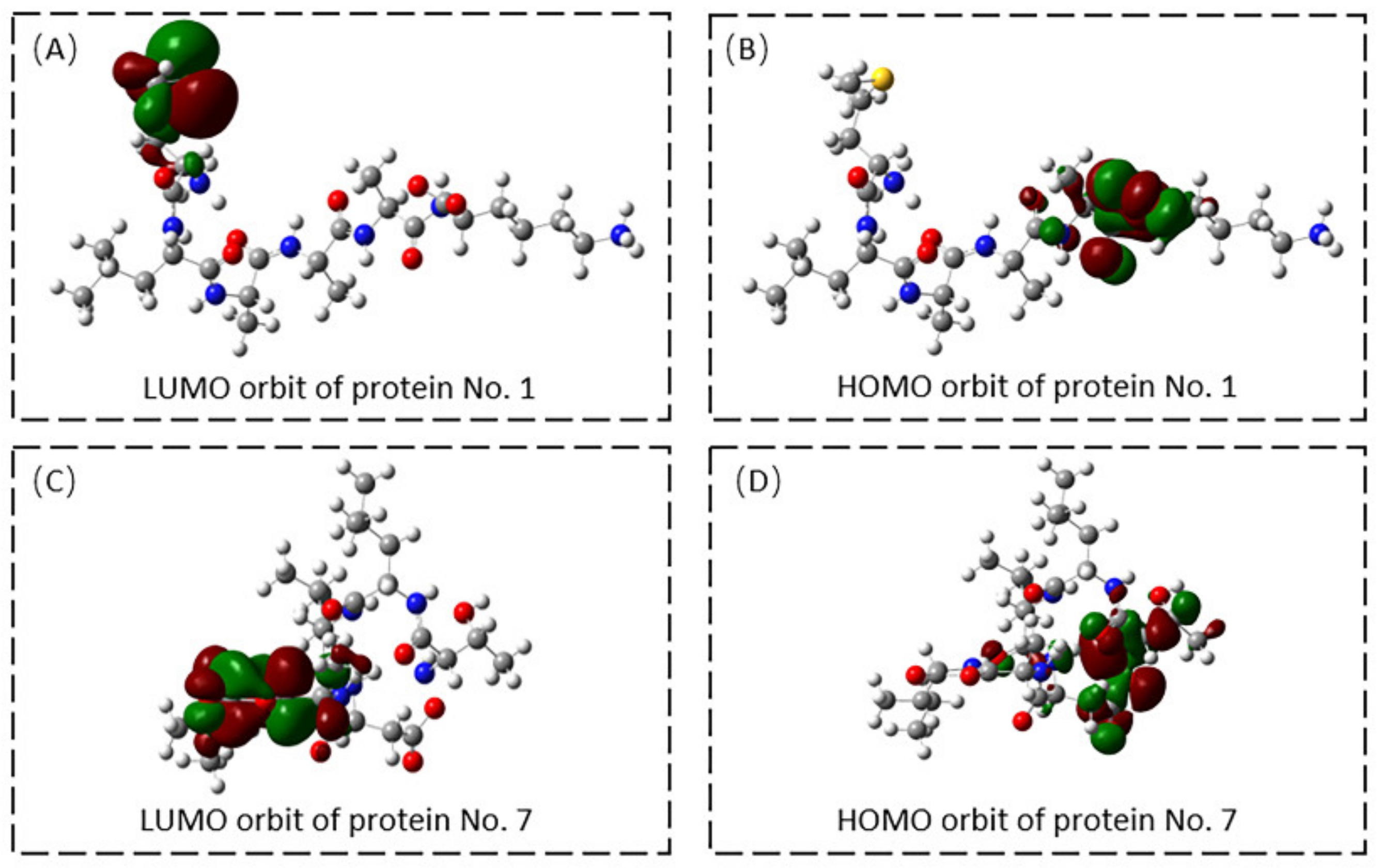
2.2.2. Molecular Docking of Receptors and Target Proteins and Quantum Chemical Calculation
3. Discussion
4. Materials and Methods
4.1. Anticancer Possibility Prediction of Peptides
4.2. In Silico Hydrolysis of the Parent Protein
4.3. Characteristic Prediction of Anticancer Peptide
4.4. Anticancer Peptide Database
4.5. Anticancer Peptide Structure Prediction
4.6. Reverse Docking
4.7. Quantitative Calculation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, L.-W.; Ghobad-Nejhad, M.; Tian, X.-M.; Wang, Y.-F.; Wu, F. Current Status of ‘Sanghuang’ as a Group of Medicinal Mushrooms and Their Perspective in Industry Development. Food Rev. Int. 2022, 38, 589–607. [Google Scholar] [CrossRef]
- He, P.-Y.; Hou, Y.-H.; Yang, Y.; Li, N. The anticancer effect of extract of medicinal mushroom Sanghuangprous vaninii against human cervical cancer cell via endoplasmic reticulum stress-mitochondrial apoptotic pathway. J. Ethnopharmacol. 2021, 279, 114345. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-K.; She, X.; Peterson, E.C.; Wang, Y.Z.; Zheng, P.; Ma, H.; Zhang, K.; Liang, J. A newly characterized exopolysaccharide from Sanghuangporus sanghuang. J. Microbiol. 2019, 57, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Zhou, L.; Fu, Y.; Wang, T.; Li, Z.; Zhou, L.; Zhang, G.; Tian, X. Chemical characterization of two fractions from Sanghuangporus sanghuang and evaluation of antidiabetic activity. J. Funct. Foods 2021, 87, 104825. [Google Scholar] [CrossRef]
- Ikekawa, T.; Nakanishi, M.; Uehara, N.; Chihara, G.; Fukuoka, F. Antitumor action of some basidiomycetes, especially Phellinus linteus. GANN Jpn. J. Cancer Res. 1968, 59, 155–157. [Google Scholar] [CrossRef]
- Plumb, J.A.; Venugopal, B.; Oun, R.; Gomez-Roman, N.; Kawazoe, Y.; Venkataramanan, N.S.; Wheate, N.J. Cucurbit[7]uril encapsulated cisplatin overcomes cisplatin resistance via a pharmacokinetic effect. Metallomics 2012, 4, 561–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabernet, G.; Müller, A.T.; Hiss, J.A.; Schneider, G. Membranolytic anticancer peptides. MedChemComm 2016, 7, 2232–2245. [Google Scholar] [CrossRef]
- Thankappan, B.; Sivakumar, J.; Asokan, S.; Ramasamy, M.; Pillai, M.M.; Selvakumar, R.; Angayarkanni, J. Dual antimicrobial and anticancer activity of a novel synthetic α-helical antimicrobial peptide. Eur. J. Pharm. Sci. 2021, 161, 105784. [Google Scholar] [CrossRef]
- Xie, M.; Liu, D.; Yang, Y. Anti-cancer peptides: Classification, mechanism of action, reconstruction and modification. Open Biol. 2020, 10, 200004. [Google Scholar] [CrossRef]
- Zheng, S.; Zhu, N.; Shi, C.; Zheng, H. Genomic data mining approaches for the discovery of anticancer peptides from Ganoderma sinense. Phytochemistry 2020, 179, 112466. [Google Scholar] [CrossRef]
- Wu, X.; Zeng, W.; Lin, F.; Xu, P.; Li, X. Anticancer Peptide Prediction via Multi-Kernel CNN and Attention Model. Front. Genet. 2022, 13, 887894. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.-C. mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; He, H.; Zhang, T.; Jiang, J. Application of Reverse Docking in the Research of Small Molecule Drugs and Traditional Chinese Medicine. Biol. Pharm. Bull. 2022, 45, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Hassabis, D. Protein structure predictions to atomic accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Li, N.; Duan, Z.; Wang, L.; Guo, C.; Zhang, H.; Gu, Z.; Gong, Q.; Luo, K. An Amphiphilic PEGylated Peptide Dendron-Gemcitabine Prodrug-Based Nanoagent for Cancer Therapy. Macromol. Rapid Commun. 2021, 42, 2100111. [Google Scholar] [CrossRef]
- Ortuso, F.; Paduano, F.; Carotenuto, A.; Gomez-Monterrey, I.; Bilotta, A.; Gaudio, E.; Sala, M.; Artese, A.; Vernieri, E.; Dattilo, V.; et al. Discovery of PTPRJ Agonist Peptides That Effectively Inhibit in Vitro Cancer Cell Proliferation and Tube Formation. ACS Chem. Biol. 2013, 8, 1497–1506. [Google Scholar] [CrossRef]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.A.; Müller, S.; Knapp, S. Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.-S.; Mason, K.; Ramyar, K.X.; Stanley, A.M.; Gabelli, S.B.; Denney, D.W., Jr.; Leahy, D.J. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003, 421, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Siwek, W.; Czapinska, H.; Bochtler, M.; Bujnicki, J.M.; Skowronek, K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 2012, 40, 7563–7572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malashkevich, V.N.; Strokopytov, B.V.; Borisov, V.V.; Dauter, Z.; Wilson, K.S.; Torchinsky, Y.M. Crystal Structure of the Closed Form of Chicken Cystosolic Aspartate Aminotransferase at 1.9 Å Resolution. J. Mol. Biol. 1995, 247, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jogl, G.; Rozovsky, S.; McDermott, A.E.; Tong, L. Optimal alignment for enzymatic proton transfer: Structure of the Michaelis complex of triosephosphate isomerase at 1.2-Å resolution. Proc. Natl. Acad. Sci. USA 2003, 100, 50–55. [Google Scholar] [CrossRef] [Green Version]
- Shamoo, Y.; A Steitz, T. Building a Replisome from Interacting Pieces: Sliding Clamp Complexed to a Peptide from DNA Polymerase and a Polymerase Editing Complex. Cell 1999, 99, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Mylona, A.; Fernández-Tornero, C.; Legrand, P.; Haupt, M.; Sentenac, A.; Acker, J.; Müller, C.W. Structure of the τ60/Δτ91 Subcomplex of Yeast Transcription Factor IIIC: Insights into Preinitiation Complex Assembly. Mol. Cell 2006, 24, 221–232. [Google Scholar] [CrossRef]
- Goihberg, E.; Dym, O.; Tel-Or, S.; Shimon, L.; Frolow, F.; Peretz, M.; Burstein, Y. Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution. Proteins Struct. Funct. Bioinform. 2008, 72, 711–719. [Google Scholar] [CrossRef]
- Harms, J.M.; Wilson, D.N.; Schluenzen, F.; Connell, S.R.; Stachelhaus, T.; Zaborowska, Z.; Spahn, C.M.; Fucini, P. Translational Regulation via L11: Molecular Switches on the Ribosome Turned On and Off by Thiostrepton and Micrococcin. Mol. Cell 2008, 30, 26–38. [Google Scholar] [CrossRef]
- Lingyun, H.; Ailing, L.; Yali, L.; Yanqin, Y.; Jing, N. Expression of CUE domain containing 2 protein in serous ovarian cancer tissue: Predicting disease-free and overall survival of patients. J. Int. Med. Res. 2020, 48, 0300060520954770. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Heightman, T.D.; Vasella, A.; McCarter, J.D.; Mackenzie, L.; Withers, A.S.G.; Matthews, B.W. A Structural View of the Action of Escherichia coli (lacZ) β-Galactosidase. Biochemistry 2001, 40, 14781–14794. [Google Scholar] [CrossRef] [PubMed]
- Shima, S.; Krueger, M.; Weinert, T.; Demmer, U.; Kahnt, J.; Thauer, R.K.; Ermler, U. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 2012, 481, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, W.; Wu, J.; Shi, Y. Solution Structure of the First HMG Box Domain in Human Upstream Binding Factor. Biochemistry 2002, 41, 5415–5420. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.K.; Cherney, M.M.; Vázquez, A.L.; Machı́n, A.; Alonso, J.M.M.; Parra, F.; James, M.N.G. Crystal Structures of Active and Inactive Conformations of a Caliciviral RNA-dependent RNA Polymerase. J. Biol. Chem. 2002, 277, 1381–1387. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Kamtekar, S.; Xiong, Y.; Sarkis, G.J.; Grindley, N.D.F.; Steitz, T.A. Structure of a Synaptic γδ Resolvase Tetramer Covalently Linked to Two Cleaved DNAs. Science 2005, 309, 1210–1215. [Google Scholar] [CrossRef]
- Chen, R.; Neill, J.D.; Estes, M.K.; Prasad, B.V.V. X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 8048–8053. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, S.; Clarke Miller, M.; Maliekal, J.; Bates, P.J.; Trent, J.O.; Lane, A.N. Solution structure of the RBD1,2 domains from human nucleolin. J. Biomol. NMR 2010, 47, 79–83. [Google Scholar] [CrossRef]
- Radaev, S.; Rostro, B.; Brooks, A.G.; Colonna, M.; Sun, P.D. Conformational Plasticity Revealed by the Cocrystal Structure of NKG2D and Its Class I MHC-like Ligand ULBP3. Immunity 2001, 15, 1039–1049. [Google Scholar] [CrossRef] [Green Version]
- Ren, B.; Kühn, J.; Meslet-Cladiere, L.; Briffotaux, J.; Norais, C.; Lavigne, R.; Flament, D.; Ladenstein, R.; Myllykallio, H. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 2009, 28, 2479–2489. [Google Scholar] [CrossRef]
- Gangwer, K.A.; Mushrush, D.J.; Stauff, D.L.; Spiller, B.; McClain, M.S.; Cover, T.L.; Lacy, D.B. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl. Acad. Sci. USA 2007, 104, 16293–16298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Kaur, H.; Kehinde, B.A.; Chhikara, N.; Sharma, D.; Panghal, A. Food-Derived Anticancer Peptides: A Review. Int. J. Pept. Res. Ther. 2021, 27, 55–70. [Google Scholar] [CrossRef]
- Cheng, J.; Song, J.; Wang, Y.; Wei, H.; He, L.; Liu, Y.; Ding, H.; Huang, Q.; Hu, C.; Huang, X.; et al. Conformation and anticancer activity of a novel mannogalactan from the fruiting bodies of Sanghuangporus sanghuang on HepG2 cells. Food Res. Int. 2022, 156, 111336. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharmacal Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Scotti, L.; Scotti, M.T. Recent Advancement in Computer-Aided Drug Design. Curr. Pharm. Des. 2020, 26, 1635–1636. [Google Scholar] [CrossRef]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef]
- Baig, M.H.; Ahmad, K.; Roy, S.; Ashraf, J.M.; Adil, M.; Siddiqui, M.H.; Khan, S.; Kamal, M.A.; Provazník, I.; Choi, I. Computer Aided Drug Design: Success and Limitations. Curr. Pharm. Des. 2016, 22, 572–581. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Li, B.; Lu, C.; Yang, S.; Long, J.; Chen, H.; Huang, J.; He, B. A database of anti-coronavirus peptides. Sci. Data 2022, 9, 1–9. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Han, W.; Zhu, J.; Wang, S.; Xu, D. Understanding the Phosphorylation Mechanism by Using Quantum Chemical Calculations and Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 3565–3573. [Google Scholar] [CrossRef] [PubMed]


| No. | Sequence | Possibility | Charge | Hydrophobicity | Hydropathicity |
|---|---|---|---|---|---|
| 1 | MLAAAK | 0.98 | 1.0 | 0.07 | 1.20 |
| 2 | YIGAGLACSGLIGAGAGIGLIFSSLIASTAR | 0.85 | 1.0 | 0.20 | 1.33 |
| 3 | GNISLRS | 0.80 | 1.0 | −0.21 | −0.24 |
| 4 | MDTTK | 0.83 | 0.0 | −0.38 | −1.38 |
| 5 | VGYNPK | 0.72 | 1.0 | −0.18 | −1.08 |
| 6 | AGVVK | 0.95 | 1.0 | 0.08 | 1.18 |
| 7 | TLLDAI | 0.61 | −1.0 | 0.19 | 1.62 |
| 8 | FYITK | 0.97 | 1.0 | 0.02 | 0.28 |
| 9 | HALLAFTLGVR | 0.75 | 1.5 | 0.10 | 1.20 |
| Sequence | Hit Sequence | Score | E Value |
|---|---|---|---|
| MLAAAK | GLLPCAESCVYIPCLTTVIGCSCKSKVCYKN | 14 | 29 |
| TLLDAI | MDSNKDERAYAQWVIIILHNVGSSPFKIANLGLSWGKLYADGNKDKEVYP | 16 | 5 |
| SDYNGKTVGPDEKIQINSCGRENASSGTEGSFDIVDPNDGNKTIRHFYW | |||
| ECPWGSKRNTWTPSGSNTKWMVEWSGQNLDSGALGTITVDVLRKGN | |||
| HALLAFTLGVR | KKWβ2,2WKK | 13 | 110 |
| YIGAGLACSGLIGAGAGIGLIFSSLIASTAR | CHHNLTAAC | 18 | 3.3 |
| Rank | PDB ID | Target Name | Organism(s) | Function or Related Diseases | Fit Score | Normalized Fit Scores |
|---|---|---|---|---|---|---|
| 1 | 4ESJ | NONE | Streptococcus pneumoniae TIGR4 | DNA methylation-dependent restriction enzymes [24] | 2.99 | 0.75 |
| 2 | 2CST | Aspartate aminotransferase, cytoplasmic | Gallus | related to liver function [25] | 2.98 | 0.75 |
| 3 | 1NF0 | Triosephosphate isomerase | Saccharomyces cerevisiae | glycolysis [26] | 2.98 | 0.74 |
| 4 | 1B8H | DNA polymerase processivity component | Escherichia phage RB69 | DNA replication [27] | 2.98 | 0.74 |
| 5 | 2J04 | Transcription factor tau 60 kDa subunit | Saccharomyces cerevisiae | transcription [28] | 2.97 | 0.74 |
| 6 | 2NVB | NADP-dependent alcohol dehydrogenase | Thermoanaerobacter brockii | ethanol metabolism [29] | 2.96 | 0.74 |
| 7 | 2ZJP | 50S ribosomal protein L33 | Deinococcus radiodurans, Streptomyces actuosus | translational regulation [30] | 2.95 | 0.74 |
| 8 | 1N8Z | Receptor tyrosine-protein kinase erbB-2 | Mus musculus, Homo sapiens | breast cancer [23] | 2.95 | 0.74 |
| 9 | 2DHY | CUE domain-containing protein 1 | Homo sapiens | ovarian cancer [31] | 2.95 | 0.74 |
| 10 | 1JZ2 | Ribonuclease HI | Escherichia coli | β-galactosidase reaction [32] | 2.94 | 0.74 |
| Rank | PDB ID | Target Name | Organism(s) | Field of Application or Related Diseases | Fit Score | Normalized Fit Scores |
|---|---|---|---|---|---|---|
| 1 | 3SQG | NONE | Uncultured archaeon | anaerobic oxidation of methane with sulphate [33] | 7.07 | 0.79 |
| 2 | 1K99 | Nucleolar transcription factor 1 | Homo sapiens | transcription [34] | 2.99 | 0.75 |
| 3 | 1KHV | Genome polyprotein | Rabbit hemorrhagic disease virus | rabbit hemorrhagic disease [35] | 2.98 | 0.74 |
| 4 | 1N8Z | Receptor tyrosine-protein kinase erbB-2 | Mus musculus, Homo sapiens | breast cancer [23] | 2.98 | 0.74 |
| 5 | 1ZR4 | Transposon gamma-delta resolvase | Escherichia coli | site-specific recombination [36] | 2.96 | 0.74 |
| 6 | 2GH8 | Capsid protein | San Miguel sea lion virus 4 | a native calicivirus [37] | 2.95 | 0.74 |
| 7 | 2YRV | AT-rich interactive domain-containing protein 4A | Homo sapiens | retinoblastoma [38] | 2.95 | 0.74 |
| 8 | 1KCG | NKG2D ligand 3 | Homo sapiens | trigger the NK cell lysis of various tumor and virally infected cells [39] | 2.94 | 0.74 |
| 9 | 2VLD | UPF0286 protein PYRAB01260 | Pyrococcus abyssi | endonuclease [40] | 2.94 | 0.74 |
| 10 | 2QV3 | Vacuolating cytotoxin | Helicobacter pylori | candidate antigens for Helicobacter pylori vaccines [41] | 2.94 | 0.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Lv, J.; Chen, L.; Li, W.; Han, W. In Silico Discovery of Anticancer Peptides from Sanghuang. Int. J. Mol. Sci. 2022, 23, 13682. https://doi.org/10.3390/ijms232213682
Liu M, Lv J, Chen L, Li W, Han W. In Silico Discovery of Anticancer Peptides from Sanghuang. International Journal of Molecular Sciences. 2022; 23(22):13682. https://doi.org/10.3390/ijms232213682
Chicago/Turabian StyleLiu, Minghao, Jiachen Lv, Liyuan Chen, Wannan Li, and Weiwei Han. 2022. "In Silico Discovery of Anticancer Peptides from Sanghuang" International Journal of Molecular Sciences 23, no. 22: 13682. https://doi.org/10.3390/ijms232213682





