Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight
Abstract
:1. Introduction
2. Results
2.1. Agronomic Trait Differences between T75 and Z9
2.2. Whole-Genome Sequencing of T75 and Z9
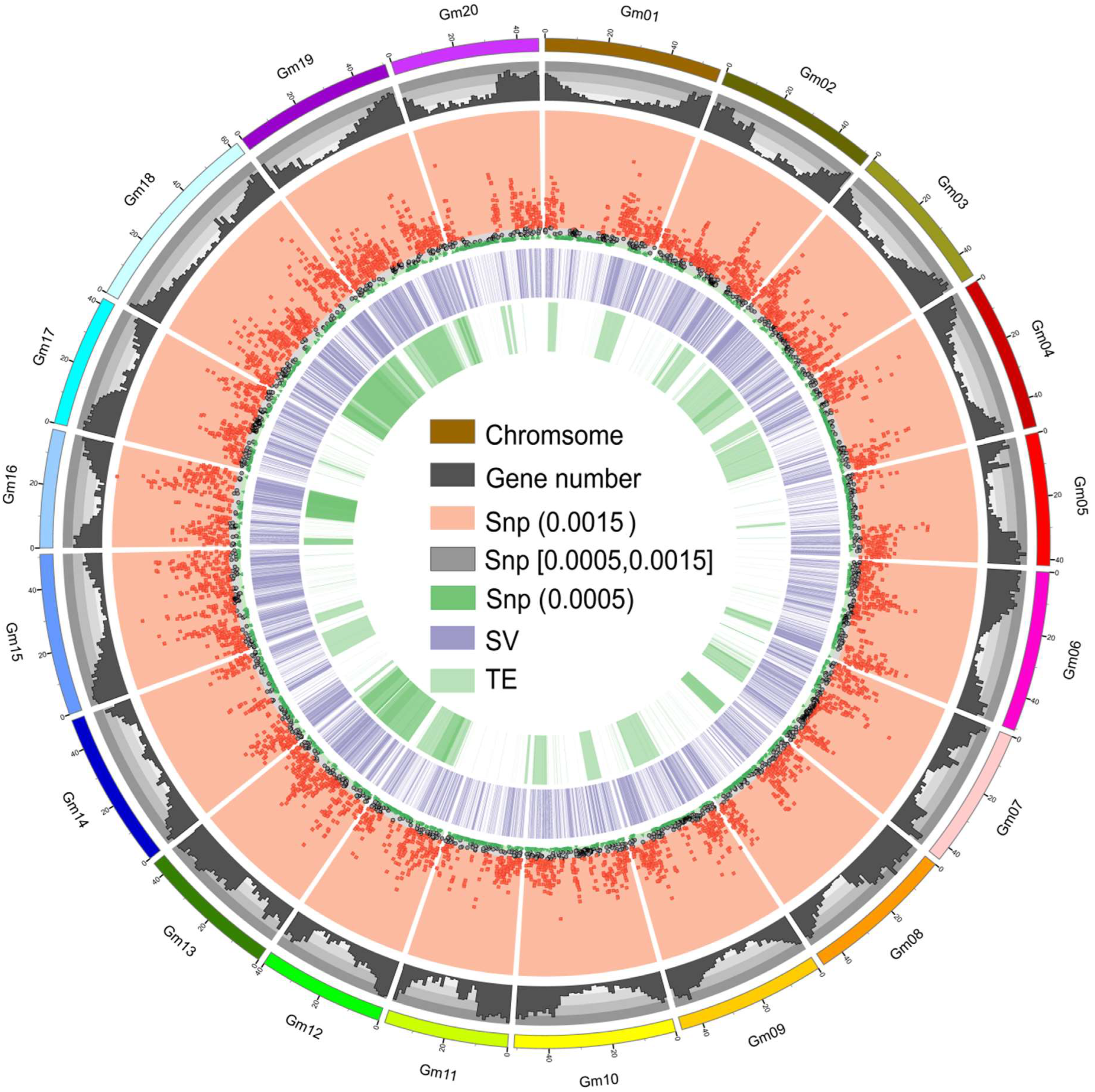
2.3. Structural Variations between T75 and Z9
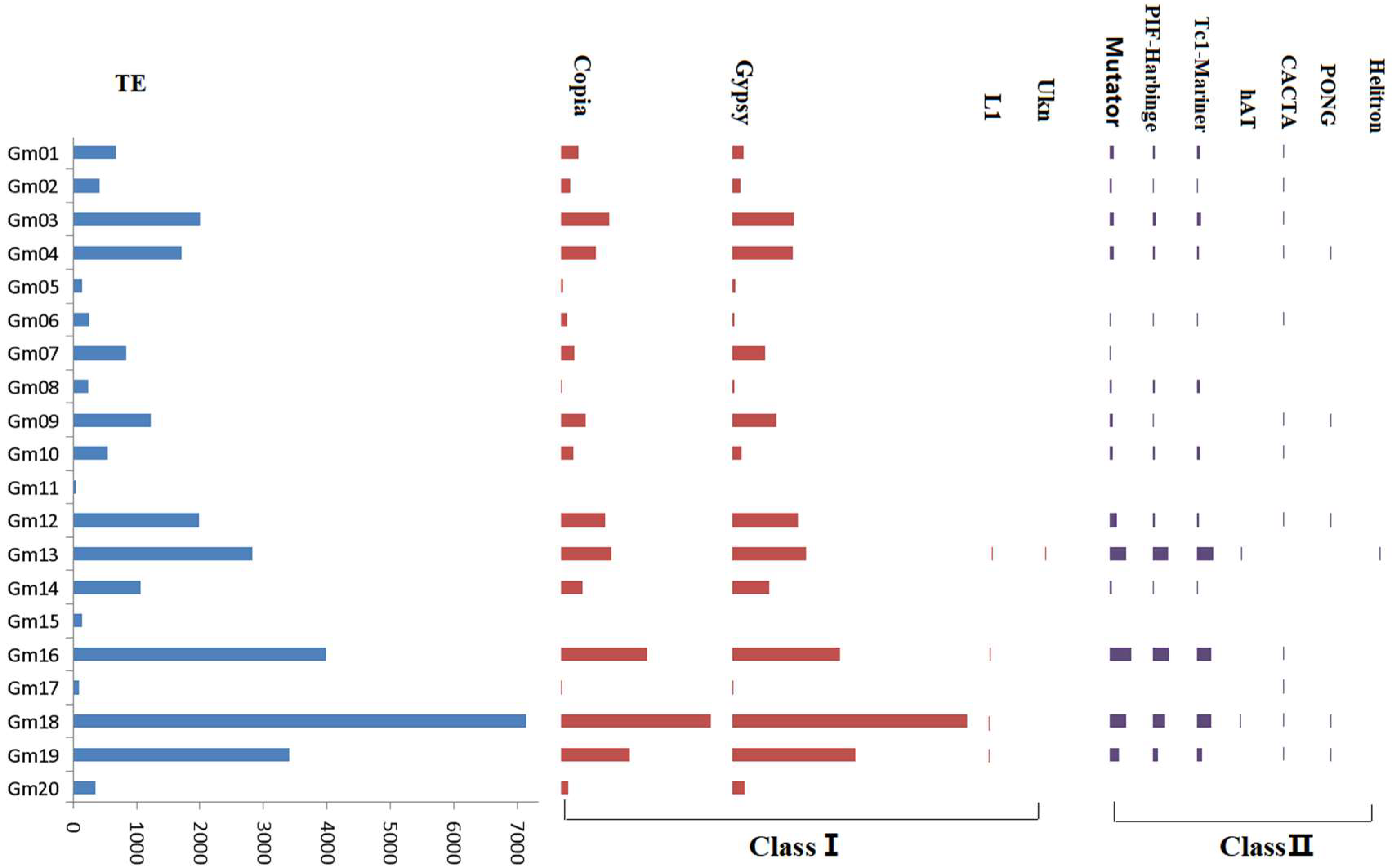
2.4. Discovery of Transposon Alterations between T75 and Z9
2.5. Genes Associated with SVs and TE Variations between T75 and Z9
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Field Experiments
4.2. DNA Sequencing of T75 and Z9
4.3. Analysis of SNPs and InDels
4.4. Identification of Structural Variations
4.5. Transposable Element Analysis
4.6. Identification of SVs and TE-Related Genes, and PCR Validation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dui-Cai, L.; Huang, Z.-X.; Zhao, Y.-L.; Guo, H.-J.; Wang, G.-L.; Jia, X.-H.; Li, C.-H.; Zhang, L.; Liu, L.-X. Space Radiation Measurement of Plant Seeds Boarding on the Shijian-8 Satellite. J. Nucl. Agric. Sci. 2008, 22, 5–8. [Google Scholar]
- Pan, D.; Zhang, Y.; Wang, J.; Gao, C.; Zhang, R.; Li, D.; Shen, Z.; Wang, D.; Liu, L. Mutagenic Effects and Variation of Leymus chinensis Embarked on Shijian-8 Satellite. J. Nucl. Agric. Sci. 2015, 29, 1233–1238. [Google Scholar]
- Fu, X.; Yang, Q.; Yuan, F.; Yu, X.; Jin, H.; Zhu, S.; Zhu, D. Breeding of Zhexian No.9 by Space Mutation and Variation Analysis of Its Character. J. Nucl. Agric. Sci. 2019, 33, 841–847. [Google Scholar]
- Jin, H.; Guo, D.; Yang, Q.; Yu, X.; Fu, X.; Yuan, F. Comprehensive evaluation of salt tolerance in soybean germination period by fuzzy membership function method. Mol. Plant Breed. 2021, 19, 8265–8271. [Google Scholar]
- Johnson, C.M.; Subramanian, A.; Pattathil, S.; Correll, M.J.; Kiss, J.Z. Comparative transcriptomics indicate changes in cell wall organization and stress response in seedlings during spaceflight. Am. J. Bot. 2017, 104, 1219–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, L.; Ou, X.; Liu, J.; Lin, X.; Sheng, L.; Liu, B. The spaceflight environment can induce transpositional activation of multiple endogenous transposable elements in a genotype-dependent manner in rice. J. Plant Physiol. 2009, 166, 2035–2045. [Google Scholar] [CrossRef]
- Ou, X.; Long, L.; Wu, Y.; Yu, Y.; Lin, X.; Qi, X.; Liu, B. Spaceflight-induced genetic and epigenetic changes in the rice (Oryza sativa L.) genome are independent of each other. Genome 2010, 53, 524–532. [Google Scholar]
- Verhage, L. Can transposable elements rewire transcriptional networks in the developing rice endosperm? Plant J. 2022, 109, 1033–1034. [Google Scholar] [CrossRef]
- Baduel, P.; Colot, V. The epiallelic potential of transposable elements and its evolutionary significance in plants. Philos. Trans. R Soc. Lond. B Biol. Sci. 2021, 376, 20200123. [Google Scholar] [CrossRef]
- González, J.; Karasov, T.L.; Messer, P.W.; Petrov, D.A. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 2010, 6, e1000905. [Google Scholar] [CrossRef] [Green Version]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Xie, J.; Xu, W.; Chen, S.; Zhang, X.; Liu, S.; Wu, J.; El-Kassaby, Y.A.; Zhang, D. Transposable Elements: Distribution, Polymorphism, and Climate Adaptation in Populus. Front. Plant Sci. 2022, 13, 814718. [Google Scholar] [CrossRef] [PubMed]
- Wos, G.; Choudhury, R.R.; Kolář, F.; Parisod, C. Transcriptional activity of transposable elements along an elevational gradient in Arabidopsis arenosa. Mob. DNA 2021, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, H.; Jin, J.; Cai, W. Single-base resolution methylome analysis shows epigenetic changes in Arabidopsis seedlings exposed to microgravity spaceflight conditions on board the SJ-10 recoverable satellite. NPJ Microgravity 2018, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef]
- Miousse, I.R.; Chalbot, M.-C.G.; Lumen, A.; Ferguson, A.; Kavouras, I.G.; Koturbash, I. Response of transposable elements to environmental stressors. Mutat. Res. Rev. Mutat. Res. 2015, 765, 19–39. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Grant, D.; Tian, Z.; Nelson, R.T.; Zhu, L.; Shoemaker, R.C.; Ma, J. SoyTEdb: A comprehensive database of transposable elements in the soybean genome. BMC Genom. 2010, 11, 113. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.M.; Subramanian, A.; Edelmann, R.E.; Kiss, J.Z. Morphometric analyses of petioles of seedlings grown in a spaceflight experiment. J. Plant. Res. 2015, 128, 1007–1016. [Google Scholar] [CrossRef]
- Grandbastien, M.-A. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta 2015, 1849, 403–416. [Google Scholar] [CrossRef]
- Springer, N.M.; Napoli, C.A.; Selinger, D.A.; Pandey, R.; Cone, K.C.; Chandler, V.L.; Kaeppler, H.F.; Kaeppler, S. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003, 132, 907–925. [Google Scholar] [CrossRef] [Green Version]
- Pontvianne, F.; Blevins, T.; Pikaard, C.S. Arabidopsis Histone Lysine Methyltransferases. Adv. Bot. Res. 2010, 53, 1–22. [Google Scholar] [PubMed] [Green Version]
- Cheng, K.; Xu, Y.; Yang, C.; Ouellette, L.; Niu, L.; Zhou, X.; Chu, L.; Zhuang, F.; Liu, J.; Wu, H.; et al. Histone tales: Lysine methylation, a protagonist in Arabidopsis development. J. Exp. Bot. 2020, 71, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, D.; Xu, W.; Kong, L.; Ye, X.; Zhuang, Q.; Fan, D.; Luo, K. Histone methyltransferase ATX1 dynamically regulates fiber secondary cell wall biosynthesis in Arabidopsis inflorescence stem. Nucleic Acids Res. 2021, 49, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Yushkova, E. Contribution of transposable elements to transgenerational effects of chronic radioactive exposure of natural populations of Drosophila melanogaster living for a long time in the zone of the Chernobyl nuclear disaster. J. Environ. Radioact. 2022, 251–252, 106945. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, T.; Mao, L.; Yan, S.; Chen, X.; Wu, Z.; Chen, R.; Luo, X.; Xie, J.; Gao, S. Genome-wide analysis of Dongxiang wild rice (Oryza rufipogon Griff.) to investigate lost/acquired genes during rice domestication. BMC Plant Biol. 2016, 16, 103. [Google Scholar]
- Phan, T.D.; Bo, W.; West, G.; Lycett, G.W.; Tucker, G.A. Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol. 2007, 144, 1960–1967. [Google Scholar] [CrossRef]
- Ramírez-Rodríguez, E.A.; McFarlane, H.E. Insights from the Structure of a Plant Cellulose Synthase Trimer. Trends Plant Sci. 2021, 26, 4–7. [Google Scholar] [CrossRef]
- Lou, H.; Tucker, M.R.; Shirley, N.J.; Lahnstein, J.; Yang, X.; Ma, C.; Schwerdt, J.; Fusi, R.; Burton, R.A.; Band, L.R.; et al. The cellulose synthase-like F3 (CslF3) gene mediates cell wall polysaccharide synthesis and affects root growth and differentiation in barley. Plant J. 2022, 110, 1681–1699. [Google Scholar] [CrossRef]
- Watanabe, Y.; Meents, M.J.; McDonnell, L.M.; Barkwill, S.; Sampathkumar, A.; Cartwright, H.N.; Demura, T.; Ehrhardt, D.W.; Samuels, A.; Mansfield, S.D. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 2015, 350, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, C.; Fujii, N.; Miyazawa, Y.; Kamada, M.; Kasahara, H.; Osada, I.; Shimazu, T.; Fusejima, Y.; Higashibata, A.; Yamazaki, T.; et al. The gravity-induced re-localization of auxin efflux carrier CsPIN1 in cucumber seedlings: Spaceflight experiments for immunohistochemical microscopy. NPJ Microgravity 2016, 2, 16030. [Google Scholar] [CrossRef] [Green Version]
- Gräfe, K.; Schmitt, L. The ABC transporter G subfamily in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [Green Version]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [PubMed]



| Sample ID | Clean_Data (Gb) | Total Read | Total UnMapped | Total Mapped |
|---|---|---|---|---|
| T75-1 | 35.64 | 253,778,974 | 1.44% | 98.56% |
| T75-2 | 27.62 | 196,657,892 | 1.61% | 98.39% |
| Z9-1 | 28.06 | 199,679,784 | 1.39% | 98.61% |
| Z9-2 | 43.22 | 307,682,454 | 1.23% | 98.77% |
| Sample | Downstream | Exonic | Exonic; Splicing | Intergenic | Intronic | Splicing | Upstream | Upstream; Downstream | UTR3 | UTR5 | UTR5; UTR3 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T75-1 | 98,838 | 66,681 | 17 | 1,160,059 | 150,634 | 704 | 109,347 | 9302 | 13,749 | 6402 | 69 | 1,615,802 |
| T75-2 | 98,393 | 66,482 | 17 | 1,155,317 | 150,316 | 701 | 108,614 | 9201 | 13,713 | 6394 | 73 | 1,609,221 |
| Z9-1 | 108,812 | 74,277 | 18 | 1,239,329 | 167,829 | 796 | 119,011 | 10,310 | 15,678 | 7157 | 103 | 1,743,320 |
| Z9-2 | 112,418 | 74,665 | 18 | 1,272,167 | 169,955 | 802 | 124,783 | 10,682 | 15,810 | 7204 | 111 | 1,788,615 |
| T75 vs. Z9 | 52,826 | 31,016 | 8 | 507,051 | 82,154 | 373 | 56,340 | 5281 | 8172 | 3792 | 59 | 747,072 |
| Sample | BND | DEL | DUP | INS | INV | CNS | Total |
|---|---|---|---|---|---|---|---|
| T75-1 | 22,468 | 7620 | 1049 | 2596 | 845 | 6459 | 41,037 |
| T75-2 | 21,930 | 7185 | 1032 | 2432 | 821 | 6406 | 39,806 |
| Z9-1 | 21,578 | 7720 | 1004 | 2844 | 880 | 6552 | 40,578 |
| Z9-2 | 23,676 | 8718 | 1092 | 3185 | 944 | 6654 | 44,269 |
| T75 vs. Z9 | 6428 | 3097 | 307 | 1332 | 295 | 569 | 12,028 |
| Primer | Gene Name | Product Size (bp) | Sequence (5′-3′) |
|---|---|---|---|
| G1-F | Glyma16g22390 | 4399 | GCCAAAGGGAAGCTTGGAGA |
| G1-R | GGCGTCCTACATGTTGCCTA | ||
| G2-F | Glyma02g38740 | 2313 | CGCTAGCTCTGCGATCATGT |
| G2-R | TTGTACCGCTGCTGAGAACA | ||
| G3-F | Glyma07g19820 | 1509 | TCAGACGAGCTGTACCCATC |
| G3-R | CATGTGTTTCGCCCTTGTGG | ||
| G4-F | Glyma07g13230 | 7711 | TTGTTGAGACGCGCTTTGTG |
| G4-R | TTGCTGGAAGGAGCCAAGAG | ||
| G5-F | Glyma06g02600 | 9392 | ACAATCACCGTTCGTCGGAA |
| G5-R | AGAACGGAAGTGAACGCAGA | ||
| G6-F | Glyma07g26460 | 5019 | AGTGGCACATCCGAAGTGAG |
| G6-R | ATGGTTTTCTTGGTGGGGCA | ||
| G7-F | Glyma02g02860 | 6462 | TTGCTCCGTGTTTGACCTGT |
| G7-R | TCAAACCAGGGTGCTTCGTT | ||
| G8-F | Glyma15g26810 | 6338 | TGGCAACTACGCACAATCCT |
| G8-R | ACCACCACATTCCTCGTGAC | ||
| G9-F | Glyma18g07740 | 757 | GGCCACGATGTGGAAGAGAC |
| G9-R | TGCATAGCCCTCCACATTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, H.; Fu, X.; Yu, X.; Zhu, L.; Yang, Q.; Yuan, F. Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight. Int. J. Mol. Sci. 2022, 23, 13721. https://doi.org/10.3390/ijms232213721
Jin H, Fu X, Yu X, Zhu L, Yang Q, Yuan F. Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight. International Journal of Molecular Sciences. 2022; 23(22):13721. https://doi.org/10.3390/ijms232213721
Chicago/Turabian StyleJin, Hangxia, Xujun Fu, Xiaomin Yu, Longming Zhu, Qinghua Yang, and Fengjie Yuan. 2022. "Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight" International Journal of Molecular Sciences 23, no. 22: 13721. https://doi.org/10.3390/ijms232213721
APA StyleJin, H., Fu, X., Yu, X., Zhu, L., Yang, Q., & Yuan, F. (2022). Genome-Wide Comparison of Structural Variations and Transposon Alterations in Soybean Cultivars Induced by Spaceflight. International Journal of Molecular Sciences, 23(22), 13721. https://doi.org/10.3390/ijms232213721





