Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review
Abstract
1. Introduction
2. Resveratrol
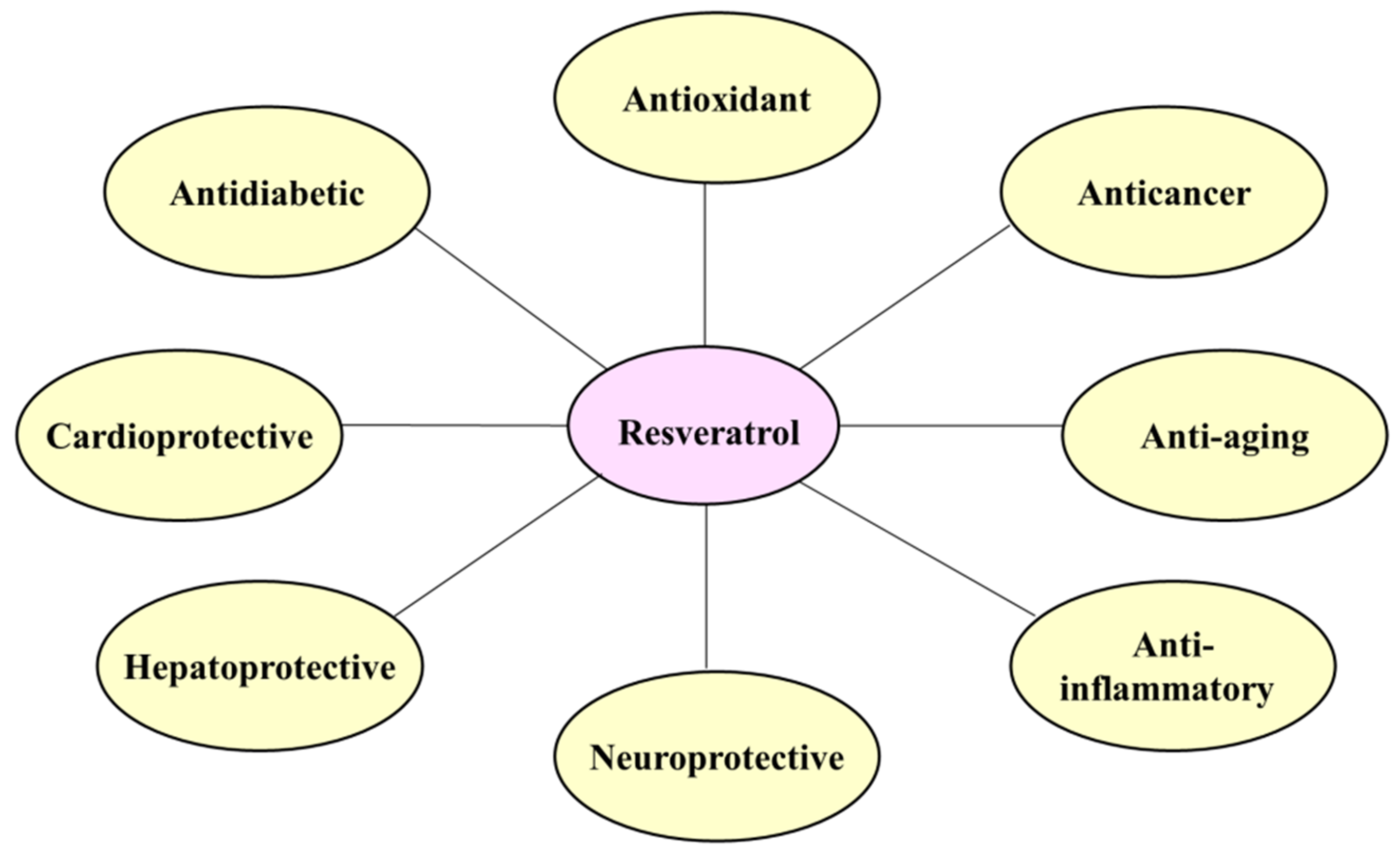
3. Physiological Functions of Resveratrol
4. Resveratrol in Cancer Therapy
5. Effect of Resveratrol on Apoptosis
5.1. Apoptosis
5.2. Types of Apoptosis
5.2.1. Intrinsic (Mitochondrial) Pathway
5.2.2. Extrinsic (Death Receptor) Pathway
5.3. Induction of Apoptosis by Resveratrol
5.3.1. Effect of Resveratrol on Tumor Suppressor p53
5.3.2. Effect of Resveratrol on AKT
5.3.3. Effect of Resveratrol on SIRT
6. Effect of Resveratrol on Autophagy
6.1. Autophagy
6.2. Types of Autophagy
6.3. Induction of Autophagy by Resveratrol
6.3.1. Effect of Resveratrol on AMPK
6.3.2. Effect of Resveratrol on p62
7. Induction of Necroptosis by Resveratrol
8. Cell Death Mechanism of Synthetic Resveratrol Derivatives and Analogues in Cancer
8.1. Programmed Cell Death Induced by Synthetic Resveratrol Derivatives
8.2. Programmed Cell Death Induced by Synthetic Resveratrol Analogues
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Song, W.; Li, D.; Cai, L.; Zhao, Y. Resveratrol as a natural regulator of autophagy for prevention and treatment of cancer. Onco Targets Ther. 2019, 12, 8601–8609. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; Alsahli, M.A.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Babiker, A.Y.; Khan, A.A.; Rahmani, A.H. Potential therapeutic targets of resveratrol, a plant polyphenol, and its role in the therapy of various types of cancer. Molecules 2022, 27, 2665. [Google Scholar] [CrossRef] [PubMed]
- Figueiró, F.; Bernardi, A.; Frozza, R.L.; Terroso, T.; Zanotto-Filho, A.; Jandrey, E.H.; Moreira, J.C.; Salbego, C.G.; Edelweiss, M.I.; Pohlmann, A.R.; et al. Resveratrol-loaded lipid-core nanocapsules treatment reduces in vitro and in vivo glioma growth. J. Biomed. Nanotechnol. 2013, 9, 516–526. [Google Scholar] [CrossRef]
- Hu, Z.; Liang, W.; Yang, Y.; Keefe, D.; Ma, Y.; Zhao, Y.; Xue, C.; Huang, Y.; Zhao, H.; Chen, L.; et al. Personalized estimate of chemotherapy-induced nausea and vomiting: Development and external validation of a nomogram in cancer patients receiving highly/moderately emetogenic chemotherapy. Medicine 2016, 95, e2476. [Google Scholar] [CrossRef]
- Ko, J.H.; Sethi, G.; Um, J.Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The role of resveratrol in cancer therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef]
- Elshaer, M.; Chen, Y.; Wang, X.J.; Tang, X. Resveratrol: An overview of its anti-cancer mechanisms. Life Sci. 2018, 207, 340–349. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Taeb, S.; Haghi-Aminjan, H.; Afrashi, S.; Moloudi, K.; Musa, A.E.; Najafi, M.; Farhood, B. Resveratrol as an enhancer of apoptosis in cancer: A mechanistic review. Anticancer Agents Med. Chem. 2021, 21, 2327–2336. [Google Scholar] [CrossRef]
- Takaoka, M. Resveratrol, a new phenolic compound, from Veratrum grandiflorum. Nippon Kagaku Kaishi 1939, 60, 1090–1100. [Google Scholar] [CrossRef]
- Guthrie, A.R.; Chow, H.S.; Martinez, J.A. Effects of resveratrol on drug- and carcinogen-metabolizing enzymes, implications for cancer prevention. Pharmacol. Res. Perspect. 2017, 5, e00294. [Google Scholar] [CrossRef]
- Yousef, M.; Vlachogiannis, I.A.; Tsiani, E. Effects of resveratrol against lung cancer: In vitro and in vivo studies. Nutrients 2017, 9, 1231. [Google Scholar] [CrossRef] [PubMed]
- Di, S.; Zhang, Z.; Wang, Y.; Shi, W. Analysis for four isomers of resveratrol in red wine by high performance liquid chromatography. Se Pu 2004, 22, 424–427. [Google Scholar] [PubMed]
- Cvejic, J.M.; Djekic, S.V.; Petrovic, A.V.; Atanackovic, M.T.; Jovic, S.M.; Brceski, I.D.; Gojkovic-Bukarica, L.C. Determination of trans- and cis-resveratrol in Serbian commercial wines. J. Chromatogr. Sci. 2010, 48, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Weiskirchen, S.; Weiskirchen, R. Resveratrol: How much wine do you have to drink to stay healthy? Adv. Nutr. 2016, 7, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Farneti, B.; Masuero, D.; Costa, F.; Magnago, P.; Malnoy, M.; Costa, G.; Vrhovsek, U.; Mattivi, F. Is there room for improving the nutraceutical composition of apple? J. Agric. Food Chem. 2015, 63, 2750–2759. [Google Scholar] [CrossRef]
- Ragab, A.S.; Van Fleet, J.; Jankowski, B.; Park, J.H.; Bobzin, S.C. Detection and quantitation of resveratrol in tomato fruit (Lycopersicon esculentum Mill.). J. Agric. Food Chem. 2006, 54, 7175–7179. [Google Scholar] [CrossRef]
- Sanders, T.H.; McMichael, R.W., Jr.; Hendrix, K.W. Occurrence of resveratrol in edible peanuts. J. Agric. Food Chem. 2000, 48, 1243–1246. [Google Scholar] [CrossRef]
- Chiva-Blanch, G.; Urpi-Sarda, M.; Rotchés-Ribalta, M.; Zamora-Ros, R.; Llorach, R.; Lamuela-Raventós, R.M.; Estruch, R.; Andrés-Lacueva, C. Determination of resveratrol and piceid in beer matrices by solid-phase extraction and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2011, 1218, 698–705. [Google Scholar] [CrossRef]
- Hurst, W.J.; Glinski, J.A.; Miller, K.B.; Apgar, J.; Davey, M.H.; Stuart, D.A. Survey of the trans-resveratrol and trans-piceid content of cocoa-containing and chocolate products. J. Agric. Food Chem. 2008, 56, 8374–8378. [Google Scholar] [CrossRef]
- Burns, J.; Yokota, T.; Ashihara, H.; Lean, M.E.; Crozier, A. Plant foods and herbal sources of resveratrol. J. Agric. Food Chem. 2002, 50, 3337–3340. [Google Scholar] [CrossRef]
- Almeida, T.C.; Guerra, C.C.C.; De Assis, B.L.G.; de Oliveira Aguiar Soares, R.D.; Garcia, C.C.M.; Lima, A.A.; da Silva, G.N. Antiproliferative and toxicogenomic effects of resveratrol in bladder cancer cells with different TP53 status. Environ. Mol. Mutagen. 2019, 60, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, X.; Lv, J.; Sun, H.; Zhou, F. Resveratrol induces p53 in colorectal cancer through SET7/9. Oncol. Lett. 2019, 17, 3783–3789. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Li, L.; Zhou, E.; Li, H.; Wu, S.; Cao, Z. Resveratrol downregulates miR-155-5p to block the malignant behavior of gastric cancer cells. BioMed Res. Int. 2022, 2022, 6968641. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Park, S.; Kim, H.J.; Lee, K.J.; Kim, D.H.; Baek, S.H.; Hong, S.T. The resveratrol rice DJ526 callus significantly increases the lifespan of drosophila (resveratrol rice DJ526 callus for longevity). Nutrients 2019, 11, 983. [Google Scholar] [CrossRef]
- de la Lastra, C.A.; Villegas, I. Resveratrol as an anti-inflammatory and anti-aging agent: Mechanisms and clinical implications. Mol. Nutr. Food Res. 2005, 49, 405–430. [Google Scholar] [CrossRef]
- Tresguerres, I.F.; Tamimi, F.; Eimar, H.; Barralet, J.; Torres, J.; Blanco, L.; Tresguerres, J.A. Resveratrol as anti-aging therapy for age-related bone loss. Rejuvenation Res. 2014, 17, 439–445. [Google Scholar] [CrossRef]
- Meng, T.; Xiao, D.; Muhammed, A.; Deng, J.; Chen, L.; He, J. Anti-inflammatory action and mechanisms of resveratrol. Molecules 2021, 26, 229. [Google Scholar] [CrossRef]
- de Sá Coutinho, D.; Pacheco, M.T.; Frozza, R.L.; Bernardi, A. Anti-inflammatory effects of resveratrol: Mechanistic insights. Int. J. Mol. Sci. 2018, 19, 1812. [Google Scholar] [CrossRef]
- Gomes, B.A.Q.; Silva, J.P.B.; Romeiro, C.F.R.; Dos Santos, S.M.; Rodrigues, C.A.; Gonçalves, P.R.; Sakai, J.T.; Mendes, P.F.S.; Varela, E.L.P.; Monteiro, M.C. Neuroprotective mechanisms of resveratrol in alzheimer’s disease: Role of SIRT1. Oxidative Med. Cell. Longev. 2018, 2018, 8152373. [Google Scholar] [CrossRef]
- Rao, Y.L.; Ganaraja, B.; Joy, T.; Pai, M.M.; Ullal, S.D.; Murlimanju, B.V. Neuroprotective effects of resveratrol in Alzheimer’s disease. Front. Biosci. 2020, 12, 139–149. [Google Scholar] [CrossRef]
- Bastianetto, S.; Ménard, C.; Quirion, R. Neuroprotective action of resveratrol. Biochim. Biophys. Acta 2015, 1852, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Tejada, S.; Capó, X.; Mascaró, C.M.; Monserrat-Mesquida, M.; Quetglas-Llabrés, M.M.; Pons, A.; Tur, J.A.; Sureda, A. Hepatoprotective effects of resveratrol in non-alcoholic fatty live disease. Curr. Pharm. Des. 2021, 27, 2558–2570. [Google Scholar] [CrossRef] [PubMed]
- Chupradit, S.; Bokov, D.; Zamanian, M.Y.; Heidari, M.; Hakimizadeh, E. Hepatoprotective and therapeutic effects of resveratrol: A focus on anti-inflammatory and antioxidative activities. Fundam. Clin. Pharmacol. 2022, 36, 468–485. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Li, Y.; Cao, L.; Du, J.; Zheng, T.; Qian, H.; Gu, Z.; Jeney, G.; Xu, P.; Yin, G. Antioxidative, anti-inflammatory and hepatoprotective effects of resveratrol on oxidative stress-induced liver damage in tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 215, 56–66. [Google Scholar] [CrossRef]
- Wu, J.M.; Hsieh, T.C. Resveratrol: A cardioprotective substance. Ann. N. Y. Acad. Sci. 2011, 1215, 16–21. [Google Scholar] [CrossRef]
- Salehi, B.; Mishra, A.P.; Nigam, M.; Sener, B.; Kilic, M.; Sharifi-Rad, M.; Fokou, P.V.T.; Martins, N.; Sharifi-Rad, J. Resveratrol: A double-edged sword in health benefits. Biomedicines 2018, 6, 91. [Google Scholar] [CrossRef]
- Kazemirad, H.; Kazerani, H.R. Cardioprotective effects of resveratrol following myocardial ischemia and reperfusion. Mol. Biol. Rep. 2020, 47, 5843–5850. [Google Scholar] [CrossRef]
- Yang, D.K.; Kang, H.S. Anti-diabetic effect of cotreatment with quercetin and resveratrol in streptozotocin-induced diabetic rats. Biomol. Ther. 2018, 26, 130–138. [Google Scholar] [CrossRef]
- Jeong, G.H.; Park, E.K.; Kim, T.H. Anti-diabetic effects of trans-resveratrol byproducts induced by plasma treatment. Food Res. Int. 2019, 119, 119–125. [Google Scholar] [CrossRef]
- Szkudelska, K.; Deniziak, M.; Sassek, M.; Szkudelski, I.; Noskowiak, W.; Szkudelski, T. Resveratrol affects insulin signaling in type 2 diabetic Goto-Kakizaki rats. Int. J. Mol. Sci. 2021, 22, 2469. [Google Scholar] [CrossRef]
- Szkudelska, K.; Okulicz, M.; Hertig, I.; Szkudelski, T. Resveratrol ameliorates inflammatory and oxidative stress in type 2 diabetic Goto-Kakizaki rats. Biomed. Pharmacother. 2020, 125, 110026. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhou, H.; Wang, X.; Mai, K.; He, G. Resveratrol attenuates oxidative stress and inflammatory response in turbot fed with soybean meal based diet. Fish Shellfish Immunol. 2019, 91, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhou, X.; Zeng, X.; Hu, O.; Yi, L.; Mi, M. Resveratrol attenuates oxidative injury in human umbilical vein endothelial cells through regulating mitochondrial fusion via TyrRS-PARP1 pathway. Nutr. Metab. 2019, 16, 9. [Google Scholar] [CrossRef]
- Renaud, S.; de Lorgeril, M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet 1992, 339, 1523–1526. [Google Scholar] [CrossRef]
- Chen, W.M.; Shaw, L.H.; Chang, P.J.; Tung, S.Y.; Chang, T.S.; Shen, C.H.; Hsieh, Y.Y.; Wei, K.L. Hepatoprotective effect of resveratrol against ethanol-induced oxidative stress through induction of superoxide dismutase in vivo and in vitro. Exp. Ther. Med. 2016, 11, 1231–1238. [Google Scholar] [CrossRef]
- Kitada, M.; Kume, S.; Imaizumi, N.; Koya, D. Resveratrol improves oxidative stress and protects against diabetic nephropathy through normalization of Mn-SOD dysfunction in AMPK/SIRT1-independent pathway. Diabetes 2011, 60, 634–643. [Google Scholar] [CrossRef]
- Sharma, S.; Anjaneyulu, M.; Kulkarni, S.K.; Chopra, K. Resveratrol, a polyphenolic phytoalexin, attenuates diabetic nephropathy in rats. Pharmacology 2006, 76, 69–75. [Google Scholar] [CrossRef]
- Gupta, S.C.; Kannappan, R.; Reuter, S.; Kim, J.H.; Aggarwal, B.B. Chemosensitization of tumors by resveratrol. Ann. N. Y. Acad. Sci. 2011, 1215, 150–160. [Google Scholar] [CrossRef]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G.; et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef]
- Galiniak, S.; Aebisher, D.; Bartusik-Aebisher, D. Health benefits of resveratrol administration. Acta Biochim Pol. 2019, 66, 13–21. [Google Scholar] [CrossRef]
- Szende, B.; Tyihák, E.; Király-Véghely, Z. Dose-dependent effect of resveratrol on proliferation and apoptosis in endothelial and tumor cell cultures. Exp. Mol. Med. 2000, 32, 88–92. [Google Scholar] [CrossRef] [PubMed]
- San Hipólito-Luengo, Á.; Alcaide, A.; Ramos-González, M.; Cercas, E.; Vallejo, S.; Romero, A.; Talero, E.; Sánchez-Ferrer, C.F.; Motilva, V.; Peiró, C. Dual effects of resveratrol on cell death and proliferation of colon cancer cells. Nutr. Cancer 2017, 69, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Dudley, J.I.; Das, D.K. Dose-dependency of resveratrol in providing health benefits. Dose Response 2010, 8, 478–500. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.; Chung, H.Y.; Kim, N.D. Role of apigenin in cancer prevention via the induction of apoptosis and autophagy. J. Cancer Prev. 2016, 21, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Sung, B.; Kim, N.D. Role of induced programmed cell death in the chemopreventive potential of apigenin. Int. J. Mol. Sci. 2022, 23, 3757. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Luedde, T. Apoptosis and necroptosis in the liver: A matter of life and death. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 738–752. [Google Scholar] [CrossRef]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef]
- Mishra, A.P.; Salehi, B.; Sharifi-Rad, M.; Pezzani, R.; Kobarfard, F.; Sharifi-Rad, J.; Nigam, M. Programmed cell death, from a cancer perspective: An overview. Mol. Diagn. Ther. 2018, 22, 281–295. [Google Scholar] [CrossRef]
- Danial, N.N.; Korsmeyer, S.J. Cell death: Critical control points. Cell 2004, 116, 205–219. [Google Scholar] [CrossRef]
- Elena-Real, C.A.; Díaz-Quintana, A.; González-Arzola, K.; Velázquez-Campoy, A.; Orzáez, M.; López-Rivas, A.; Gil-Caballero, S.; De la Rosa, M.; Díaz-Moreno, I. Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition. Cell Death Dis. 2018, 9, 365. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in cell death, inflammation, and disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; Gores, G.J. Life and death by death receptors. FASEB J. 2009, 23, 1625–1637. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Boyce, M.; Yuan, J. A decade of caspases. Oncogene 2003, 22, 8543–8567. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ding, J.; Wu, Y. Resveratrol induces apoptosis of bladder cancer cells via miR-21 regulation of the Akt/Bcl-2 signaling pathway. Mol. Med. Rep. 2014, 9, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Mao, Q.Q.; Qin, J.; Zheng, X.Y.; Wang, Y.B.; Yang, K.; Shen, H.F.; Xie, L.P. Resveratrol induces apoptosis and cell cycle arrest of human T24 bladder cancer cells in vitro and inhibits tumor growth in vivo. Cancer Sci. 2010, 101, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.C.; Chen, Y.J.; Huang, K.H.; Kuo, K.L.; Yang, T.H.; Huang, K.Y.; Wang, C.C.; Tang, C.H.; Yang, R.S.; Liu, S.H. Induction of sirtuin-1 signaling by resveratrol induces human chondrosarcoma cell apoptosis and exhibits antitumor activity. Sci. Rep. 2017, 7, 3180. [Google Scholar] [CrossRef]
- Jin, H.; Chen, H.; Yu, K.; Zhang, J.; Li, B.; Cai, N.; Pan, J. Resveratrol inhibits phosphorylation within the signal transduction and activator of transcription 3 signaling pathway by activating sirtuin 1 in SW1353 chondrosarcoma cells. Mol. Med. Rep. 2016, 14, 2685–2690. [Google Scholar] [CrossRef][Green Version]
- Venkatadri, R.; Muni, T.; Iyer, A.K.; Yakisich, J.S.; Azad, N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Dis. 2016, 7, e2104. [Google Scholar] [CrossRef]
- Kumar, B.; Iqbal, M.A.; Singh, R.K.; Bamezai, R.N. Resveratrol inhibits TIGAR to promote ROS induced apoptosis and autophagy. Biochimie 2015, 118, 26–35. [Google Scholar] [CrossRef]
- Gogada, R.; Prabhu, V.; Amadori, M.; Scott, R.; Hashmi, S.; Chandra, D. Resveratrol induces p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated Bax protein oligomerization on mitochondria to initiate cytochrome c release and caspase activation. J. Biol. Chem. 2011, 286, 28749–28760. [Google Scholar] [CrossRef]
- Li, D.; Wang, G.; Jin, G.; Yao, K.; Zhao, Z.; Bie, L.; Guo, Y.; Li, N.; Deng, W.; Chen, X.; et al. Resveratrol suppresses colon cancer growth by targeting the AKT/STAT3 signaling pathway. Int. J. Mol. Med. 2019, 43, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Ye, Y.; Zhu, G.; Xu, Y.; Sun, J.; Wu, H.; Feng, F.; Wen, Z.; Jiang, S.; Li, Y.; et al. Resveratrol induces human colorectal cancer cell apoptosis by activating the mitochondrial pathway via increasing reactive oxygen species. Mol. Med Rep. 2021, 23. [Google Scholar] [CrossRef]
- Yan, C.; Li, F.; Zhang, Y.; Li, Y.; Li, M.; Wang, F.; Zhang, G.; Li, Y.; Li, B.; Zhao, X. Effects of As2O3 and resveratrol on the proliferation and apoptosis of colon cancer cells and the hERG-mediated potential mechanisms. Curr. Pharm. Des. 2019, 25, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Zhong, L.X.; Zhan, Z.Y.; Huang, Z.H.; Xiong, J.P. Resveratrol treatment inhibits proliferation of and induces apoptosis in human colon cancer cells. Med. Sci. Monit. 2016, 22, 1101–1108. [Google Scholar] [CrossRef]
- Buhrmann, C.; Shayan, P.; Popper, B.; Goel, A.; Shakibaei, M. Sirt1 is required for resveratrol-mediated chemopreventive effects in colorectal cancer cells. Nutrients 2016, 8, 145. [Google Scholar] [CrossRef]
- Miki, H.; Uehara, N.; Kimura, A.; Sasaki, T.; Yuri, T.; Yoshizawa, K.; Tsubura, A. Resveratrol induces apoptosis via ROS-triggered autophagy in human colon cancer cells. Int. J. Oncol. 2012, 40, 1020–1028. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; She, G.; Zheng, X.; Shao, L.; Wang, P.; Pang, M.; Xie, S.; Sun, Y. Resveratrol induces cervical cancer HeLa cell apoptosis through the activation and nuclear translocation promotion of FOXO3a. Pharmazie 2020, 75, 250–254. [Google Scholar] [CrossRef]
- García-Zepeda, S.P.; García-Villa, E.; Díaz-Chávez, J.; Hernández-Pando, R.; Gariglio, P. Resveratrol induces cell death in cervical cancer cells through apoptosis and autophagy. Eur. J. Cancer Prev. 2013, 22, 577–584. [Google Scholar] [CrossRef]
- Mineda, A.; Nishimura, M.; Kagawa, T.; Takiguchi, E.; Kawakita, T.; Abe, A.; Irahara, M. Resveratrol suppresses proliferation and induces apoptosis of uterine sarcoma cells by inhibiting the Wnt signaling pathway. Exp. Ther. Med. 2019, 17, 2242–2246. [Google Scholar] [CrossRef]
- Fukuda, T.; Oda, K.; Wada-Hiraike, O.; Sone, K.; Inaba, K.; Ikeda, Y.; Makii, C.; Miyasaka, A.; Kashiyama, T.; Tanikawa, M.; et al. Autophagy inhibition augments resveratrol-induced apoptosis in Ishikawa endometrial cancer cells. Oncol. Lett. 2016, 12, 2560–2566. [Google Scholar] [CrossRef]
- Tang, Q.; Li, G.; Wei, X.; Zhang, J.; Chiu, J.F.; Hasenmayer, D.; Zhang, D.; Zhang, H. Resveratrol-induced apoptosis is enhanced by inhibition of autophagy in esophageal squamous cell carcinoma. Cancer Lett. 2013, 336, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, Y.; Zhu, B.; Liu, Q.; Yao, Q.; Zhao, G. Resveratrol induces apoptosis in SGC-7901 gastric cancer cells. Oncol. Lett. 2018, 16, 2949–2956. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, X.; Chen, S.; Ma, G.; Zhu, M.; Yan, F.; Yu, J. Resveratrol induced apoptosis in human gastric carcinoma SGC-7901 cells via activation of mitochondrial pathway. Asia Pac. J. Clin. Oncol. 2018, 14, e317–e324. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, X.; Cheng, L.; Li, C.; Wu, Z.; Luo, Y.; Zhou, K.; Li, Y.; Zhao, Q.; Huang, Y. Modulation of lncRNA H19 enhances resveratrol-inhibited cancer cell proliferation and migration by regulating endoplasmic reticulum stress. J. Cell Mol. Med. 2022, 26, 2205–2217. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Ko, J.H.; Lee, H.; Jung, J.; Kong, M.; Lee, J.W.; Lee, J.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; et al. Resveratrol inhibits STAT3 signaling pathway through the induction of SOCS-1: Role in apoptosis induction and radiosensitization in head and neck tumor cells. Phytomedicine 2016, 23, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Takashina, M.; Inoue, S.; Tomihara, K.; Tomita, K.; Hattori, K.; Zhao, Q.L.; Suzuki, T.; Noguchi, M.; Ohashi, W.; Hattori, Y. Different effect of resveratrol to induction of apoptosis depending on the type of human cancer cells. Int. J. Oncol. 2017, 50, 787–797. [Google Scholar] [CrossRef]
- Siedlecka-Kroplewska, K.; Wozniak, M.; Kmiec, Z. The wine polyphenol resveratrol modulates autophagy and induces apoptosis in MOLT-4 and HL-60 human leukemia cells. J. Physiol. Pharmacol. 2019, 70, 825–838. [Google Scholar] [CrossRef]
- Khanzadeh, T.; Hagh, M.F.; Talebi, M.; Yousefi, B.; Azimi, A.; Hossein Pour Feizi, A.A.; Baradaran, B. Investigation of BAX and BCL2 expression and apoptosis in a resveratrol- and prednisolone-treated human T-ALL cell line, CCRF-CEM. Blood Res. 2018, 53, 53–60. [Google Scholar] [CrossRef]
- Tian, H.; Yu, Z. Resveratrol induces apoptosis of leukemia cell line K562 by modulation of sphingosine kinase-1 pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 2755–2762. [Google Scholar]
- Fan, Y.; Chiu, J.F.; Liu, J.; Deng, Y.; Xu, C.; Zhang, J.; Li, G. Resveratrol induces autophagy-dependent apoptosis in HL-60 cells. BMC Cancer 2018, 18, 581. [Google Scholar] [CrossRef]
- Chai, R.; Fu, H.; Zheng, Z.; Liu, T.; Ji, S.; Li, G. Resveratrol inhibits proliferation and migration through SIRT1 mediated post-translational modification of PI3K/AKT signaling in hepatocellular carcinoma cells. Mol. Med. Rep. 2017, 16, 8037–8044. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, Y.; Zhang, Y.; Liu, Y.; Yu, Y.; Ma, M. Resveratrol induces autophagy and apoptosis in non-small-cell lung cancer cells by activating the NGFR-AMPK-mTOR pathway. Nutrients 2022, 14, 2413. [Google Scholar] [CrossRef] [PubMed]
- Rasheduzzaman, M.; Jeong, J.K.; Park, S.Y. Resveratrol sensitizes lung cancer cell to TRAIL by p53 independent and suppression of Akt/NF-κB signaling. Life Sci. 2018, 208, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Chen, T. Resveratrol induces apoptosis via a Bak-mediated intrinsic pathway in human lung adenocarcinoma cells. Cell. Signal. 2012, 24, 1037–1046. [Google Scholar] [CrossRef]
- Li, X.; Wang, D.; Zhao, Q.C.; Shi, T.; Chen, J. Resveratrol inhibited non-small cell lung cancer through inhibiting STAT-3 signaling. Am. J. Med. Sci. 2016, 352, 524–530. [Google Scholar] [CrossRef]
- Fan, Y.; Li, J.; Yang, Y.; Zhao, X.; Liu, Y.; Jiang, Y.; Zhou, L.; Feng, Y.; Yu, Y.; Cheng, Y. Resveratrol modulates the apoptosis and autophagic death of human lung adenocarcinoma A549 cells via a p53-dependent pathway: Integrated bioinformatics analysis and experimental validation. Int. J. Oncol. 2020, 57, 925–938. [Google Scholar] [CrossRef]
- Wright, C.; Iyer, A.K.V.; Yakisich, J.S.; Azad, N. Anti-tumorigenic effects of resveratrol in lung cancer cells through modulation of c-FLIP. Curr. Cancer Drug Targets 2017, 17, 669–680. [Google Scholar] [CrossRef]
- Heo, J.R.; Kim, S.M.; Hwang, K.A.; Kang, J.H.; Choi, K.C. Resveratrol induced reactive oxygen species and endoplasmic reticulum stress-mediated apoptosis, and cell cycle arrest in the A375SM malignant melanoma cell line. Int. J. Mol. Med. 2018, 42, 1427–1435. [Google Scholar] [CrossRef]
- Ma, R.; Yu, D.; Peng, Y.; Yi, H.; Wang, Y.; Cheng, T.; Shi, B.; Yang, G.; Lai, W.; Wu, X.; et al. Resveratrol induces AMPK and mTOR signaling inhibition-mediated autophagy and apoptosis in multiple myeloma cells. Acta Biochim. Biophys. Sin. 2021, 53, 775–783. [Google Scholar] [CrossRef]
- Chang, C.H.; Lee, C.Y.; Lu, C.C.; Tsai, F.J.; Hsu, Y.M.; Tsao, J.W.; Juan, Y.N.; Chiu, H.Y.; Yang, J.S.; Wang, C.C. Resveratrol-induced autophagy and apoptosis in cisplatin-resistant human oral cancer CAR cells: A key role of AMPK and Akt/mTOR signaling. Int. J. Oncol. 2017, 50, 873–882. [Google Scholar] [CrossRef]
- Kim, S.E.; Shin, S.H.; Lee, J.Y.; Kim, C.H.; Chung, I.K.; Kang, H.M.; Park, H.R.; Park, B.S.; Kim, I.R. Resveratrol induces mitochondrial apoptosis and inhibits epithelial-mesenchymal transition in oral squamous cell carcinoma cells. Nutr. Cancer 2018, 70, 125–135. [Google Scholar] [CrossRef]
- Chen, L.; Xia, J.S.; Wu, J.H.; Chen, Y.G.; Qiu, C.J. Resveratrol inhibits oral squamous cell carcinoma cells proliferation while promoting apoptosis through inhibition of CBX7 protein. Environ. Toxicol. 2020, 35, 1234–1240. [Google Scholar] [CrossRef]
- Lang, F.; Qin, Z.; Li, F.; Zhang, H.; Fang, Z.; Hao, E. Apoptotic cell death induced by resveratrol is partially mediated by the autophagy pathway in human ovarian cancer Cells. PLoS ONE 2015, 10, e0129196. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, L.; Luo, Y.; Li, X.; Chen, G.; Wang, Y. Resveratrol inhibits the proliferation and induces the apoptosis in ovarian cancer cells via inhibiting glycolysis and targeting AMPK/mTOR signaling pathway. J. Cell. Biochem. 2018, 119, 6162–6172. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Gao, M.; Wang, Z.; Wang, W.; Zhan, L.; Wei, B. Upregulation of microRNA-34a sensitizes ovarian cancer cells to resveratrol by targeting Bcl-2. Yonsei Med. J. 2021, 62, 691–701. [Google Scholar] [CrossRef]
- Qin, Y.; Ma, Z.; Dang, X.; Li, W.; Ma, Q. Effect of resveratrol on proliferation and apoptosis of human pancreatic cancer MIA PaCa-2 cells may involve inhibition of the Hedgehog signaling pathway. Mol. Med. Rep. 2014, 10, 2563–2567. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Banerjee, S.; Acosta, E.P.; Lillard, J.W.; Singh, R. Resveratrol induces cell cycle arrest and apoptosis with docetaxel in prostate cancer cells via a p53/p21WAF1/CIP1 and p27KIP1 pathway. Oncotarget 2017, 8, 17216–17228. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ganapathy, S.; Singh, K.P.; Shankar, S.; Srivastava, R.K. Resveratrol induces growth arrest and apoptosis through activation of FOXO transcription factors in prostate cancer cells. PLoS ONE 2010, 5, e15288. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, L.; Tong, S.; Hu, X.; Zu, X.; Li, Y.; He, W.; Liu, L.; Chen, M.; Qi, L. Resveratrol suppresses the epithelial-to-mesenchymal transition in PC-3 cells by down-regulating the PI3K/AKT signaling pathway. Anim. Cells Syst. 2016, 20, 77–85. [Google Scholar] [CrossRef]
- Shih, A.; Davis, F.B.; Lin, H.Y.; Davis, P.J. Resveratrol induces apoptosis in thyroid cancer cell lines via a MAPK- and p53-dependent mechanism. J. Clin. Endocrinol. Metab. 2002, 87, 1223–1232. [Google Scholar] [CrossRef]
- Zheng, X.; Jia, B.; Tian, X.T.; Song, X.; Wu, M.L.; Kong, Q.Y.; Li, H.; Liu, J. Correlation of reactive oxygen species levels with resveratrol sensitivities of anaplastic thyroid cancer cells. Oxidative Med. Cell. Longev. 2018, 2018, 6235417. [Google Scholar] [CrossRef]
- Zhao, L.; Sanyal, S. p53 isoforms as cancer biomarkers and therapeutic targets. Cancers 2022, 14, 3145. [Google Scholar] [CrossRef]
- Ou, A.; Zhao, X.; Lu, Z. The potential roles of p53 signaling reactivation in pancreatic cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188662. [Google Scholar] [CrossRef]
- Feng, J. The p53 pathway related genes predict the prognosis of colon cancer. Int. J.Gen. Med. 2022, 15, 169–177. [Google Scholar] [CrossRef]
- Revathidevi, S.; Munirajan, A.K. Akt in cancer: Mediator and more. Semin. Cancer Biol. 2019, 59, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.H. AKT as a therapeutic target for cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef]
- Kang, Y.J.; Jang, J.Y.; Kwon, Y.H.; Lee, J.H.; Lee, S.; Park, Y.; Jung, Y.S.; Im, E.; Moon, H.R.; Chung, H.Y.; et al. MHY2245, a sirtuin inhibitor, induces cell cycle arrest and apoptosis in HCT116 human colorectal cancer cells. Int. J. Mol. Sci. 2022, 23, 1590. [Google Scholar] [CrossRef] [PubMed]
- Ong, A.L.C.; Ramasamy, T.S. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res. Rev. 2018, 43, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Im, E.; Choi, Y.H.; Kim, N.D. Mechanism of bile acid-induced programmed cell death and drug discovery against cancer: A review. Int. J. Mol. Sci. 2022, 23, 7184. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Kaplan, V.; Livneh, I.; Avni, N.; Cohen-Rosenzweig, C.; Ciechanover, A. The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int. J. Biochem. Cell Biol. 2016, 79, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef]
- Gentile, D.; Esposito, M.; Grumati, P. Metabolic adaption of cancer cells toward autophagy: Is there a role for ER-phagy? Front. Mol. Biosci. 2022, 9, 930223. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Zhang, Y.; Liu, Q.; Zhang, M.; Tu, K. The crosstalk between sonodynamic therapy and autophagy in cancer. Front. Pharmacol. 2022, 13, 961725. [Google Scholar] [CrossRef]
- Fu, Y.; Chang, H.; Peng, X.; Bai, Q.; Yi, L.; Zhou, Y.; Zhu, J.; Mi, M. Resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/β-catenin signaling pathway. PLoS ONE 2014, 9, e102535. [Google Scholar] [CrossRef]
- Puissant, A.; Robert, G.; Fenouille, N.; Luciano, F.; Cassuto, J.P.; Raynaud, S.; Auberger, P. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010, 70, 1042–1052. [Google Scholar] [CrossRef]
- Zhang, B.; Yin, X.; Sui, S. Resveratrol inhibited the progression of human hepatocellular carcinoma by inducing autophagy via regulating p53 and the phosphoinositide 3-kinase/protein kinase B pathway. Oncol. Rep. 2018, 40, 2758–2765. [Google Scholar] [CrossRef]
- Zhang, J.; Chiu, J.; Zhang, H.; Qi, T.; Tang, Q.; Ma, K.; Lu, H.; Li, G. Autophagic cell death induced by resveratrol depends on the Ca(2+)/AMPK/mTOR pathway in A549 cells. Biochem. Pharmacol. 2013, 86, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Cao, N.; Li, Z.; Han, J.; Li, L. Resveratrol, an activator of SIRT1, induces protective autophagy in non-small-cell lung cancer via inhibiting Akt/mTOR and activating p38-MAPK. Onco Targets Ther. 2018, 11, 7777–7786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, K.; Qi, T.; Wei, X.; Zhang, Q.; Li, G.; Chiu, J.F. P62 regulates resveratrol-mediated Fas/Cav-1 complex formation and transition from autophagy to apoptosis. Oncotarget 2015, 6, 789–801. [Google Scholar] [CrossRef]
- Ferraresi, A.; Phadngam, S.; Morani, F.; Galetto, A.; Alabiso, O.; Chiorino, G.; Isidoro, C. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Mol. Carcinog. 2017, 56, 1164–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.X.; Zhang, Y.; Wu, M.L.; Liu, Y.N.; Zhang, P.; Chen, X.Y.; Kong, Q.Y.; Liu, J.; Li, H. Resveratrol and STAT inhibitor enhance autophagy in ovarian cancer cells. Cell Death Discov. 2016, 2, 15071. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, Y.; Wang, J.; Gu, A.; Li, Q.; Mao, D.; Guo, L. Effect of autophagy on the resveratrol-induced apoptosis of ovarian cancer SKOV3 cells. J. Cell. Biochem. 2018, 120, 7788–7793. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y. AMPK and Autophagy. In Autophagy: Biology and Diseases: Basic Science; Qin, Z.-H., Ed.; Springer: Singapore, 2019; pp. 85–108. [Google Scholar]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef]
- Islam, M.A.; Sooro, M.A.; Zhang, P. Autophagic regulation of p62 is critical for cancer therapy. Int. J. Mol. Sci. 2018, 19, 1405. [Google Scholar] [CrossRef]
- Cha-Molstad, H.; Yu, J.E.; Feng, Z.; Lee, S.H.; Kim, J.G.; Yang, P.; Han, B.; Sung, K.W.; Yoo, Y.D.; Hwang, J.; et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 2017, 8, 102. [Google Scholar] [CrossRef]
- Yan, G.; Elbadawi, M.; Efferth, T. Multiple cell death modalities and their key features. World Acad. Sci. J. 2020, 2, 39–48. [Google Scholar] [CrossRef]
- Zanetti, L.C.; Weinlich, R. Necroptosis, the other main caspase-independent cell death. Adv. Exp. Med. Biol. 2021, 1301, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Weinlich, R.; Oberst, A.; Beere, H.M.; Green, D.R. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kos, R.; Garssen, J.; Redegeld, F. Molecular insights into the mechanism of necroptosis: The necrosome as a potential therapeutic target. Cells 2019, 8, 1486. [Google Scholar] [CrossRef] [PubMed]
- Seifert, L.; Miller, G. Molecular pathways: The necrosome-a target for cancer therapy. Clin. Cancer Res. 2017, 23, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; He, W.; Sun, L. Necrosome core machinery: MLKL. Cell. Mol. Life Sci. 2016, 73, 2153–2163. [Google Scholar] [CrossRef]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.G.; Liu, Z.G. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef]
- Li, X.; Guo, J.; Ding, A.P.; Qi, W.W.; Zhang, P.H.; Lv, J.; Qiu, W.S.; Sun, Z.Q. Association of mixed lineage kinase domain-like protein expression with prognosis in patients with colon cancer. Technol. Cancer Res. Treat. 2017, 16, 428–434. [Google Scholar] [CrossRef]
- Seifert, L.; Werba, G.; Tiwari, S.; Giao Ly, N.N.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016, 532, 245–249. [Google Scholar] [CrossRef]
- Ertao, Z.; Jianhui, C.; Kang, W.; Zhijun, Y.; Hui, W.; Chuangqi, C.; Changjiang, Q.; Sile, C.; Yulong, H.; Shirong, C. Prognostic value of mixed lineage kinase domain-like protein expression in the survival of patients with gastric caner. Tumour Biol. 2016, 37, 13679–13685. [Google Scholar] [CrossRef]
- Fulda, S. Targeting apoptosis for anticancer therapy. Semin. Cancer Biol. 2015, 31, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Ma, D.; Tan, Y.X.; Wang, H.Y.; Cai, Z. The role of necroptosis in cancer: A double-edged sword? Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.J. Synergistic anticancer activity of resveratrol in combination with docetaxel in prostate carcinoma cells. Nutr. Res. Pract. 2021, 15, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.S.; Kim, S.D.; Jeong, S.K.; Oh, S.J.; Par, K.M.; Lee, C.G.; Kang, Y.R.; Jeong, M.H. Resveratrol analogue, HS-1793, inhibits inflammatory mediator release from macrophages by interfering with the TLR4 mediated NF-κB activation. Food Sci. Biotechnol. 2022, 31, 433–441. [Google Scholar] [CrossRef]
- Feng, Y.; Clayton, J.; Yake, W.; Li, J.; Wang, W.; Winne, L.; Hong, M. Resveratrol derivative, trans-3, 5, 4′-trimethoxystilbene sensitizes osteosarcoma cells to apoptosis via ROS-induced caspases activation. Oxidative Med. Cell. Longev. 2021, 2021, 8840692. [Google Scholar] [CrossRef]
- Walle, T.; Hsieh, F.; DeLegge, M.H.; Oatis, J.E., Jr.; Walle, U.K. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos. 2004, 32, 1377–1382. [Google Scholar] [CrossRef]
- Chimento, A.; Sirianni, R.; Saturnino, C.; Caruso, A.; Sinicropi, M.S.; Pezzi, V. Resveratrol and its analogues as antitumoral agents for breast cancer treatment. Mini Rev. Med. Chem. 2016, 16, 699–709. [Google Scholar] [CrossRef]
- Athar, M.; Back, J.H.; Tang, X.; Kim, K.H.; Kopelovich, L.; Bickers, D.R.; Kim, A.L. Resveratrol: A review of preclinical studies for human cancer prevention. Toxicol. Appl. Pharmacol. 2007, 224, 274–283. [Google Scholar] [CrossRef]
- Chimento, A.; Santarsiero, A.; Iacopetta, D.; Ceramella, J.; De Luca, A.; Infantino, V.; Parisi, O.I.; Avena, P.; Bonomo, M.G.; Saturnino, C.; et al. A phenylacetamide resveratrol derivative exerts inhibitory effects on breast cancer cell growth. Int. J. Mol. Sci. 2021, 22, 5255. [Google Scholar] [CrossRef]
- Li, H.; Wu, W.K.; Zheng, Z.; Che, C.T.; Yu, L.; Li, Z.J.; Wu, Y.C.; Cheng, K.W.; Yu, J.; Cho, C.H.; et al. 2,3′,4,4′,5′-Pentamethoxy-trans-stilbene, a resveratrol derivative, is a potent inducer of apoptosis in colon cancer cells via targeting microtubules. Biochem. Pharmacol. 2009, 78, 1224–1232. [Google Scholar] [CrossRef]
- Park, J.W.; Choi, W.G.; Lee, P.J.; Chung, S.W.; Kim, B.S.; Chung, H.T.; Cho, S.; Kim, J.H.; Kang, B.H.; Kim, H.; et al. The novel resveratrol derivative 3,5-diethoxy-3′,4′-dihydroxy-trans-stilbene induces mitochondrial ROS-mediated ER stress and cell death in human hepatoma cells in vitro. Acta Pharmacol. Sin. 2017, 38, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, W.K.; Zheng, Z.; Che, C.T.; Li, Z.J.; Xu, D.D.; Wong, C.C.; Ye, C.G.; Sung, J.J.; Cho, C.H.; et al. 3,3′,4,5,5′-Pentahydroxy-trans-stilbene, a resveratrol derivative, induces apoptosis in colorectal carcinoma cells via oxidative stress. Eur. J. Pharmacol. 2010, 637, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Mikuła-Pietrasik, J.; Sosińska, P.; Murias, M.; Wierzchowski, M.; Brewińska-Olchowik, M.; Piwocka, K.; Szpurek, D.; Książek, K. High potency of a novel resveratrol derivative, 3,3′,4,4′-tetrahydroxy-trans-stilbene, against ovarian cancer is associated with an oxidative stress-mediated imbalance between DNA damage accumulation and repair. Oxidative Med. Cell. Longev. 2015, 2015, 135691. [Google Scholar] [CrossRef]
- Fan, X.X.; Yao, X.J.; Xu, S.W.; Wong, V.K.; He, J.X.; Ding, J.; Xue, W.W.; Mujtaba, T.; Michelangeli, F.; Huang, M.; et al. (Z)3,4,5,4′-trans-tetramethoxystilbene, a new analogue of resveratrol, inhibits gefitinb-resistant non-small cell lung cancer via selectively elevating intracellular calcium level. Sci. Rep. 2015, 5, 16348. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Gopalakrishnan, V.; Hegde, M.; Kumar, S.; Karki, S.S.; Raghavan, S.C.; Choudhary, B. A novel resveratrol based tubulin inhibitor induces mitotic arrest and activates apoptosis in cancer cells. Sci. Rep. 2016, 6, 34653. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.H.; Yang, H.H.; Gan, Y.H.; Meng, Y.L.; Li, Y.P.; Ge, L.P.; Zhang, C.H.; Liu, L.N.; Kang, Y.F. Finding a resveratrol analogue as potential anticancer agent with apoptosis and cycle arrest. J. Pharmacol. Sci. 2020, 143, 238–241. [Google Scholar] [CrossRef]
- Duan, J.J.; Yue, W.; Jian, E.J.; Malhotra, J.; Lu, S.-E.; Gu, J.; Xu, F.; Tan, X.-L. In vitro comparative studies of resveratrol and triacetylresveratrol on cell proliferation, apoptosis, and STAT3 and NFκB signaling in pancreatic cancer cells. Sci. Rep. 2016, 6, 31672. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, F.; Sheng, P.L.; Chen, Z.Q.; Xu, Q.P.; Guo, Y.Q. Resveratrol analogue 3,4,4′-trihydroxy-trans-stilbene induces apoptosis and autophagy in human non-small-cell lung cancer cells in vitro. Acta Pharmacol. Sin. 2015, 36, 1256–1265. [Google Scholar] [CrossRef]
- Savio, M.; Ferraro, D.; Maccario, C.; Vaccarone, R.; Jensen, L.D.; Corana, F.; Mannucci, B.; Bianchi, L.; Cao, Y.; Stivala, L.A. Resveratrol analogue 4,4′-dihydroxy-trans-stilbene potently inhibits cancer invasion and metastasis. Sci. Rep. 2016, 6, 19973. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, D.H.; Hossain, M.A.; Kim, M.Y.; Sung, B.; Yoon, J.H.; Suh, H.; Jeong, T.C.; Chung, H.Y.; Kim, N.D. HS-1793, a resveratrol analogue, induces cell cycle arrest and apoptotic cell death in human breast cancer cells. Int. J. Oncol. 2014, 44, 473–480. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, M.J.; Sung, B.; Suh, H.; Jung, J.H.; Chung, H.Y.; Kim, N.D. Resveratrol analogue, HS-1793, induces apoptotic cell death and cell cycle arrest through downregulation of AKT in human colon cancer cells. Oncol. Rep. 2017, 37, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Um, H.J.; Bae, J.H.; Park, J.W.; Suh, H.; Jeong, N.Y.; Yoo, Y.H.; Kwon, T.K. Differential effects of resveratrol and novel resveratrol derivative, HS-1793, on endoplasmic reticulum stress-mediated apoptosis and Akt inactivation. Int. J. Oncol. 2010, 36, 1007–1013. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, S.H.; Jo, W.S.; Song, S.; Suh, H.; Seol, S.Y.; Leem, S.H.; Kwon, T.K.; Yoo, Y.H. A novel resveratrol derivative, HS1793, overcomes the resistance conferred by Bcl-2 in human leukemic U937 cells. Biochem. Pharmacol. 2009, 77, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Saiko, P.; Graser, G.; Giessrigl, B.; Steinmann, M.T.; Schuster, H.; Lackner, A.; Grusch, M.; Krupitza, G.; Jaeger, W.; Somepalli, V.; et al. Digalloylresveratrol, a novel resveratrol analog inhibits the growth of human pancreatic cancer cells. Investig. New Drugs 2013, 31, 1115–1124. [Google Scholar] [CrossRef]
- Piotrowska, H.; Myszkowski, K.; Amarowicz, R.; Murias, M.; Kulcenty, K.; Wierzchowski, M.; Jodynis-Liebert, J. Different susceptibility of colon cancer DLD-1 and LOVO cell lines to apoptosis induced by DMU-212, a synthetic resveratrol analogue. Toxicol. In Vitro 2013, 27, 2127–2134. [Google Scholar] [CrossRef]
- Piotrowska, H.; Myszkowski, K.; Ziółkowska, A.; Kulcenty, K.; Wierzchowski, M.; Kaczmarek, M.; Murias, M.; Kwiatkowska-Borowczyk, E.; Jodynis-Liebert, J. Resveratrol analogue 3,4,4′,5-tetramethoxystilbene inhibits growth, arrests cell cycle and induces apoptosis in ovarian SKOV-3 and A-2780 cancer cells. Toxicol. Appl. Pharmacol. 2012, 263, 53–60. [Google Scholar] [CrossRef]
- Jozkowiak, M.; Skupin-Mrugalska, P.; Nowicki, A.; Borys-Wojcik, S.; Wierzchowski, M.; Kaczmarek, M.; Ramlau, P.; Jodynis-Liebert, J.; Piotrowska-Kempisty, H. The effect of 4′-hydroxy-3,4,5-trimetoxystilbene, the metabolite of resveratrol analogue DMU-212, on growth, cell cycle and apoptosis in DLD-1 and LOVO colon cancer cell lines. Nutrients 2020, 12, 1327. [Google Scholar] [CrossRef]
- Ronghe, A.; Chatterjee, A.; Bhat, N.K.; Padhye, S.; Bhat, H.K. Tamoxifen synergizes with 4-(E)-{(4-hydroxyphenylimino)-methylbenzene, 1,2-diol} and 4-(E)-{(p-tolylimino)-methylbenzene-1,2-diol}, novel azaresveratrol analogues, in inhibiting the proliferation of breast cancer cells. Oncotarget 2016, 7, 51747–51762. [Google Scholar] [CrossRef]
- Zielińska-Przyjemska, M.; Kaczmarek, M.; Krajka-Kuźniak, V.; Wierzchowski, M.; Baer-Dubowska, W. Effect of methoxy stilbenes-analogues of resveratrol-on the viability and induction of cell cycle arrest and apoptosis in human myeloid leukemia cells. Mol. Cell. Biochem. 2020, 474, 113–123. [Google Scholar] [CrossRef]
- Lin, M.H.; Hung, C.F.; Sung, H.C.; Yang, S.C.; Yu, H.P.; Fang, J.Y. The bioactivities of resveratrol and its naturally occurring derivatives on skin. J. Food Drug Anal. 2021, 29, 15–38. [Google Scholar] [CrossRef]
- Li, H.; Wu, W.K.; Li, Z.J.; Chan, K.M.; Wong, C.C.; Ye, C.G.; Yu, L.; Sung, J.J.; Cho, C.H.; Wang, M. 2,3′,4,4′,5′-Pentamethoxy-trans-stilbene, a resveratrol derivative, inhibits colitis-associated colorectal carcinogenesis in mice. Br. J. Pharmacol. 2010, 160, 1352–1361. [Google Scholar] [CrossRef] [PubMed]



| Cancer/Cell Lines | Upregulation | Downregulation | Refs. |
|---|---|---|---|
| Bladder | |||
| RT4 | AKT, DNMT1, PLK1, mTOR, SRC | [21] | |
| 5637 | caspase-3 activity | PLK1, p-AKT, Bcl-2 | [21,64] |
| SV-HUC-1 | caspase-3 activity | miR-21 | [64] |
| T24 | RASSF1A, p21, AKT, p-p38, Bax, caspase-3, -9, and PARP cleavage, caspase-3 activity | HOXB3, PLK1, cyclin D1, CDK4, p-Rb, p-AKT(Ser473), Bcl-2, Bcl-xL, p-Bad(Ser112), p-Bad(Ser136), procaspase-3 and -9, VEGF, FGF-2, miR-21 | [21,64,65] |
| T24 xenograft | VEGF, FGF-2 | [65] | |
| Bone | |||
| JJ012 | SIRT1, caspase-3 cleavage | Ac-p65 | [66] |
| JJ012 xenograft | SIRT1, caspase-3 cleavage | [66] | |
| SW1353 | Bax, caspase-3 cleavage, SIRT1 | Bcl-2, p-STAT3 | [67] |
| Breast | |||
| MCF-7 | caspase-8 and -9 activity, PARP cleavage | caspase-3, -8, and -9, Bcl-2, XIAP, CDK2, CDK4, CDK6, GSH, TIGAR, p-S6 | [68,69] |
| MDA-MB-231 | caspase-3, caspase-8 and -9 activity, Cyt c(cytosol), Bax(mitochondrial), Bim(mitochondrial), t-Bid(mitochondrial) | caspase-3, -8, and -9, Bcl-2, XIAP, CDK2, CDK4, CDK6, Bax(cytosol) | [68,70] |
| Colon | |||
| CO115 | SET7/9, Me-p53 at K372, PARP cleavage, p53 | [22] | |
| DLD1 | p53, Bax | cyclin D1, cyclin E1, Bcl-2, p-STAT3(Tyr705) | [71] |
| HCT116 | Bax, Cyt c, caspase-3 and -9 cleavage, SET7/9, Me-p53 at K372, PARP cleavage, p53, SIRT1 | Bcl-2, procaspase-3 and -9, hERG, COX-2, EP1, EP4 | [22,70,72,73,74,75] |
| SW480 | SET7/9, Me-p53 at K372, PARP cleavage, p53, SIRT1 | COX-2, EP1, EP4 | [22,74,75] |
| SW620 | Bax, Cyt c, caspase-3 and -9 cleavage | Bcl-2, procaspase-3 and -9 | [72] |
| HT15 | p53, Bax | cyclin D1, cyclin E1, Bcl-2, p-STAT3(Tyr705) | [71] |
| HT29 | PARP, caspase-8, and caspase-3 cleavage, LC3-I, LC3-II | COX-2, EP1, EP4 | [74,76] |
| COLO 201 | PARP, caspase-8, and caspase-3 cleavage | [76] | |
| Cervical | |||
| HeLa | FOXO3a, Bim | p-FOXO3a, p-ERK, ∆Ψm, p53, p65 | [77,78] |
| MES-SA | β-catenin, c-myc | [79] | |
| CaSki | p53, p65 | [78] | |
| C33A | p53 | p65 | [78] |
| CaLo | p53, p65 | [78] | |
| Ishikawa | p-AMPKα, p-ERK, LC3, PARP cleavage | [80] | |
| Esophageal | |||
| EC109 | caspase-3 cleavage, Bax, caspase-3 activity, Beclin-1, Atg5, LC3-II, p-LKB1, p-AMPK | Bcl-2, LC3-I, p-Rapter | [81] |
| EC9706 | caspase-3 cleavage, Bax, Beclin-1, Atg5, LC3-II | Bcl-2, LC3-I | [81] |
| Gastric | |||
| SGC7901 | Bax, Bak, caspase-3, -8, and PARP1 cleavage, Cyt c(cytosol), MLKL, p62, VDAC1, LC3-II, Atg3, Atg5, Beclin-1, p-ERK, p-p38, BiP, CHOP, BAP31 | miR-155-5p, c-Myc, cyclin B1, cyclin D1, claudin 1, Bcl-2, caspase-3 and -9, procaspase-3 and -8, p-NF-κB p65(cytoplasm), p-NF-κB p65(nucleus), p-NF-κB p65(total), ∆Ψm, Cyt c(mitochondria), p21, p-mTOR, p-AKT, β-catenin, Wnt3a, ZO-1, fibronectin, α-SMA, Vimentin, MMP-2 | [23,82,83,84] |
| SGC7901 xenograft | caspase-3 cleavage | Bcl-2 | [83] |
| MGC803 | miR-155-5p | [23] | |
| Head and neck | |||
| HN3 | p-STAT3(Tyr705) | [85] | |
| FaDu | SOCS-1, caspase-3, -9, and PARP cleavage | p-STAT3(Tyr705), p-STAT3(Ser727), p-JAK2(Thy1007/1008), STAT3, Bcl-2, Bcl-xL, Survivin, IAP-1, cyclin D1, VEGF, MMP-2 and -9, procaspase-3 and -9 | [85] |
| Leukemia | |||
| U937 | p-AMPK, Bax(mitochondrial) | p-AKT, Bax(cytosol), H-Ras | [86] |
| MOLT-4 | p-AMPK, p62, LC3-II, PARP1 cleavage, caspase-3 activity, ROS | p-AKT, H-Ras, ∆Ψm | [86,87] |
| CCRF-CEM | Bax | Bcl-2 | [88] |
| K562 | SphK1(cytosol), ceramide | SphK1(membrane), SphK activity, S1P | [89] |
| HL-60 | p62, LC3-I, LC3-II, caspase-3, -8, and PARP1 cleavage, caspase-3 activity, Bax, Bad, Fas, Bid, Atg5, Beclin-1, LC3II, p-AMPK, p-LKB1, p-Raptor | Bcl-2, ROS, ∆Ψm, p-Bad, FasL, PI3K(p85), p-AKT, p-p70S6K | [87,90] |
| Liver | |||
| HepG2 | Bax, PARP1 cleavage, SIRT1, SIRT1 activity, DCL1 | PCNA, Bcl-2, Bcl-2/Bax ratio, PARP, caspase-3 and -7, p-PI3K, p-AKT, Ac-FOXO1, p-FOXO3a | [91] |
| Bel-7402 | Bax, PARP1 cleavage, SIRT1, DCL1 | Bcl-2, Bcl-2/Bax ratio, PARP, caspase-3 and -7, p-FOXO3a | [91] |
| SMMC-7721 | Bax, PARP1 cleavage, SIRT1, DCL1 | Bcl-2, Bcl-2/Bax ratio, PARP, caspase-3 and -7, p-FOXO3a | [91] |
| HL-7702 | SIRT1 | [91] | |
| Lung | |||
| A549 | Beclin-1, LC3-I, Bax, NGFR, Ac-p53, p53, PUMA, Cyt c, Bak, AIF(cytosol), caspase-3 and -9 activity, Bim-L, caspase-3 cleavage, LC3-II, p62 | Bcl-2, p-mTOR, p-AKT, p-NF-κB p62, Bcl-xL, ∆Ψm, AIF(mitochondrial), STAT3, p-STAT3, procaspase-3, p62, p-MDM2(Ser166), p-AKT(Ser473) | [92,93,94,95,96] |
| ASTC-a-1 | Bak, AIF(cytosol), caspase-3 and -9 activity, Bim-L | ∆Ψm, AIF(mitochondrial), Bcl-xL | [94] |
| H1299 | PARP cleavage, LC3-II, caspase-3 cleavage | GSH, TIGAR, p-S6, caspase-3 | [69] |
| H460 | caspase-8 activity | c-FLIP, VEGF | [97] |
| Melanoma | |||
| A375SM | p21, p27, ROS, p-eIF2α, CHOP, p-p38, p53, Bax | cyclin E, cyclin B, Nrf2, Bcl-2 | [98] |
| Multiple myeloma | |||
| U266 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| RPMI-8226 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| NCI-H929 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| Oral | |||
| CAR | AMPKα, p-AMPKα(Thr172), Atg5, Atg7, Atg12, Atg14, Atg16L1, Beclin-1, PI3K class III, LC3-II, caspase-3 and -9 cleavage, Cyt c, Apaf-1, AIF, Endo G, Bax, Bad, caspase-3 and -9 activity | p-AKT(Ser473), p-mTOR(Ser2448), Rubicon, Bcl-2, p-Bad(Ser136) | [100] |
| CAL27 | Bak, Bax, Apaf-1, caspase-3, ICAD and PARP cleavage, E-cadherin, N-cadherin | Bcl-2, Bcl-xL, procaspase-3 and -9, Snail, Slug, Smad2/3 | [101] |
| HSC-3 | PARP and caspase-3 cleavage, caspase-3 and -9 activity, p16 | CBX7, p-AKT | [102] |
| Ovarian | |||
| OVCAR-3 | Atg5, p62, LC3-II, caspase-3 and PARP activity, ROS | ∆Ψm | [103] |
| Caov-3 | LC3-II, p62 | [103] | |
| A2780 | p-AMPK, caspase-3 cleavage | p-mTOR | [104] |
| SKOV3 | p-AMPK, caspase-3 cleavage, miR-34a, Bax | p-mTOR, Bcl-2 | [104,105] |
| SKOV3 xenograft | Ki-67 index, Metastasis index | [104] | |
| OV-90 | miR-34a, Bax, caspase-3 cleavage | Bcl-2 | [105] |
| Pancreatic | |||
| MIA PaCa-2 | IHH, Ptch, SMO | [106] | |
| Prostate | |||
| C4-2B | PARP, Bax, Bid, Bak, p27, p53 | Bcl-2, Mcl-1, Bcl-xL, CDK1, CDK2, CDK4, cyclin D1, cyclin E1 | [107] |
| DU145 | PARP, Bax, Bid, p27, p53 | Bcl-2, Mcl-1, Bcl-xL, CDK1, CDK2, CDK4, cyclin E1, PCNA, cyclin B1 | [107] |
| LNCaP | caspase-3 activity | p-PI3K, p-AKT, p-mTOR, p-FKHR, p-FKHRL1, p-AFX | [70,108] |
| PC-3 | Bax, caspase-3 and -9 cleavage, E-cadherin, caspase-3 activity | Bcl-2, ∆Ψm, Vimentin, p-AKT, pAKT/AKT | [70,109] |
| Thyroid | |||
| BHP 18–21 | p-ERK1, p-ERK2, p53, p-p53(ser15), p21, c-Fos | [110] | |
| BHP 2–7 | p-ERK1, p-ERK2, p53, p21, c-Fos, c-Jun | [110] | |
| FTC 236 | p-ERK1, p-ERK2, p53, p-p53(ser15), c-Fos, p21 | [110] | |
| FTC 238 | c-Fos, p53, p21 | [110] | |
| THJ-16T | caspase-3 and -9 activity | SOD2, CAT, procaspase-3 and -9 | [111] |
| THJ-11T | SULT1A1 | [111] |
| Cancer/Cell Lines | Upregulation | Downregulation | Refs. |
|---|---|---|---|
| Breast | |||
| MCF-7 | PARP cleavage, Beclin-1, Atg7, LC3-II | GHS, TIGAR, p-S6, LC3-I, β-catenin, cyclin D1 | [69,128] |
| SUM-159 | Beclin-1, Atg7, LC3-II | LC3-I, cyclin D1 | [128] |
| Colon | |||
| HT29 | caspase-3, -8, and PARP cleavage, LC3-I, LC3-II | [76] | |
| COLO 201 | caspase-3, -8, and PARP cleavage | [76] | |
| Cervical | |||
| Ishikawa | p-AMPKα, p-ERK, LC3, PARP cleavage | [80] | |
| Esophageal | |||
| EC109 | caspase-3 cleavage, Bax, caspase-3 activity, Beclin-1, Atg5, LC3-II, p-LKB1, p-AMPK | Bcl-2, LC3-I, p-Rapter | [81] |
| EC9706 | caspase-3 cleavage, Bax, Beclin-1, Atg5, LC3-II | Bcl-2, LC3-I | [81] |
| Gastric | |||
| SGC7901 | Bak, Bax, MLKL, p62, VDAC1, LC3-II, Atg3, Beclin-1, p-ERK, p-p38, BiP, CHOP, BAP31 | cyclin B1, p21, p-mTOR, p-AKT, β-catenin, Wnt3a, ZO-1, fibronectin, α-SMA, Vimentin, MMP-2 | [84] |
| Leukemia | |||
| MOLT-4 | p62, LC3-II, PARP1 cleavage, caspase-3 activity, ROS | ∆Ψm | [87] |
| HL-60 | p62, LC3-I, LC3-II, caspase-3, -8, and PARP1 cleavage, caspase-3 activity, Bax, Bad, Fas, Bid, Atg5, Beclin-1, LC3II, p-AMPK, p-LKB1, p-Raptor | Bcl-2, ROS, ∆Ψm, p-Bad, FasL, PI3K(p85), p-AKT, p-p70S6K | [87,90] |
| K562 | p62, LC3-I, LC3-II, p-JNK2/3(Thr 183/Tyr 185), p-c-Jun(Ser 63), p-AMPKα(Thr 172) | p-mTOR(Ser )2448, p-p70/85-S6K(Thr389 ), p-S6 ribo(Ser 235/236), p-4EBP1(Thr 37/46) | [129] |
| Liver | |||
| MHCC-97H | Beclin-1, LC3-II, LC3-II/I, p53 | p62, p-AKT, p-AKT/AKT | [130] |
| Lung | |||
| A549 | Beclin-1, LC3-I, LC3-II, Bax, NGFR, caspase-3 and -8 cleavage, p53, Ac-p53, p53, PUMA, Cyt c, Cyt c(cytosol), Atg5, p-Raptor, p-AMPK, p62, SIRT1, LC3-II/LC3-I, p-p38, p-p38/p38, caspase-3 activity, Fas, Cav-1 | Bcl-2, p-mTOR, procaspase-3, p62, p-MDM2(Ser166), p-AKT(Ser473), p62, Bcl-xL, LC3-I, p-p70S6K, p-AKT/AKT, p-p70S6K/p70S6K, p-mTOR/mTOR, Cyt c(mitochondrial), | [92,93,96,131,132,133] |
| H1299 | PARP cleavage, LC3-II, caspase-3 cleavage | GSH, TIGAR, p-S6, caspase-3 | [69] |
| Multiple myeloma | |||
| U266 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| RPMI-8226 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| NCI-H929 | Beclin-1, LC3-I, LC3-II, caspase-3 and PARP cleavage, p-AMPKα | Survivin, p-mTOR, p-p70S6K, p-4EBP1 | [99] |
| Oral | |||
| CAR | AMPKα, p-AMPKα(Thr172), Atg5, Atg7, Atg12, Atg14, Atg16L1, Beclin-1, PI3K class III, LC3-II, caspase-3 and -9 cleavage, Cyt c, Apaf-1, AIF, Endo G, Bax, Bad, caspase-3 and -9 activity | p-AKT(Ser473), p-mTOR(Ser2448), Rubicon, Bcl-2, p-Bad(Ser136) | [100] |
| Ovarian | |||
| OVCAR-3 | Atg5, p62, LC3-II, caspase-3 and PARP activity, ROS, ARHI, Beclin-1 | ∆Ψm, p-AKT(Ser473), p-S6 (Ser235/236), p-STAT3(Thy705) | [103,134,135] |
| Caov-3 | LC3-II, p62 | p-STAT3 | [103,135] |
| SKOV-3 | Beclin-1, LC3-II | [136] |
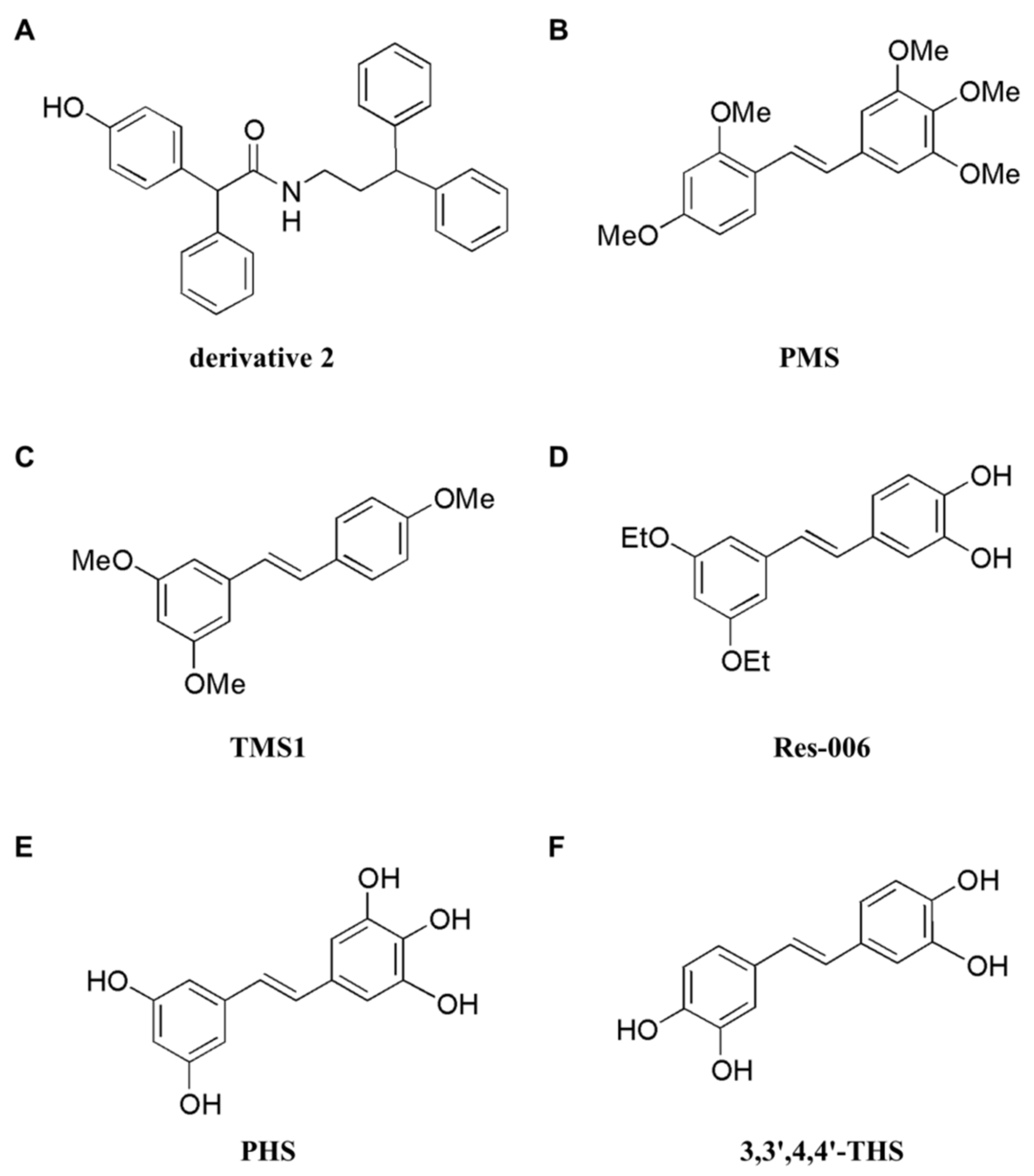
| Type | Name | Mechanism | Upregulation | Downregulation | Cancer/Cell Lines | Refs. |
|---|---|---|---|---|---|---|
| Derivatives | derivative 2 | apoptosis | Bax, PARP cleavage | cyclin D1, CDK4, Bcl-2 | Breast (MCF-7, MDA-MB-231) | [160] |
| PMS | apoptosis | caspase-3, -7, -9, and PARP cleavage, Bad, Bik, Bok, Bim, PUMA, Bcl-2, Bcl-2(Thr56), Bcl-2(Ser70) | Mcl-1 | Colon (HT29) | [161] | |
| TMS1 | apoptosis | p53, p-H2AX(Ser139), p-p53(Ser15), p-p53(Ser46), p- p53(Ser392), PUMA | Osteosarcoma (143B cell) | [156] | ||
| Res-006 | apoptosis | caspase-3 and PARP cleavage, p-IRE1α, p-JNK, XBP1, ERdj4, p-PERK, p-elF2α, CHOP, 3XFlag-ATF6α∆C, GRP78 | Liver (HepG2) | [162] | ||
| PHS | apoptosis | caspase-3, -9, and PARP cleavage, Bad | p-AKT(Ser473), GSH | Colon (HT29) | [163] | |
| 3,3′,4,4′ -THS | apoptosis | ROS, 8-OH-dG, hOGG1, caspase-3, -8, and -9 activity, SA-β-gal activity, p-p38/p38 | SOD, CAT | Ovarian (A2780, OVCAR-3, SKOV-3) | [164] | |
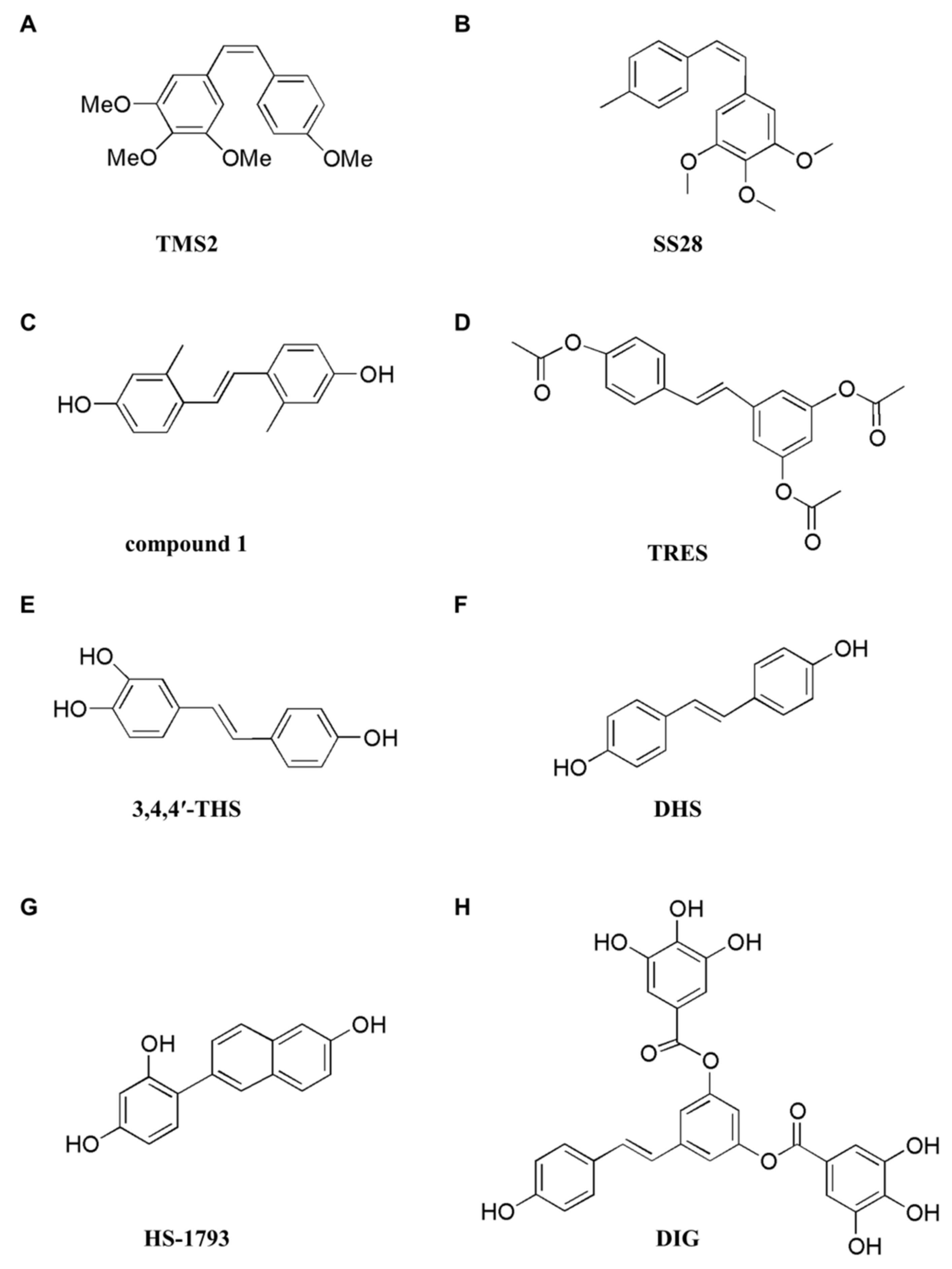
| Analogues | TMS2 | apoptosis, autophagy | caspase-3 and PARP cleavage, p-PERK, CHOP, p-eIF2α, p-AMPK, LC3-II, p-JNK | Bcl-2, p-AKT, p-p70S6K, p-S6, p-ACC, p-EGFR, p-PI3K, p-ERK | Lung (H1975) | [165] |
| SS28 | apoptosis | caspase-3, -9, and PARP cleavage | ∆Ψm, cyclin B1, CDK6 | Lung (A549), Leukemia (CEM) | [166] | |
| compound 1 | apoptosis | ROS, p53, p21, Bax | cyclin A1, cyclin A2, Bcl-2 | Cervical (HeLa) | [167] | |
| TRES | apoptosis | caspase-3 and PARP cleavage, Bim, PUMA, p-STAT3(cytoplasm), p-NF-κB(cytoplasm) | Mcl-1, p-STAT3, p-NF-κB, p-STAT3(nucleus), p-NF-κB(nucleus) | Pancreatic (PANC-1, BxPC-3) | [168] | |
| 3,4,4′-THS | apoptosis, autophagy | caspase-3, -9, and PARP cleavage, Bax, LC3-II, ROS | Bcl-2, Survivin, p62, p-p70S6K, p-4EBP1 | Lung (A549) | [169] | |
| DHS | apoptosis | PARP-1 cleavage | Lung (LLC) | [170] | ||
| HS-1793 | apoptosis | p53, p21, Fas-L, Fas, PARP cleavage, Bax, caspase-3, -8, and -9 activity, ERK, p-ERK, JNK, p-JNK | MDM2, cyclin D1, CDK4, cyclin B1, Cdc2, Cdc25C, Bcl-2 | Breast (MCF-7, MDA-MB-231) | [171] | |
| apoptosis | caspase-3, -8, and PARP cleavage, Bax, Cyt c(cytosol) | procaspase-3, -8, and -9, Bcl-2, Cyt c(mitochondria), cyclin B1, Cdc2, Cdc25C, CDK2, CDK4, CDK6, p-AKT, p-p38, p-ERK1/2, p-JNK | Colon (HCT116) | [172] | ||
| apoptosis | PARP cleavage, CHOP, GRP78 | procaspase-3, XBP1, p-AKT | Colon (HT29) | [173] | ||
| apoptosis | PARP cleavage | caspase-3, -6, Mcl-1, Bcl- 2, Bcl-xL, XIAP, 14-3-3,p-Bad, (Ser136), p-Bad(Ser155) | Leukemia (U937) | [174] | ||
| DIG | apoptosis | p-Chk2(Thr68), p-Cdc25A(Ser177), p-ATM(Ser1981), p-p38(Thr180/Tyr182), p-AKT (Ser473) | Pancreatic (AsPC-1) | [175] | ||
| DMU-212 | apoptosis | EF value, caspase-3/7 and -9 activity, Bax, Apaf-1, p53, Bad, Bak1, Bik, Bok, Noxa, PARP-1 cleavage | Bag1, Bcl-2, Bcl-xL, CYP1A1, CYP1B1 | Colon (DLD-1, LOVO) | [176] | |
| apoptosis | EF value, caspase-3/7 activity, Fas, FasL, TNF, TNFRF10A, TNFRSF21, TNFRSF16, Bax, Apaf-1, p53 | TRAF-1, -3, -5, and -7, BIRC-2, Bcl-2, Bcl2l10, CYP1A1, CYP1B1 | Ovarian (A-2780, SKOV-3) | [177] | ||
| DMU-281 | apoptosis | caspase-3/7, -8, and -9 activity, Bik, Bad, Bak1, Fas, TNFSRF10B, TNFSRF11B, TNFSF8, FADD, HSP60 | Bcl-2, Bcl2L1, HMGB1, STAT5A, STAT5b, HSP27 | Colon (LoVo) | [178] | |
| apoptosis | Bik, TNF, caspase-3/7, -8, and -9 activity, Smac/Diablo | Bcl-2, Bcl-xL, BIRC2, HMGB1, STAT5b, TNFRSF10C, TNFRSF11B, TRAF-1, -3, and -5 procaspase-3, HSP27 | Colon (DLD-1) | [178] | ||
| HPIMBD | apoptosis, autophagy | Beclin-1, LC3-I, LC3-II | ERα, c-Myc | Breast (MDA-MB-231, T47D) | [179] | |
| TIMBD | apoptosis, autophagy | Beclin-1, LC3-I, LC3-II | ERα, c-Myc | Breast (MDA-MB-231, T47D) | [179] | |
| 3,4,4ʹ-tri-MS, 3,4,2ʹ,4ʹ-tetra-MS | apoptosis | p53, Bax | Bcl-xL | Leukemia (HL-60, THP-1) | [180] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, J.Y.; Im, E.; Kim, N.D. Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review. Int. J. Mol. Sci. 2022, 23, 13689. https://doi.org/10.3390/ijms232213689
Jang JY, Im E, Kim ND. Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review. International Journal of Molecular Sciences. 2022; 23(22):13689. https://doi.org/10.3390/ijms232213689
Chicago/Turabian StyleJang, Jung Yoon, Eunok Im, and Nam Deuk Kim. 2022. "Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review" International Journal of Molecular Sciences 23, no. 22: 13689. https://doi.org/10.3390/ijms232213689
APA StyleJang, J. Y., Im, E., & Kim, N. D. (2022). Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review. International Journal of Molecular Sciences, 23(22), 13689. https://doi.org/10.3390/ijms232213689







