Abstract
Non-coding RNAs (ncRNAs) regulate cell proliferation, migration, differentiation, inflammation, metabolism of clinically important biomolecules, and other cellular processes. They do not encode proteins but are involved in the regulatory network of various proteins that are directly related to the pathogenesis of diseases. Little is known about the ncRNA-associated mechanisms of atherosclerosis and related cardiovascular disorders. Remodeling of the extracellular matrix (ECM) is critical in the pathogenesis of atherosclerosis and related disorders; however, its regulatory proteins are the potential subjects to explore with special emphasis on epigenetic regulatory components. The activity of regulatory proteins involved in ECM remodeling is regulated by various ncRNA molecules, as evident from recent research. Thus, it is important to critically evaluate the existing literature to enhance the understanding of nc-RNAs-regulated molecular mechanisms regulating ECM components, remodeling, and progression of atherosclerosis. This is crucial since deregulated ECM remodeling contributes to atherosclerosis. Thus, an in-depth understanding of ncRNA-associated ECM remodeling may identify novel targets for the treatment of atherosclerosis and other cardiovascular diseases.
1. Introduction
Cardiovascular disease has been a major cause of death worldwide, posing serious concerns to human health for the past 20 years [1]. The pathophysiological basis for many prevalent cardiovascular disorders is atherosclerosis, a chronic inflammatory disease [2]. Lipoproteins deposit in the subintimal region, and subsequent oxidative responses mediate the process of plaque formation and the progression of atherosclerosis, which is more common in big or medium-sized arteries [3]. This is accompanied by macrophage recruitment and foam cell formation, movement of vascular smooth muscle cells (VSMCs) to the intima, and progression of atherosclerotic plaque. Plaques that extend into the artery cause duct stenosis, and rupture of plaque gives rise to emboli causing adverse ischemic events [4]. Remodeling of the extracellular matrix (ECM) is a major process involved in atherogenesis that changes the vasculature and affects its regulation. The early change in the atherosclerosis starts with deposition of fibronectin which is then predominantly occupied by deposition of collagen and cross-linking [5]. Subsequently, various complex changes like degradation of ECM proteins begin that lead to rupture of the plaque [6]. Although the pathophysiology of atherosclerosis and other cardiovascular diseases have been extensively studied, epigenetic regulation is largely not well studied. Therefore, it is critical to explore the epigenetic regulation of various molecular mechanisms involved in the pathogenesis of atherosclerosis to identify novel therapeutic strategies.
The discovery of non-coding ribonucleic acids (ncRNAs) changed our understanding of the post-translational, post-transcriptional, and epigenetic regulation of gene expression in controlling cellular homeostasis in various diseases. Recent breakthroughs in the field of genomics, facilitated by technologies such as chromatin immune-precipitation RNA sequencing (ChIP RNA Seq), Assay for Transposase-Accessible Chromatin (ATAC) seq, transcriptome analysis, and next-generation sequencing (NGS), have provided fresh insights and fundamentally altered our knowledge of small ncRNA molecules, which were long regarded as “junk DNA.” The fact that about 99% of the genome consists of non-coding DNA and approximately 1% codes for functional proteins demonstrates the intricacy and significance of ncRNAs in regulating gene expression [7,8,9]. Regulatory ncRNAs such as microRNAs (miRNAs; miRs) and long non-coding RNAs (lncRNAs) have had a profound impact on research in numerous domains, like cancer [10,11,12], cardiovascular diseases [13,14,15], and diabetes [16,17,18]. The epigenetic regulation of these ncRNAs is crucial in both the early development and the etiology of heart diseases [19,20,21].
Emerging approaches based on genomic data have changed diagnostic and therapeutic procedures, allowing for the early detection of problems, and providing hope for more successful treatments. The purpose of this article is to offer an up-to-date account of the involvement of noncoding RNAs (ncRNAs) in cardiovascular diseases with an emphasis on the regulation of extracellular matrix remodeling in atherosclerosis.
2. Extracellular Matrix
The extracellular matrix comprises various cells and cellular structures that constitute atherosclerotic plaque scaffolding. It is made up of different structural components that are regulated by a class of different regulatory factors. Collagens, hyaluronan, elastic fibers, proteoglycans, and many glycoproteins are essential elements of vascular ECMs that are all coupled in a complex dynamic 3D matrix system. This link controls the biomechanical properties of arteries and the phenotype of the cells such as ECs, VSMCs, adventitial fibroblasts, and immune cells invading circulation. VSMCs are the predominant cell types identified in terms of their ability to produce ECM macromolecules [22]. The development of atherosclerosis commences with focal endothelial cell injury in arteries, which enhances the invasion of freely circulating monocytes and T lymphocytes [23]. Monocytes differentiate into macrophages in the subendothelial intima, where they release cytokines and aggravate the inflammatory environment and endocytose LDL fragments and then become lipid-laden foam cells. Concurrently, the SMCs from the medial layer migrate into the intima, proliferate, and make collagen fibers to form a fibrous cap that stabilizes the intima. In contrast, forming a lipid-rich malignant core destabilizes the lesion, eventually leading to erosion in high-risk, rupture-prone plaques, causing thrombosis, which could develop arterial obstruction, resulting in myocardial infarction (MI) [24,25].
ECM Remodelling and Atherosclerosis
Throughout life, the structure and function of the vasculature are determined by the interaction between various ECM components. In the early phase of atherosclerosis, proteoglycans comprise the majority of the ECM, but as the disease progresses, collagens become the predominant ECM component, accompanied by a decrease in elastic fibers, glycoproteins, and proteoglycans [26]. Moreover, elastin and collagen are the most thoroughly researched ECM proteins in the etiology of atherosclerosis. Uncontrolled degradation of these proteins increases the course of atherosclerosis because it permits transendothelial migration of leukocytes, VSMC migration and proliferation, neovascularization, vascular cell death, neointima formation, and ultimately the rupture of the arterial wall [27]. Elastic fibers, composed of elastin components, stabilize collagen; hence, damaged elastin (seen in plaques), resulting in the inability to generate a stable matrix, which leads to plaque rupture [28].
Lipoproteins containing apo-B penetrate the arterial intimal endothelial layer and concentrate in the subendothelial region, where they are endocytosed by intimal macrophages. Simultaneously, local blood flow disruptions (non-linear flow) in atherosclerosis-prone locations (e.g., arterial branches) result in decreased shear stress, which is recognized by endothelial cells during the mechano-transduction process. These processes alter the microenvironment of the arterial wall intima, promoting subsequent changes such as foam cell generation, VSMC migration and conversion from contractile to synthetic phenotypes, ECM matrix remodeling, and necrotic core formation and calcifications [25,29]. Furthermore, the immune system is critical in the pathogenesis of atherogenesis [30]. Many plaques mature into stable structures that cause the chronic coronary syndrome, while some of them undergo ultrastructural changes that make them prone to rupture. These plaques are referred to as ‘unstable’ or ‘susceptible’ [31]. According to the reported characteristics, susceptible plaque is a thin cap fibroatheroma (TCFA) with a necrotic core and an overlaying fibrous cap of 65µm thickness [32]. The presence of susceptible plaques is required for the occurrence of significant cardiovascular adverse effects [33]. As a result, medications aiming to stabilize atherosclerotic plaque are required. However, due to clinical denial, ideal therapy focused on stabilizing plaques must focus on the advancement of molecular stabilizing paths ‘in general’ [34,35,36,37,38,39,40] instead of on the stabilization of specific atherosclerotic lesions.
3. Non-Coding RNA
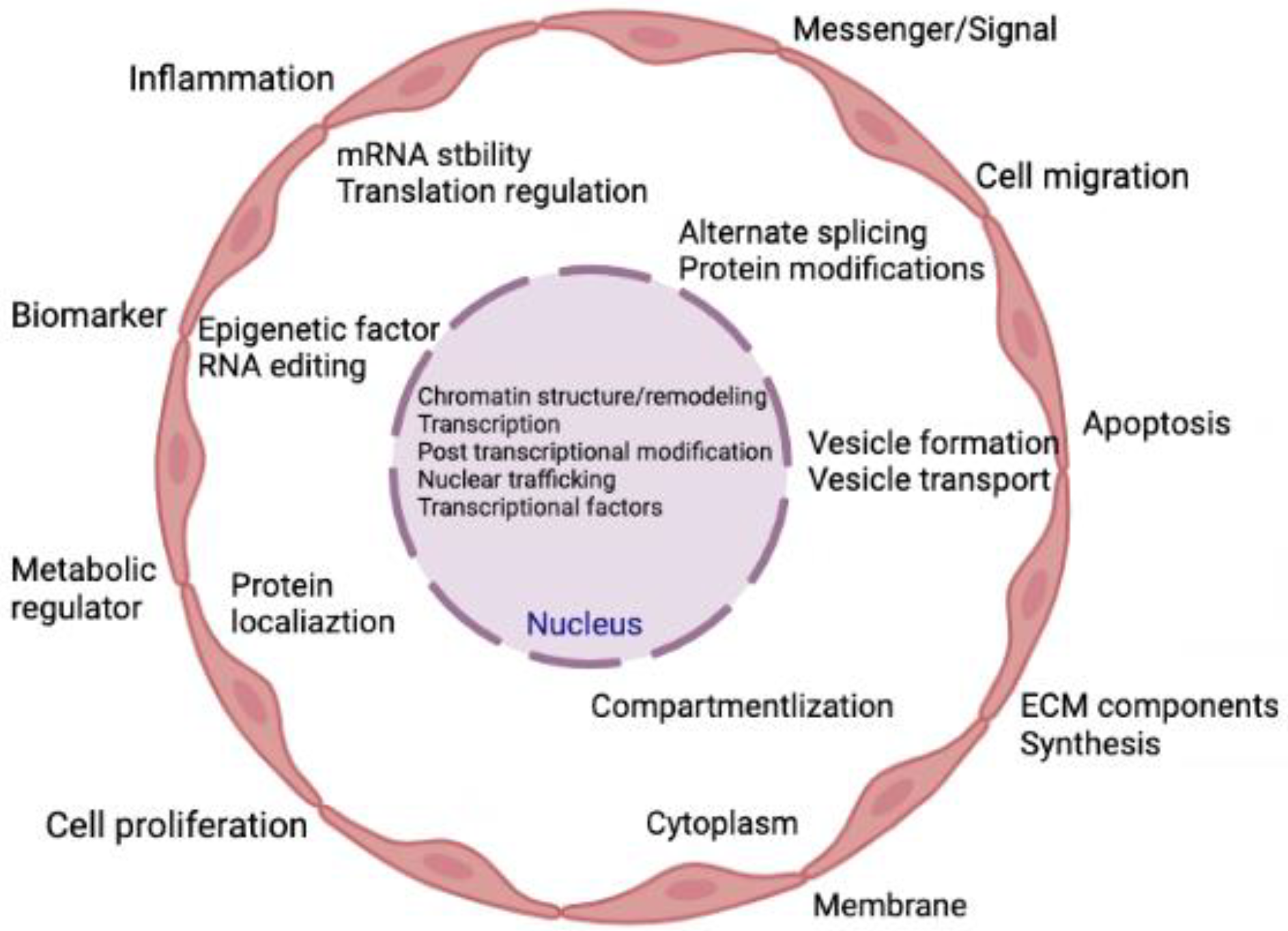
Transcripts that are not translated into polypeptides or proteins are known as non-coding RNAs (ncRNAs). Approximately 1–2% of genes are responsible for making proteins, suggesting that there are many non-coding genes with unidentified roles [41]. These ncRNAs demonstrate various biological roles and directly participate in several physiological processes [42]. Prior research on ncRNAs focused mostly on their regulatory functions within cells. Later, extracellular vesicles (EVs) were thoroughly investigated, and it was discovered that these EVs contain lipids, proteins, messenger RNAs (mRNAs), and ncRNAs with biological functions [43,44]. The ncRNAs typically exist in EVs or attach to proteins or lipids to avoid ribonuclease-mediated destruction [45]. The majority of ncRNAs in the blood are either contained in EVs [46] or are protein-bound, such as with lipoproteins [47], argonaute protein (AGO2), and nucleophosphoprotein 1 (NPM1) [48]. Recent research revealed that extracellular ncRNAs control intracellular gene expression, mediate intercellular communication, and are intimately connected to numerous pathogenic processes [49,50,51,52,53]. Figure 1 represents the various cellular processes being controlled by the action of various ncRNAs.
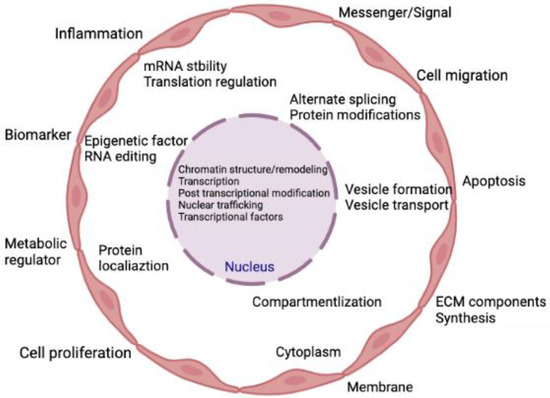
Figure 1.
Summary of the cellular activities that are regulated by ncRNAs. Indeed, ncRNAs can directly and simultaneously modulate multiple targets and are involved in both gene expression and genome remodeling. Thus, ncRNAs control cellular functions directly or indirectly in both physiological and pathological conditions.
According to recent findings, extracellular ncRNAs are thought to be closely associated with atherosclerosis. Extracellular ncRNAs are crucial regulators of many cells, including immune cells, macrophages, and endothelial cells. They play a role in atherosclerotic processes such as angiogenesis, foam cell formation, and atherosclerotic plaque progression and rupture [41,54,55]. Extracellular ncRNAs serve as a useful diagnostic indicator and potential treatment target in atherosclerosis [56]. The association of ncRNAs to disease development, diagnosis, therapy, and incidence, in the past few years has seen a major advancement in the study of ncRNAs related to cardiovascular disorders. This review article critically discusses the specific function, putative mechanism, and prospective applications of ncRNAs in atherosclerosis, emphasizing miRNAs, circRNAs, and lncRNAs-mediated ECM remodeling in atherosclerosis.
3.1. miRNAs and Atherosclerosis
The miRNAs are genetically conserved, containing 18–24 nucleotides, small single-stranded non-coding RNAs that regulate gene expression at the post-transcriptional level by binding to the 3′-untranslated region of certain target mRNA sequences, thereby reducing protein synthesis by inhibiting mRNA translation [10,57,58,59]. There are more than 60% of human protein-coding genes have miRNA target sites in their 3′-UTR, and various studies have shown the involvement of miRNA/mRNA interactions as the key regulatory network in different biological processes [60,61,62]. With the unique characteristics of miRNAs, these have been extensively used as key regulators of mRNA and protein expression in many diseases, including cardiovascular diseases [61,62,63,64]. Various studies (Table 1) have analyzed the role of miRNAs in atherosclerosis and ECM remodeling.

Table 1.
miRNAs, their target, and functions.
The expression of MiR-1a-3p, miR-1b-5p, and miR-1 was found to be the most prominently increased in different diseases related to subclinical atherosclerosis. The miR-1 mimics can activate endothelial inflammation through increased production of E-selectin, intercellular adhesion molecule (ICAM)-1, and vascular cell adhesion molecule (VCAM)-1 at both the mRNA and protein levels. The in-vivo findings showed that miR-1 knockdown by antagomiR-1 reversed the endothelial and inflammatory activation at the lesion site, revealing a novel therapeutic target for atherosclerosis [19]. Wu et al. [111] demonstrated that miR-142-5p targeted myocardin-like protein 2 to drive the amplification and migration of human aortic smooth muscle cells, promoting atherosclerosis. Su et al. [112] reported the presence of miR-181a-5p and miR-181a-3p in atherosclerotic lesions of ApoE mice fed with a high-fat diet and in the plasma of patients with coronary artery disease (CAD). These findings indicate the potential role of these two miRNAs in atherogenesis. Also, the overexpression of miR-181a-5p and miR-181a-3p in ApoE mice decreased the plaque size. In contrast, the gain-of-function mutation decreased inflammatory genes like ICAM-1 and VCAM-1 and leukocyte infiltration in the aortic intima.
Raitoharju et al. [113] reported 58 miRNAs that were differentially expressed between atherosclerotic plaques and non-atherosclerotic left internal thoracic arteries. Of these, up-regulated five miRs viz. miR-21, miR-34a, miR-146a, miR-146b-5p, and miR-2010 were involved in the regulation of 187 mRNAs in atherosclerotic plaques. The proteins translated from these genes were involved in signal transduction, transcription control, and vesicular transport. In another study, it was reported that miR-130a expression was increased in atherosclerotic mice. In an in-vitro model of atherosclerosis, miR-130a overexpression enhanced inflammatory factors like tumor necrosis factor (TNF)-α and interleukin (IL)-1, IL-6, and IL-8 and its downregulation reduced the inflammation by attenuating TNF-α, IL-1, IL-6, and IL-8. Furthermore, in the in-vitro model, over-expression of miR-130a might reduce peroxisome proliferator-activated receptor (PPAR) protein expression while inducing NF-κB protein expression. Still, its suppression promoted PPAR protein expression while suppressing NF-κB protein expression. PPAR activation inhibited the pro-inflammatory effects of miR-130a in an atherosclerosis-induced in-vitro model [114]. Polyakova et al. [115] reported that the SYNTAX (tool to score complexity of CAD) score I index and serum miR-203 expression level exhibited a positive association in patients with CAD. In the atrial myocardium of patients with triple vessel disease, miR-27a, miR-133a, and miR-203 expressions were substantially greater than those of patients with 1–2 vessel disease. This association was also observed for miR-27a, miR-133a, and miR-203 expressions in the blood. Another study revealed that targeting miR-33 in atherosclerotic macrophages by anti-miR-33 conjugated pH low-insertion peptide (pHLIP) constructs to inhibit miR-33 improves collagen formation and reduces lipid accumulation, thereby improving atherosclerotic regression. Additionally, a single-cell RNA sequencing study showed that macrophages from atherosclerotic lesions targeted by pHLIP-anti-miR-33 had lower levels of matrix metalloproteinase (MMP)-12 and greater levels of fibrotic genes (Col2a1, Col3a1, Col1a2, Fn1, etc.) and tissue inhibitor of metalloproteinase (TIMP)-3 [116].
Another study reported that the expression of three miRs, miR-129-1-3p, miR-4312, and miR-5196-3p differed significantly between the acute ischemic stroke and atherosclerosis/healthy control groups. Twelve pathways were affected by the miR-129-1-3p target genes, three of which were related to axonal and synaptic function: sphingolipid signaling, retrograde endocannabinoid signaling, and axon guidance. Cortical neurite length and Runx2 levels were considerably reduced by miR-129-1-3p mimics, whereas Runx2 expression was elevated, and neurite growth was boosted by miR-129-1-3p inhibitors [117].
Egea et al. [118] demonstrated that the treatment of human mesenchymal stem cells (hMSCs) with LL-37 boosted let-7f and N-formyl peptide receptor 2 (FPR2) production, which ultimately helped in the stabilization of atherosclerotic plaque. Circulating hMSCs attach to athero-prone endothelium more frequently in an ApoE animal model of atherosclerosis. High levels of let-7f in the hMSCs, as determined by two-photon laser scanning imaging and ex-vivo artery perfusion, contributed to increased attachment of MSCs. Additionally, the exposure of hMSCs to homogenized human atheromatous plaque material significantly increased the production of different cytokines, chemokines, MMPs, and TIMPs. Moreover, the exposure of hMSCs to human plaque extracts causes hMSCs to differentiate into cells of the myogenic lineage, indicating a potential stabilizing influence on the plaque.
3.2. circRNAs and Atherosclerosis
A group of RNA molecules known as circRNAs is produced through exon reverse splicing or intron lariat. Due to their closed ring structure, which shields them from the effects of RNA exonuclease, circRNA production is generally stable and tissue- and developmental stage-specific [119]. Due to the self-regulating, transposing, and other salient features of circRNAs, many studies have been recently conducted to investigate the role of circRNAs in the initiation and progression of atherosclerotic plaque and other cardiovascular diseases [120,121]. The studies have proposed the diagnostic value of different circRNAs in preventing and treating atherosclerosis (Table 2).

Table 2.
Circular RNAs, their targets, and functions.
To discover circRNAs involved in atherosclerosis, human umbilical vein endothelial cells (HUVECs) stimulated with oxidized low-density lipoprotein (ox-LDL) were subjected to circRNA microarray analysis, where Hsa circ 0003575 showed the highest upregulation among all the circRNAs. Loss-of-function tests demonstrated that Hsa circ 0003575 inhibits endothelial cell (EC) growth, promotes apoptosis, and may act as a sponge for miRs miR-199-3p, miR-9-5p, miR-377-3p, and miR-141-3 [140]. Some circRNAs, such as ANRIL and LincP21, have significantly higher circulating levels and are associated with the severity of atherosclerosis [141,142].
CircRNAs play a major role in atherosclerosis and CAD [143,144,145,146]. In patients with CAD, nine circRNAs were reported by Pan et al. [147]. Ox-LDL treatment of HUVECs and feeding a high-fat diet to mice resulted in a downregulation of circHIPK3 expression, whereas overexpressing circHIPK3 increased autophagy, which was inhibited in atherosclerosis [143]. The expression of circHIPK3 was downregulated in mice on a high-fat diet and in ox-LDL-treated HUVECs. The level of autophagy was decreased in atherosclerosis, which was reversed by the overexpression of circHIPK3. Meanwhile, forced expression of circHIPK3 would reduce the accumulation of lipids in HUVECs.
In an atherosclerotic rabbit model, analysis of a variably expressed circRNA-miR-mRNA triple network showed that competition among circRNAs and their mRNAs might be a key factor in the onset of atherosclerosis [16]. When Hsa circ 0030042 was overexpressed, it acted as an internal eukaryotic initiation factor, inhibiting ox-LDL-induced aberrant autophagy in HUVECs, and sustaining plaque stability in-vivo. Furthermore, Hsa circ 0030042 inhibited autophagy by sponging eIF4A3 and preventing its recruitment to the mRNAs for beclin1 and forkhead box O1 (FOXO1), though the suppression of beclin1 and FOXO1 caused by Hsa circ 0030042 was offset by increased eIF4A3 expression or decreased Hsa circ 0030042 interaction. In ApoE−/− rats fed a high-fat diet, Hsa circ 0030042 also increased plaque stabilization and reversed eIF4A3-induced plaque instability [148]. A microarray examining the circRNAs in the peripheral blood of CAD patients showed a strong correlation of hsa-circRNA11783-2 with the condition, and Hsa circ 0008507, Hsa circ 0001946, and Hsa circ 0000284 are independent risk factors for CAD [149]. Wang et al. [150] revealed that in CAD patients, 624 circRNAs and 171 circRNAs were significantly elevated and downregulated, respectively, compared to controls. In large cohorts, Hsa circ 0001879 and Hsa circ 0004104 were shown to be considerably elevated. The combination of Hsa circ 0001879 and Hsa circ 0004104, along with CAD risk variables, performed best in distinguishing CAD patients from healthy controls. Additionally, two non-coding RNA, namely, ANRIL (antisense non-coding RNA at the INK4 locus) and circANRIL (circular ANRIL), transcribed at the chromosome 9p21 region, were found to be associated with a high risk of cardiovascular disease. However, it was discovered that they had opposing effects on the onset of CAD. Although upregulation of circANRIL prevented the onset of CAD [151], upregulation of ANRIL was linked to an increase in the incidence of atherosclerosis [141].
The circRNAs are crucial in controlling the stability of atherosclerotic plaques, as in acutely ruptured carotid plaques. It was discovered that circRNA-16 was elevated while miR-221, which is linked to VSMC proliferation and death, was downregulated [152]. Axis inhibition protein 2 was another target of miR-221-3p, which enhanced the proliferation of pulmonary arterial smooth muscle cells. Therefore, through the miR-221/Ets-1 and AXIN2 axes, circRNA-16 may play a significant regulatory function in the stability of arterial plaques [153].
3.3. lncRNA and Atherosclerosis
Several lncRNAs with a role in atherosclerosis have been identified. lncRNAs are expressed in different cell types, present in atherosclerotic lesions, and have been implicated in several atherogenic processes, such as endothelial dysfunction and lipid deposition [154]. Some of the lncRNAs, their targets, and their functions are listed in Table 3.

Table 3.
Long noncoding RNAs, their targets, and functions.
LncRNAs are more than 200 nucleotides long and account for the majority of ncRNA [179,180]. However, less than 5% have been characterized to date, owing in part to poor conservation among species [181,182,183]. Although lncRNAs lack functional initiation codon and termination codons [184], some lncRNAs have been shown to translate into micropeptides [185]. In a study conducted by Ann et al. [186], it was shown that among the 380 RNAs that differed in expression between plaque and control tissues, lncRNA HSPA7 was increased by oxidized low-density lipoprotein (oxLDL). HSPA7 knockdown decreased human aortic smooth muscle cell migration as well as IL-1 and IL-6 secretion and expression. However, HSPA7 knockdown reversed the oxLDL-induced reduction in contractile marker expression. HSPA7 had an effect on miR-223 via an AGO2-dependent mechanism. HSPA7 is variably expressed in human atheroma and promotes transdifferentiation of contractile VSMCs phenotype to inflammatory de-differentiated/secretory phenotype through sponging miR-223. Li et al. [162], examining the serum samples of 38 patients with atherosclerosis, found that the level of the lncRNA TUG1 had dramatically increased in atherosclerotic plaques and VSMC damage models, and the expression of the lncRNA TUG1 was likewise elevated. A study by Hu et al. [187] demonstrated significant downregulation of the NEXN gene, lncRNA gene, and NEXN-AS1 in atherosclerotic lesions. An in-vivo experiment showed that the lncRNA NEXN-AS1 could increase the expression of NEXN in ECs and that NEXN-AS1 overexpression decreased endothelial inflammatory activation by blocking the NF-κB pathway [187]. It is widely accepted that oxLDL is one of the most potent inflammatory triggers for atherosclerosis and that autophagy is the survival mechanism for cells under stress. Studies demonstrated that oxLDL lowered the number and activity of mature-Cathepsin D, resulting in decreased lysosome activity, which largely contributed to impaired autophagic flux and decreased cell survival during atherogenesis [188,189].
In another study conducted by Vacante et al. [190], it was demonstrated that lncRNA CARMN and related microRNAs were downregulated in advanced versus early atherosclerotic lesions in humans and animals. Under homeostatic settings, CARMN deletion affected the expression of miR-143 and miR-145. When atherosclerosis was produced in mice, CARMN deletion increased the volume, size, and content of proinflammatory Lgals3 (galectin 3)-expressing cells and altered plaque composition, resulting in an advanced phenotype. Wang et al. [191] reported that in atherosclerotic mice and ox-LDL-stimulated VSMCs, SNHG16 and HMGB2 expression were enhanced, but the miR-22-3p expression was decreased. SNHG16 inhibited miR-22-3p expression through direct binding, and miR-22-3p mimicked reduced proliferation, migration, and invasion in ox-LDL-treated VSMCs. In addition, because HMGB2 was a target of miR-22-3p, SNHG16 increased HMGB2 levels by functioning as a competitive endogenous RNA (ceRNA) of miR-22-3p. The sh-HMGB2 inhibited ox-LDL-induced VSMC proliferation, migration, and invasion when combined with a miR-22-2p inhibitor. Through miR-22-3p/HMGB2 axis, SNHG16 accelerated atherosclerotic plaque production and increased ox-LDL-activated VSMC proliferation and migration. It has been observed that EC pyroptosis and atherosclerotic plaque formation were greatly reduced when Gaplinc was silenced. Gaplinc may interact with SP1 to bind to the NLRP3 promoter and upregulate NLRP3 target gene expression in high-fat diet-fed animals, promoting EC pyroptosis and atherosclerotic plaque growth [192]. Ni et al. [193] studied lncRNA from smooth muscle cells, which regulates cell plasticity and atherosclerosis by interacting with serum response factors. It was observed that CARMN, a lncRNA, is a key regulator of VSMC plasticity and atherosclerosis. Moreover, it was documented that in HUVECs, plasmacytoma variant translocation (PVT)1 knockdown reduced ox-LDL-induced inflammation, apoptosis, and oxidative stress. PVT1 worked as a sponge for miR-153-3p, while growth factor receptor binding protein 2 (GRB2) was identified as a miR-153-3p target. Overexpression of MiR-153-3p reduced the effects of PVT1 on ox-LDL-induced cell injury. Overexpression of GRB2 reduced the protective effects of miR-153-3p against ox-LDL-induced damage. Inhibition of PVT1 attenuated the activation of the ERK1/2 and p38 pathways via the miR-153-3p/GRB2 axis. Furthermore, in atherosclerotic mice, silencing of the PVT1 gene reduced atherosclerotic plaques, lipid formation, inflammation, oxidative stress, and apoptosis [194].
Though most atherosclerotic plaques are therapeutically silent, inflammation and persistent monocyte mobilization lead to plaque growth and instability, which might result in potentially fatal complications like myocardial infarction (MI), dementia, and brain/cerebral edema. LncRNA CCL2 controls the expression of the CCL2 gene, which codes for monocyte chemoattractant protein 1 and increases the course of vascular inflammation [195]. lncRNA NEAT1, which also interacts with a chromatin modification and decreases the production of smooth muscle cell proteins, hence promoting the phenotypic switch of VSMCs from a contractile to a synthetic state, has also been demonstrated to enhance plaque destabilization [196].
4. Regulation of ECM Components by ncRNAs
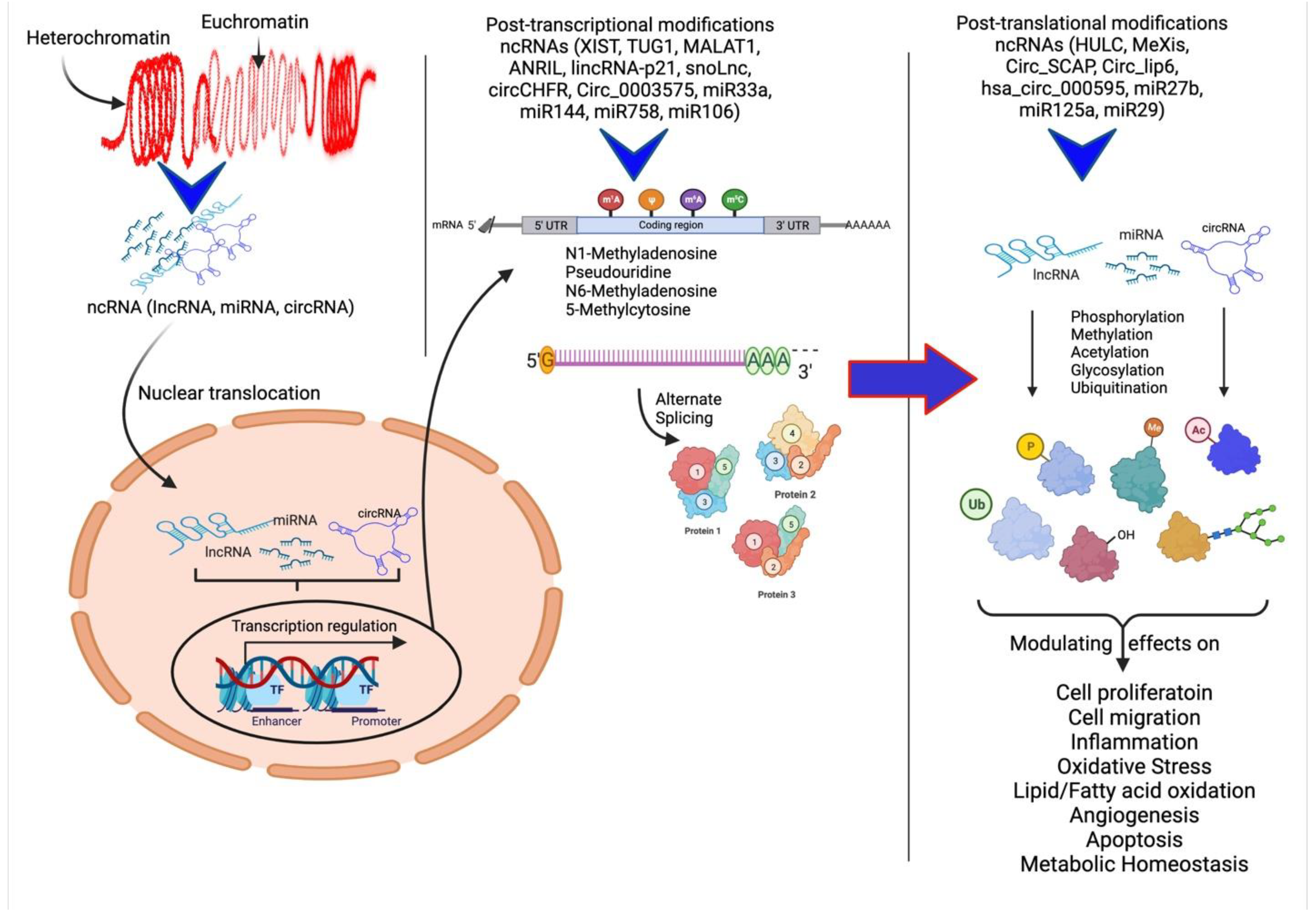
In the past few years, various studies have investigated the upregulation or downregulation of ncRNAs and their modulation in different animal models, clinical samples, and cell systems that mimic different diseases or diseased states to understand their specific role. The ncRNAs are directly or indirectly involved in the regulation of expression of different ECM components within the atherosclerotic plaque, discussed below. The ncRNAs regulate the gene expression of ECM proteins and different cellular processes and their modulatory effects on plaque pathogenesis through post-transcriptional and post-translational regulation (Figure 2).
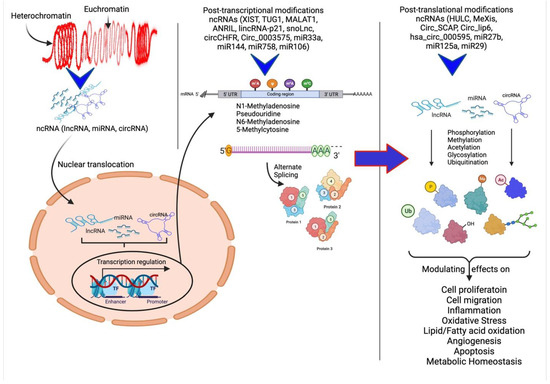
Figure 2.
Schematic representation of ncRNAs acting on various cellular processes and their modulatory effects. Non-coding RNAs (ncRNAs) regulate gene expression at the transcriptional and post-transcriptional levels and are also involved in the epigenetic regulation of various genes. The ncRNAs play a critical role in heterochromatin formation and histone modification involving methylation, acetylation, ubiquitination, citrullination, alternative splicing, and gene silencing. Modified protein structure and levels regulate various molecular mechanisms involved in angiogenesis, cell proliferation and migration, inflammation, and remodeling.
The production of type III collagen is regulated by miR-29, whose target gene is COL3A. As shown in atherosclerotic mice, chronic administration of miR-29 antagonist (LNA-miR-29) results in beneficial plaque remodeling [197]. In human leiomyomas, the miR-29 effect on collagen type III expression has also been confirmed [198]. In addition, collagen type VIII may play a crucial role in the plaque destabilization process. These short collagen fibers stimulate the formation of atherosclerotic plaques by encouraging the migration and proliferation of smooth muscle cells (SMCs). In addition, apolipoprotein E (ApoE) is an endogenous inhibitor of collagen type VIII, which may explain why ApoE−/− mice develop atherosclerosis. Lopes et al. [199] reported that double-knockout Col8−/− ApoE−/− mice display a more susceptible plaque with a thin fibrous cap than single knockout ApoE−/− mice. However, type I collagen reduces arterial flexibility. The amount of miR-145 is decreased in ApoE−/− mice, resulting in enhanced expression of the lysyl oxidase gene (LOX). Lysyl oxidase crosslinks collagen helices and strengthens collagen fibers, hence increasing the arterial rigidity of these mice [200]. The expression of elastin is controlled by microRNA from the miR-15 family (particularly miR-195) and the miR-29 family, and these inhibit the expression of collagen and proteoglycan. Antagomir-29b significantly reduces aortic aneurysm diameter in ApoE−/− mice, whereas the miR-195 serum level corresponds with the aortic aneurysm diameter in humans [201]. Surprisingly, this is an inverse association, as miR-195 inhibits elastin and collagens and the ECM-degrading enzyme MMP-9 [201]. The molecule miR-181b is an additional epigenetic regulator of elastin gene expression [202]. In ApoE−/− mice, its suppression by anti-miR-181b reduced the formation of aortic aneurysms, increased the fibrotic response, and stabilized atherosclerotic plaques or aneurysms. Decorins, a proteoglycan, are frequently used in relation to microRNA involvement. The expression of this gene is negatively regulated by miR-181b, as proven by studies on hypertrophic scars [203]. Decorin also stimulates the activation of proinflammatory macrophages via PDCD4 (programmed cell death 4) and adversely regulates miR-21 expression. Given that miR-21 is considered an oncogene (oncomir), decorin appears to inhibit cancer development [204]. However, it is thought that hyaluronic acid increases miR-10 expression. miR-10 stimulates blood vessel development by direct control of fms-related tyrosine kinase-1 (flt-1) and Mib-1 [205,206]. The significance of hyaluronic acid in the instability of atherosclerotic plaques and its regulation by microRNA molecules must be explored. Specifically, the hyaluronic acid receptor CD44 is blocked by miR-328, which has been observed in renal tubular cells [207]. Notably, proteoglycan expression can also be regulated by miR-599 in conjunction with collagen expression [208].
Peptidylarginine deiminase (PAD) plays an important role in ECM stability and remodeling. Increased levels of PAD in cardiovascular diseases (CVDs), including atherosclerosis, coronary heart disease, venous thrombosis, cardiac fibrosis, heart failure, and acute inflammation, suggesting its critical role in CVDs. PAD-mediated deamination or citrullination is involved in various physiological and pathological conditions in the body [209]. Citrullination, a post-translational process, causes the deamination of arginine (Arg) and conversion of peptidyl-based arginine to peptidyl-based citrulline. This alters the original three-dimensional structure and function of target proteins and results in dysregulated inflammatory signaling [210]. MMPs play a critical role in ECM remodeling, and along with glycosylation, nitrosylation, and proteolysis, citrullination is also involved. Hypercitrullination of MMP-9 results in a higher affinity for MMP-9 gelatin compared to control MMP-9 [211]. Further, the association of PAD-mediated citrullination of fibronectin, an important constituent of ECM, with CVDs, fibrosis, carcinogenesis, rheumatoid arthritis, alteration of integrin clustering, and focal adhesion stability suggests its role in regulating vascular remodeling because fibronectin-mediated inflammatory signaling through integrin α5 is important for vascular remodeling [212,213,214].
Collagen and elastin are the main ECM components contributing to the structural matrix and elasticity of the arteries. Collagen type I, III, IV, V, VI, XVI, XVII, nidogen, perlecan, agrin, fibronectin, laminin, and prostaglandins (PGs) are major components of the vascular wall, and type I and III fibrillar collagens, chondroitin sulfate, and dermatan sulfate PGs, and fibronectin are major ECM component in the adventitia. During remodeling, the levels of these components get altered to provide a favorable microenvironment to get a vessel to remodel during CVDs [210,215]. Various mediators regulate ECM and vascular remodeling, and post-transcriptional regulation is an important evolving aspect (Table 4). The studies presented in Table 4 suggest that IncRNAs play a regulatory role in the expression of various ECM components and the proteases modulating their expression. These findings are further supported by the involvement of ox-LDL with the inflammatory response of macrophages in atherogenesis [216], LASER, LeXis, and CHROME IncRNA in cholesterol homeostasis and foam cell formation, and MANTIS, lncRNA-CCL2, and MALAT1 in vascular inflammation [154]. Further, the functional relevance of IncRNAs with atherosclerosis [217] and the association of MALAT1, GAS5, lncRNASNP, HAND2-AS1, H19, and others, and miRNAs in atherosclerotic plaque formation [218,219] support the notion that IncRNA plays an important role in atherosclerotic plaque formation and progression. Moreover, the regulation of smooth muscle cell proliferation and calcification plays a critical role in plaque formation and regulation of MMP-16, co-expressed with MMP-2 and MMP-9 and various other MMPs by IncRNAs [220]. All these effects support the role of and warrant a further in-depth understanding of the role of lncRNA-mediated regulation of plaque formation and progression, ECM and vascular remodeling, and associated complications.

Table 4.
Various ECM components regulated by ncRNAs.
5. Translational Aspects and Clinical Significance
As discussed above, the expression levels of various components of ECM are regulated by ncRNA. However, the research studies investigating this correlation are limited in the literature. The available studies and clinical trials (NCT03603431, NCT03494712, NCT02603224, and NCT04045405) [237] have discussed the role of miR-92a, miR-29b, and miR-132 in association with cutaneous healing and cardiac fibrosis, both having similar pathogenesis of inflammation and ECM remodeling. This implies that these miRNAs may also regulate ECM remodeling during plaque formation and progression, an inflammatory pathology of the vessels. This notion is supported by the fact that miR-92a is involved in angiogenesis, vascular inflammation, and vasodilation; miR-29b regulates elastin degradation; miR-132 regulates vascular smooth muscle cell proliferation and neointimal hyperplasia [66,67,99,102,103]. Although the studies investigating ncRNA-mediated ECM remodeling are limited, the involvement of ncRNAs regulating molecular mechanisms in plaque pathogenesis warrants further research. In the context of the treatment of plaque pathology, preclinical investigations have proven that several methods have a plaque-stabilizing impact by targeting apolipoprotein E, apolipoprotein B, and LDLs in SMCs, macrophages, monocytes [238,239,240,241]. Most of these studies are in animal models; thus, the positive outcomes have not been replicated in human clinical trials [242,243,244,245,246]. This may be due to different molecular compositions (macrophage subsets), locations, pathophysiological processes involved in atherosclerotic plaque instability, the animal model used, and varying human populations [247,248,249].
The clinical trials conducted in the treatment of atherosclerosis are mainly focused on the outcomes of cardiovascular diseases and acute ischemic events. Canakinumab administered to individuals with a prior myocardial infarction resulted in a substantial decline in subsequent cardiovascular problems in comparison to placebo [250]. Accordingly, in the COLCOT study [251], colchicine administered to patients after a myocardial infarction resulted in a considerable decline in composite endpoint and a significant reduction in recurrent myocardial infarction. In comparison, an experiment called STABILITY with darapladib, which was performed on patients with stabilized cardiac artery disorder (no prior myocardial infarction), was unable to show a statistically considerable difference between the darapladib and placebo groups in terms of composite endpoint and mortality, despite showing a subtle but notable decline in significant cardiac problems [252]. Similarly, the cholesterylester transfer protein (CETP) inhibitor anacetrapib, which causes an increase in HDL, showed a minor but substantial reduction in major coronary events [253]. The results from these and other clinical trials (Table 5) suggest that these drugs mainly stabilize plaque or attenuate atherosclerosis and target the ncRNA involved in ECM remodeling, inflammation, stabilization of atherosclerotic plaque, or other related events will be of significance in the treatment of atherosclerosis. Of note, to determine whether a specific type of therapy results in atherosclerotic plaque stability, the composition and morphology of the plaque must be visualized, and their stability exponents must be assessed using intravascular ultrasonography and optical coherence tomography [254,255] to enhance the therapeutic efficacy of the agent under consideration.

Table 5.
Clinical trials in atherosclerosis.
A convergence of basic and clinical research has significantly transformed the strategies for managing atherosclerosis and involves mainly targeting inflammatory components. This was mainly due to the advancement in the approach of randomized clinical trials involving individuals with an atherosclerotic plaque at different stages and treatment strategies. Furthermore, understanding plaque pathology has also been aided by improvements in human genetic studies enabled by next-generation sequencing and other technological innovations, along with an ever-evolving toolbox in the form of genetically modified mice models allowing for gene-editing and induced pluripotential stem cell methodology [267]. Understanding the activities of ncRNAs in atherosclerosis has progressed beyond DNA and mRNA analyses because of the involvement of microRNAs and lncRNAs regulating gene transcription in atherosclerosis [67,268].
6. Conclusions
Based on the studies discussed in this article, it is evident that ECM remodeling is epigenetically regulated involving miRNAs, lncRNA, and circRNA, and these ncRNAs regulate the expression of various proteins involved during plaque formation and vulnerability. Since ECM remodeling plays a critical role in plaque vulnerability to stabilize plaque, ncRNAs can be strong contenders to target. Additionally, the levels of these change during the process of plaque formation, as evidenced by various studies. ncRNAs may also serve as diagnostic and prognostic biomarkers for atherosclerosis. Therefore, ncRNAs can be strong contenders for therapeutic targets for atherosclerosis and related disorders, and the identification and characterization of relative ncRNAs may have clinical applications, both as prognostic tools and for therapeutic targets. Further investigations are required to develop and use specific ncRNAs in diagnosis and therapeutics in patients with cardiovascular diseases. Translating these scientific advancements in therapeutics has necessitated large-scale clinical trials, which have necessitated increased creativity and money due to the success of conventional treatments. Placebo-controlled, randomized clinical trials continue to be the most reliable approach for evaluating the applicability of lab findings to patients. Indeed, the globalization of cardiovascular disease risk has raised the overall burden of atherosclerotic disease. However, the progress in laboratory and clinical research promises to provide us with methods to combat this global epidemic. To make progress in the control of atherosclerosis, a multidisciplinary collaboration of public health measures, applied behavioral psychology, risk factor control, consistent implementation of existing therapies, and the development and validation of new therapeutic approaches will be required.
Author Contributions
Concept and design: D.S., V.R. and D.K.A.; Literature Search: D.S. and V.R.; Critical review and interpretation of the findings: D.S. and V.R.; Drafting the article: D.S. and V.R.; Revising and editing the manuscript: V.R. and D.K.A.; Final approval of the article: D.S., V.R. and D.K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the research grants R01 HL144125 and R01HL147662 to DKA from the National Heart, Lung, and Blood Institute, National Institutes of Health, USA. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable since the information is gathered from published articles.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- Salekeen, R.; Haider, A.N.; Akhter, F.; Billah, M.M.; Islam, M.E.; Didarul Islam, K.M. Lipid oxidation in pathophysiology of atherosclerosis: Current understanding and therapeutic strategies. Int. J. Cardiol. Cardiovasc. Risk Prev. 2022, 14, 200143. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Maseri, A. Inflammation and atherosclerosis. Circulation 2002, 105, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. Role of Macrophages and RhoA Pathway in Atherosclerosis. Int. J. Mol. Sci. 2020, 22, 216. [Google Scholar] [CrossRef] [PubMed]
- Sapa-Wojciechowska, A.; Rak-Pasikowska, A.; Pormanczuk, K.; Czapla, B.; Bil-Lula, I. Extracellular Matrix Remodeling Factors as Markers of Carotid Artery Atherosclerosis. Cardiol. Res. Pract. 2020, 2020, 9036157. [Google Scholar] [CrossRef]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef]
- Kim, J.; Kim, K.M.; Noh, J.H.; Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Long noncoding RNAs in diseases of aging. Biochim. Biophys. Acta 2016, 1859, 209–221. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Luo, Y. Understanding the Functions of Long Non-Coding RNAs through Their Higher-Order Structures. Int. J. Mol. Sci. 2016, 17, 702. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Zumwalt, T.J.; Goel, A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, R.; Zhang, X.; Wu, Y.; Li, X.; Zhang, S.; Hou, W.; Ding, Y.; Tian, J.; Sun, L.; et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging 2018, 10, 2266–2283. [Google Scholar] [CrossRef] [PubMed]
- Rotini, A.; Martinez-Sarra, E.; Pozzo, E.; Sampaolesi, M. Interactions between microRNAs and long non-coding RNAs in cardiac development and repair. Pharm. Res. 2018, 127, 58–66. [Google Scholar] [CrossRef]
- Kreutzer, F.P.; Fiedler, J.; Thum, T. Non-coding RNAs: Key players in cardiac disease. J. Physiol. 2020, 598, 2995–3003. [Google Scholar] [CrossRef]
- Zhu, K.; Hu, X.; Chen, H.; Li, F.; Yin, N.; Liu, A.L.; Shan, K.; Qin, Y.W.; Huang, X.; Chang, Q.; et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine 2019, 49, 341–353. [Google Scholar] [CrossRef]
- Loganathan, T.S.; Sulaiman, S.A.; Abdul Murad, N.A.; Shah, S.A.; Abdul Gafor, A.H.; Jamal, R.; Abdullah, N. Interactions Among Non-Coding RNAs in Diabetic Nephropathy. Front. Pharm. 2020, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.Y.; Lu, F.H.; Huang, X.R.; Zhang, L.; Mao, W.; Yu, X.Q.; Liu, X.S.; Lan, H.Y. Non-Coding RNAs as Biomarkers and Therapeutic Targets for Diabetic Kidney Disease. Front. Pharm. 2020, 11, 583528. [Google Scholar] [CrossRef]
- Jiang, F.; Chen, Q.; Wang, W.; Ling, Y.; Yan, Y.; Xia, P. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J. Hepatol. 2020, 72, 156–166. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Martino, F.; Thum, T. Epigenetic modifications in cardiovascular disease. Basic Res. Cardiol. 2012, 107, 245. [Google Scholar] [CrossRef]
- Dong, Y.; Xu, S.; Liu, J.; Ponnusamy, M.; Zhao, Y.; Zhang, Y.; Wang, Q.; Li, P.; Wang, K. Non-coding RNA-linked epigenetic regulation in cardiac hypertrophy. Int. J. Biol. Sci. 2018, 14, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Gialeli, C.; Shami, A.; Goncalves, I. Extracellular matrix: Paving the way to the newest trends in atherosclerosis. Curr. Opin. Lipidol. 2021, 32, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.B.; Mengi, S.A.; Xu, Y.J.; Arneja, A.S.; Dhalla, N.S. Pathogenesis of atherosclerosis: A multifactorial process. Exp. Clin. Cardiol. 2002, 7, 40–53. [Google Scholar] [PubMed]
- Moore, K.J.; Sheedy, F.J.; Fisher, E.A. Macrophages in atherosclerosis: A dynamic balance. Nat. Rev. Immunol. 2013, 13, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef]
- Barallobre-Barreiro, J.; Loeys, B.; Mayr, M.; Rienks, M.; Verstraeten, A.; Kovacic, J.C. Extracellular Matrix in Vascular Disease, Part 2/4: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 2189–2203. [Google Scholar] [CrossRef]
- Goncalves, R.C.; Banfi, A.; Oliveira, M.B.; Mano, J.F. Strategies for re-vascularization and promotion of angiogenesis in trauma and disease. Biomaterials 2021, 269, 120628. [Google Scholar] [CrossRef]
- Heinz, A. Elastic fibers during aging and disease. Ageing Res. Rev. 2021, 66, 101255. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, X.; Xia, Y.; Mao, L. An update on the phenotypic switching of vascular smooth muscle cells in the pathogenesis of atherosclerosis. Cell Mol. Life Sci. 2021, 79, 6. [Google Scholar] [CrossRef]
- Libby, P.; Nahrendorf, M.; Swirski, F.K. Leukocytes Link Local and Systemic Inflammation in Ischemic Cardiovascular Disease: An Expanded “Cardiovascular Continuum”. J. Am. Coll. Cardiol. 2016, 67, 1091–1103. [Google Scholar] [CrossRef]
- Schaefer, J.R.; Klumpp, S.; Maisch, B.; Krieglstein, J. Why does atherosclerosis occur where it occurs? Atherosclerosis 2005, 180, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Wilensky, R.L.; Song, H.K.; Ferrari, V.A. Role of magnetic resonance and intravascular magnetic resonance in the detection of vulnerable plaques. J. Am. Coll. Cardiol. 2006, 47, C48–C56. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Finn, A.V.; Nakano, M.; Narula, J.; Kolodgie, F.D.; Virmani, R. Concept of vulnerable/unstable plaque. Arter. Thromb. Vasc. Biol. 2010, 30, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Rai, V.; Agrawal, D.K. Immunomodulation of IL-33 and IL-37 with Vitamin D in the Neointima of Coronary Artery: A Comparative Study between Balloon Angioplasty and Stent in Hyperlipidemic Microswine. Int. J. Mol. Sci. 2021, 22, 8824. [Google Scholar] [CrossRef] [PubMed]
- Rai, V.; Agrawal, D.K. The role of damage- and pathogen-associated molecular patterns in inflammation-mediated vulnerability of atherosclerotic plaques. Can. J. Physiol. Pharm. 2017, 95, 1245–1253. [Google Scholar] [CrossRef]
- Rao, V.H.; Rai, V.; Stoupa, S.; Subramanian, S.; Agrawal, D.K. Data on TREM-1 activation destabilizing carotid plaques. Data Brief 2016, 8, 230–234. [Google Scholar] [CrossRef]
- Gupta, G.K.; Agrawal, T.; Rai, V.; Del Core, M.G.; Hunter, W.J., 3rd; Agrawal, D.K. Vitamin D Supplementation Reduces Intimal Hyperplasia and Restenosis following Coronary Intervention in Atherosclerotic Swine. PLoS ONE 2016, 11, e0156857. [Google Scholar] [CrossRef]
- Rai, V.; Rao, V.H.; Shao, Z.; Agrawal, D.K. Dendritic Cells Expressing Triggering Receptor Expressed on Myeloid Cells-1 Correlate with Plaque Stability in Symptomatic and Asymptomatic Patients with Carotid Stenosis. PLoS ONE 2016, 11, e0154802. [Google Scholar] [CrossRef]
- Rao, V.H.; Rai, V.; Stoupa, S.; Subramanian, S.; Agrawal, D.K. Tumor necrosis factor-alpha regulates triggering receptor expressed on myeloid cells-1-dependent matrix metalloproteinases in the carotid plaques of symptomatic patients with carotid stenosis. Atherosclerosis 2016, 248, 160–169. [Google Scholar] [CrossRef]
- Rao, V.H.; Rai, V.; Stoupa, S.; Agrawal, D.K. Blockade of Ets-1 attenuates epidermal growth factor-dependent collagen loss in human carotid plaque smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1075–H1086. [Google Scholar] [CrossRef]
- Cui, Y.; Zhou, Y.; Gan, N.; Xiang, Q.; Xia, M.; Liao, W.; Zheng, X.L.; Peng, J.; Tang, Z. The Role of Extracellular Non-coding RNAs in Atherosclerosis. J. Cardiovasc. Transl. Res. 2022, 15, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell. Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Cuesta, M.; Osorio-Querejeta, I.; Otaegui, D. Extracellular Vesicles in Multiple Sclerosis: What are They Telling Us? Front. Cell Neurosci. 2014, 8, 100. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- de Jong, O.G.; Murphy, D.E.; Mager, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; van Rheenen, J.; et al. Publisher Correction: A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun. 2020, 11, 1701. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Colpaert, R.M.W.; Calore, M. MicroRNAs in Cardiac Diseases. Cells 2019, 8, 737. [Google Scholar] [CrossRef]
- Rizzacasa, B.; Amati, F.; Romeo, F.; Novelli, G.; Mehta, J.L. Epigenetic Modification in Coronary Atherosclerosis: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2019, 74, 1352–1365. [Google Scholar] [CrossRef]
- Bhattacharyya, N.; Pandey, V.; Bhattacharyya, M.; Dey, A. Regulatory role of long non coding RNAs (lncRNAs) in neurological disorders: From novel biomarkers to promising therapeutic strategies. Asian J. Pharm. Sci. 2021, 16, 533–550. [Google Scholar] [CrossRef]
- Gorabi, A.M.; Ghanbari, M.; Sathyapalan, T.; Jamialahmadi, T.; Sahebkar, A. Implications of microRNAs in the Pathogenesis of Atherosclerosis and Prospects for Therapy. Curr. Drug Targets 2021, 22, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Pu, S.D.; Li, X.; Yu, Z.W.; Zhang, Y.T.; Tong, X.W.; Shan, Y.Y.; Gao, X.Y. Exosomal ncRNAs: Novel therapeutic target and biomarker for diabetic complications. Pharm. Res. 2022, 178, 106135. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xu, L.; Geng, Z.; Liu, J.; Zhang, L.; Wu, Y.; He, D.; Qu, P. The role of non-coding RNA network in atherosclerosis. Life Sci. 2021, 265, 118756. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.W.; Paneni, F.; Jandeleit-Dahm, K.A.M. Cell-specific epigenetic changes in atherosclerosis. Clin. Sci. 2021, 135, 1165–1187. [Google Scholar] [CrossRef]
- Zhao, Z.; Guo, N.; Chen, W.; Wang, Z. Leveraging Extracellular Non-coding RNAs to Diagnose and Treat Heart Diseases. J. Cardiovasc. Transl. Res. 2022, 15, 456–468. [Google Scholar] [CrossRef]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Ooi, J.Y.; Lin, R.C.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Min, P.K.; Chan, S.Y. The biology of circulating microRNAs in cardiovascular disease. Eur. J. Clin. Investig. 2015, 45, 860–874. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Zhang, F.; Wang, D. Comparative Analysis of microRNA Binding Site Distribution and microRNA-Mediated Gene Expression Repression of Oncogenes and Tumor Suppressor Genes. Genes 2022, 13, 481. [Google Scholar] [CrossRef]
- Rottiers, V.; Naar, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Im, H.I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Nigi, L.; Grieco, G.E.; Ventriglia, G.; Brusco, N.; Mancarella, F.; Formichi, C.; Dotta, F.; Sebastiani, G. MicroRNAs as Regulators of Insulin Signaling: Research Updates and Potential Therapeutic Perspectives in Type 2 Diabetes. Int. J. Mol. Sci. 2018, 19, 3705. [Google Scholar] [CrossRef]
- Boon, R.A. Endothelial microRNA tells smooth muscle cells to proliferate. Circ. Res. 2013, 113, 7–8. [Google Scholar] [CrossRef]
- Aryal, B.; Rotllan, N.; Fernández-Hernando, C. Noncoding RNAs and atherosclerosis. Curr. Atheroscler. Rep. 2014, 16, 1–11. [Google Scholar] [CrossRef]
- Jaé, N.; Dimmeler, S. Noncoding RNAs in vascular diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Koppel, T.; Jahantigh, M.N.; Lutgens, E.; et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009, 2, ra81. [Google Scholar] [CrossRef]
- Vickers, K.C.; Shoucri, B.M.; Levin, M.G.; Wu, H.; Pearson, D.S.; Osei-Hwedieh, D.; Collins, F.S.; Remaley, A.T.; Sethupathy, P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 2013, 57, 533–542. [Google Scholar] [CrossRef]
- Rong, X.; Ge, D.; Shen, D.; Chen, X.; Wang, X.; Zhang, L.; Jia, C.; Zeng, J.; He, Y.; Qiu, H.; et al. miR-27b Suppresses Endothelial Cell Proliferation and Migration by Targeting Smad7 in Kawasaki Disease. Cell Physiol. Biochem. 2018, 48, 1804–1814. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, J.; Bhattachariya, A.; Alajbegovic, A.; Rippe, C.; Ekman, M.; Dahan, D.; Hien, T.T.; Boettger, T.; Braun, T.; Sward, K.; et al. Loss of Vascular Myogenic Tone in miR-143/145 Knockout Mice Is Associated With Hypertension-Induced Vascular Lesions in Small Mesenteric Arteries. Arter. Thromb. Vasc. Biol. 2018, 38, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Xu, T.; Huang, X.; Li, S.; Qin, W.; Chen, W.; Zhang, Z. miR-21 regulates vascular smooth muscle cell function in arteriosclerosis obliterans of lower extremities through AKT and ERK1/2 pathways. Arch. Med. Sci. 2019, 15, 1490–1497. [Google Scholar] [CrossRef]
- Chen, T.; Huang, Z.; Wang, L.; Wang, Y.; Wu, F.; Meng, S.; Wang, C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc. Res. 2009, 83, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Cui, H.; Xie, N.; Tan, Z.; Yang, S.; Icyuz, M.; Thannickal, V.J.; Abraham, E.; Liu, G. miR-125a-5p regulates differential activation of macrophages and inflammation. J. Biol. Chem. 2013, 288, 35428–35436. [Google Scholar] [CrossRef]
- Yang, K.; He, Y.S.; Wang, X.Q.; Lu, L.; Chen, Q.J.; Liu, J.; Sun, Z.; Shen, W.F. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 2011, 585, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Marquart, T.J.; Allen, R.M.; Ory, D.S.; Baldan, A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 12228–12232. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Ramirez, C.M.; Rotllan, N.; Vlassov, A.V.; Davalos, A.; Li, M.; Goedeke, L.; Aranda, J.F.; Cirera-Salinas, D.; Araldi, E.; Salerno, A.; et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 2013, 112, 1592–1601. [Google Scholar] [CrossRef]
- Kim, J.; Yoon, H.; Ramirez, C.M.; Lee, S.M.; Hoe, H.S.; Fernandez-Hernando, C.; Kim, J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp. Neurol. 2012, 235, 476–483. [Google Scholar] [CrossRef]
- Soh, J.; Iqbal, J.; Queiroz, J.; Fernandez-Hernando, C.; Hussain, M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013, 19, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Nazari-Jahantigh, M.; Wei, Y.; Noels, H.; Akhtar, S.; Zhou, Z.; Koenen, R.R.; Heyll, K.; Gremse, F.; Kiessling, F.; Grommes, J.; et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Investig. 2012, 122, 4190–4202. [Google Scholar] [CrossRef]
- Donners, M.M.; Wolfs, I.M.; Stoger, L.J.; van der Vorst, E.P.; Pottgens, C.C.; Heymans, S.; Schroen, B.; Gijbels, M.J.; de Winther, M.P. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS ONE 2012, 7, e35877. [Google Scholar] [CrossRef]
- Thulin, P.; Wei, T.; Werngren, O.; Cheung, L.; Fisher, R.M.; Grander, D.; Corcoran, M.; Ehrenborg, E. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor delta in human monocytes during the inflammatory response. Int. J. Mol. Med. 2013, 31, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, W.; Chang, G.Q.; Ye, C.S.; Ou, J.S.; Li, X.X.; Liu, Y.; Cheang, T.Y.; Huang, X.L.; Wang, S.M. MicroRNA-21 regulates vascular smooth muscle cell function via targeting tropomyosin 1 in arteriosclerosis obliterans of lower extremities. Arter. Thromb. Vasc. Biol. 2011, 31, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Small, E.M.; Sutherland, L.B.; Qi, X.; McAnally, J.; Plato, C.F.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009, 23, 2166–2178. [Google Scholar] [CrossRef] [PubMed]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef]
- Sarkar, J.; Gou, D.; Turaka, P.; Viktorova, E.; Ramchandran, R.; Raj, J.U. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am. J. Physiol. Lung Cell Mol. Physiol. 2010, 299, L861–L871. [Google Scholar] [CrossRef]
- Chen, J.; Yin, H.; Jiang, Y.; Radhakrishnan, S.K.; Huang, Z.P.; Li, J.; Shi, Z.; Kilsdonk, E.P.; Gui, Y.; Wang, D.Z.; et al. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arter. Thromb. Vasc. Biol. 2011, 31, 368–375. [Google Scholar] [CrossRef]
- Xie, C.; Huang, H.; Sun, X.; Guo, Y.; Hamblin, M.; Ritchie, R.P.; Garcia-Barrio, M.T.; Zhang, J.; Chen, Y.E. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011, 20, 205–210. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ. Res. 2009, 104, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem. 2009, 284, 3728–3738. [Google Scholar] [CrossRef] [PubMed]
- Latronico, M.V.; Catalucci, D.; Condorelli, G. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 2007, 101, 1225–1236. [Google Scholar] [CrossRef]
- Zhang, P.; Huang, A.; Ferruzzi, J.; Mecham, R.P.; Starcher, B.C.; Tellides, G.; Humphrey, J.D.; Giordano, F.J.; Niklason, L.E.; Sessa, W.C. Inhibition of microRNA-29 enhances elastin levels in cells haploinsufficient for elastin and in bioengineered vessels--brief report. Arter. Thromb. Vasc. Biol. 2012, 32, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wang, X.; Zhang, Y.; Eisner, G.M.; Asico, L.D.; Jose, P.A.; Zeng, C. Insulin promotes vascular smooth muscle cell proliferation via microRNA-208-mediated downregulation of p21. J. Hypertens 2011, 29, 1560–1568. [Google Scholar] [CrossRef]
- Yu, M.L.; Wang, J.F.; Wang, G.K.; You, X.H.; Zhao, X.X.; Jing, Q.; Qin, Y.W. Vascular smooth muscle cell proliferation is influenced by let-7d microRNA and its interaction with KRAS. Circ. J. 2011, 75, 703–709. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, N.; Zhang, J.; Tong, Y. Hsa-let-7g miRNA targets caspase-3 and inhibits the apoptosis induced by ox-LDL in endothelial cells. Int. J. Mol. Sci. 2013, 14, 22708–22720. [Google Scholar] [CrossRef]
- Chen, K.C.; Hsieh, I.C.; Hsi, E.; Wang, Y.S.; Dai, C.Y.; Chou, W.W.; Juo, S.H. Negative feedback regulation between microRNA let-7g and the oxLDL receptor LOX-1. J. Cell Sci. 2011, 124, 4115–4124. [Google Scholar] [CrossRef]
- Choe, N.; Kwon, J.S.; Kim, J.R.; Eom, G.H.; Kim, Y.; Nam, K.I.; Ahn, Y.; Kee, H.J.; Kook, H. The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis 2013, 229, 348–355. [Google Scholar] [CrossRef]
- Liao, X.B.; Zhang, Z.Y.; Yuan, K.; Liu, Y.; Feng, X.; Cui, R.R.; Hu, Y.R.; Yuan, Z.S.; Gu, L.; Li, S.J.; et al. MiR-133a modulates osteogenic differentiation of vascular smooth muscle cells. Endocrinology 2013, 154, 3344–3352. [Google Scholar] [CrossRef]
- Suarez, Y.; Wang, C.; Manes, T.D.; Pober, J.S. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: Feedback control of inflammation. J. Immunol. 2010, 184, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Davies, P.F. Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium. Arter. Thromb. Vasc. Biol. 2012, 32, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Loyer, X.; Potteaux, S.; Vion, A.C.; Guerin, C.L.; Boulkroun, S.; Rautou, P.E.; Ramkhelawon, B.; Esposito, B.; Dalloz, M.; Paul, J.L.; et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 2014, 114, 434–443. [Google Scholar] [CrossRef]
- Poliseno, L.; Tuccoli, A.; Mariani, L.; Evangelista, M.; Citti, L.; Woods, K.; Mercatanti, A.; Hammond, S.; Rainaldi, G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006, 108, 3068–3071. [Google Scholar] [CrossRef] [PubMed]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Son, D.J.; Kumar, S.; Takabe, W.; Kim, C.W.; Ni, C.W.; Alberts-Grill, N.; Jang, I.H.; Kim, S.; Kim, W.; Won Kang, S.; et al. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat. Commun. 2013, 4, 3000. [Google Scholar] [CrossRef]
- Fang, Y.; Shi, C.; Manduchi, E.; Civelek, M.; Davies, P.F. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 13450–13455. [Google Scholar] [CrossRef]
- Urbich, C.; Kaluza, D.; Fromel, T.; Knau, A.; Bennewitz, K.; Boon, R.A.; Bonauer, A.; Doebele, C.; Boeckel, J.N.; Hergenreider, E.; et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 2012, 119, 1607–1616. [Google Scholar] [CrossRef]
- Ito, T.; Yagi, S.; Yamakuchi, M. MicroRNA-34a regulation of endothelial senescence. Biochem. Biophys. Res. Commun. 2010, 398, 735–740. [Google Scholar] [CrossRef]
- Vasa-Nicotera, M.; Chen, H.; Tucci, P.; Yang, A.L.; Saintigny, G.; Menghini, R.; Mahe, C.; Agostini, M.; Knight, R.A.; Melino, G.; et al. miR-146a is modulated in human endothelial cell with aging. Atherosclerosis 2011, 217, 326–330. [Google Scholar] [CrossRef]
- Wu, W.; Shang, Y.; Dai, S.; Yu, C.; Wang, J. Downregulation of miR1425p inhibits human aortic smooth muscle cell proliferation and migration by targeting MKL2. Mol. Med. Rep. 2020, 22, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yuan, J.; Zhang, F.; Lei, Q.; Zhang, T.; Li, K.; Guo, J.; Hong, Y.; Bu, G.; Lv, X.; et al. MicroRNA-181a-5p and microRNA-181a-3p cooperatively restrict vascular inflammation and atherosclerosis. Cell Death Dis. 2019, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Raitoharju, E.; Lyytikainen, L.P.; Levula, M.; Oksala, N.; Mennander, A.; Tarkka, M.; Klopp, N.; Illig, T.; Kahonen, M.; Karhunen, P.J.; et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011, 219, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, Y.; Du, Y.; Li, Y. MiRNA-130a promotes inflammation to accelerate atherosclerosis via the regulation of proliferator-activated receptor gamma (PPARgamma) expression. Anatol. J. Cardiol. 2021, 25, 630–637. [Google Scholar] [CrossRef]
- Polyakova, E.A.; Zaraiskii, M.I.; Mikhaylov, E.N.; Baranova, E.I.; Galagudza, M.M.; Shlyakhto, E.V. Association of myocardial and serum miRNA expression patterns with the presence and extent of coronary artery disease: A cross-sectional study. Int. J. Cardiol. 2021, 322, 9–15. [Google Scholar] [CrossRef]
- Zhang, X.; Rotllan, N.; Canfran-Duque, A.; Sun, J.; Toczek, J.; Moshnikova, A.; Malik, S.; Price, N.L.; Araldi, E.; Zhong, W.; et al. Targeted Suppression of miRNA-33 Using pHLIP Improves Atherosclerosis Regression. Circ. Res. 2022, 131, 77–90. [Google Scholar] [CrossRef]
- Zhou, B.; Li, B.; Feng, P.; Wang, X.; Gao, H.; Xu, L.; Wang, T.; Guo, X. Identification of a miRNA biomarker for the large artery atherosclerosis subtype of acute ischemic stroke. Folia Neuropathol. 2022, 60, 210–220. [Google Scholar] [CrossRef]
- Egea, V.; Megens, R.T.A.; Santovito, D.; Wantha, S.; Brandl, R.; Siess, W.; Khani, S.; Soehnlein, O.; Bartelt, A.; Weber, C.; et al. Properties and fate of human mesenchymal stem cells upon miRNA let-7f-promoted recruitment to atherosclerotic plaques. Cardiovasc. Res. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Glagov, S.; Weisenberg, E.; Zarins, C.K.; Stankunavicius, R.; Kolettis, G.J. Compensatory enlargement of human atherosclerotic coronary arteries. N. Engl. J. Med. 1987, 316, 1371–1375. [Google Scholar] [CrossRef]
- Lin, X.; Lo, H.C.; Wong, D.T.; Xiao, X. Noncoding RNAs in human saliva as potential disease biomarkers. Front. Genet. 2015, 6, 175. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Dang, R.Y.; Liu, F.L.; Li, Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 2017, 490, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yao, M.D.; Li, C.P.; Shan, K.; Yang, H.; Wang, J.J.; Liu, B.; Li, X.M.; Yao, J.; Jiang, Q.; et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877. [Google Scholar] [CrossRef]
- Huang, H.S.; Huang, X.Y.; Yu, H.Z.; Xue, Y.; Zhu, P.L. Circular RNA circ-RELL1 regulates inflammatory response by miR-6873-3p/MyD88/NF-kappaB axis in endothelial cells. Biochem. Biophys. Res. Commun. 2020, 525, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, F.; Zhao, H.; Wang, M.; Zhang, Y. Circular RNA circCHFR Facilitates the Proliferation and Migration of Vascular Smooth Muscle via miR-370/FOXO1/Cyclin D1 Pathway. Mol. Ther. Nucleic Acids 2019, 16, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.Y.; Wang, J.Q.; Guo, X.X.; Bi, Y.; Wang, C.X. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem. Biophys. Res. Commun. 2018, 505, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cui, L.; Yuan, J.; Zhang, Y.; Sang, H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017, 494, 126–132. [Google Scholar] [CrossRef]
- Wang, X.; Bai, M. CircTM7SF3 contributes to oxidized low-density lipoprotein-induced apoptosis, inflammation and oxidative stress through targeting miR-206/ASPH axis in atherosclerosis cell model in vitro. BMC Cardiovasc. Disord. 2021, 21, 51. [Google Scholar] [CrossRef]
- He, Q.; Shao, D.; Hao, S.; Yuan, Y.; Liu, H.; Liu, F.; Mu, Q. CircSCAP Aggravates Oxidized Low-density Lipoprotein-induced Macrophage Injury by Upregulating PDE3B by miR-221-5p in Atherosclerosis. J. Cardiovasc. Pharm. 2021, 78, e749–e760. [Google Scholar] [CrossRef]
- Pan, L.; Lian, W.; Zhang, X.; Han, S.; Cao, C.; Li, X.; Li, M. Human circular RNA0054633 regulates high glucoseinduced vascular endothelial cell dysfunction through the microRNA218/roundabout 1 and microRNA218/heme oxygenase1 axes. Int. J. Mol. Med. 2018, 42, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Liu, C.; Liu, B.H.; Chen, X.; Dong, R.; Liu, X.; Zhang, Y.Y.; Liu, B.; Zhang, S.J.; Wang, J.J.; et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017, 136, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, R.; Liu, W.; Wang, Z.; Rong, J.; Long, X.; Liu, Z.; Ge, J.; Shi, B. Exosomal circHIPK3 Released from Hypoxia-Pretreated Cardiomyocytes Regulates Oxidative Damage in Cardiac Microvascular Endothelial Cells via the miR-29a/IGF-1 Pathway. Oxidative Med. Cell. Longev. 2019, 2019, 7954657. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Quan, A.; Sun, H.; Xu, Y.; Sun, G.; Cao, P. MicroRNA-148a-3p promotes survival and migration of endothelial cells isolated from Apoe deficient mice through restricting circular RNA 0003575. Gene 2019, 711, 143948. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Liu, Q.; Hu, N.; Zheng, F.; Zhang, X.; Ni, Y.; Liu, J. Downregulation of hsa_circ_0068087 ameliorates TLR4/NF-kappaB/NLRP3 inflammasome-mediated inflammation and endothelial cell dysfunction in high glucose conditioned by sponging miR-197. Gene 2019, 709, 1–7. [Google Scholar] [CrossRef]
- Shen, L.; Hu, Y.; Lou, J.; Yin, S.; Wang, W.; Wang, Y.; Xia, Y.; Wu, W. CircRNA0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR107. Mol. Med. Rep. 2019, 19, 3923–3932. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef]
- Zheng, C.; Niu, H.; Li, M.; Zhang, H.; Yang, Z.; Tian, L.; Wu, Z.; Li, D.; Chen, X. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 2015, 12, 6656–6662. [Google Scholar] [CrossRef]
- Hall, I.F.; Climent, M.; Quintavalle, M.; Farina, F.M.; Schorn, T.; Zani, S.; Carullo, P.; Kunderfranco, P.; Civilini, E.; Condorelli, G.; et al. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circ. Res. 2019, 124, 498–510. [Google Scholar] [CrossRef]
- Li, C.Y.; Ma, L.; Yu, B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed. Pharm. 2017, 95, 1514–1519. [Google Scholar] [CrossRef]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gabel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arter. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.Y.; Lv, R.R.; Teng, Z. Circular RNA circHIPK3 as a novel circRNA regulator of autophagy and endothelial cell dysfunction in atherosclerosis. Eur. Rev. Med. Pharm. Sci. 2020, 24, 12849–12858. [Google Scholar] [CrossRef]
- Sun, X.; Deng, K.; Zang, Y.; Zhang, Z.; Zhao, B.; Fan, J.; Huang, L. Exploring the regulatory roles of circular RNAs in the pathogenesis of atherosclerosis. Vascul. Pharm. 2021, 141, 106898. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, Y.; Zhou, H.; Li, Y. Circular RNAs in atherosclerosis. Clin. Chim. Acta 2022, 531, 71–80. [Google Scholar] [CrossRef]
- Cao, Q.; Guo, Z.; Du, S.; Ling, H.; Song, C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020, 255, 117837. [Google Scholar] [CrossRef]
- Pan, R.Y.; Liu, P.; Zhou, H.T.; Sun, W.X.; Song, J.; Shu, J.; Cui, G.J.; Yang, Z.J.; Jia, E.Z. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget 2017, 8, 60280–60290. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, Y.; Wang, Z.; Gong, W.; Zhang, C. Hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics 2021, 11, 5404–5417. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, R.; Lin, S.; Xie, X.; Ye, H.; Zheng, F.; Lin, J.; Huang, Q.; Huang, S.; Ruan, Q.; et al. Association of circular RNAs and environmental risk factors with coronary heart disease. BMC Cardiovasc. Disord. 2019, 19, 223. [Google Scholar] [CrossRef]
- Wang, L.; Shen, C.; Wang, Y.; Zou, T.; Zhu, H.; Lu, X.; Li, L.; Yang, B.; Chen, J.; Chen, S.; et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 2019, 286, 88–96. [Google Scholar] [CrossRef]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Bazan, H.A.; Lightell, D.; Sternbergh, W.C., III; Woods, T.C. Recently ruptured carotid plaques have increased levels of circular RNA-16, which negatively regulates the proproliferative and antiapoptotic microRNA-221: A novel mediator of carotid plaque rupture. Arterioscler. Thromb. Vasc. Biol. 2014, 34, A123. [Google Scholar] [CrossRef]
- Nie, X.; Chen, Y.; Tan, J.; Dai, Y.; Mao, W.; Qin, G.; Ye, S.; Sun, J.; Yang, Z.; Chen, J. MicroRNA-221-3p promotes pulmonary artery smooth muscle cells proliferation by targeting AXIN2 during pulmonary arterial hypertension. Vasc. Pharm. 2019, 116, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Josefs, T.; Boon, R.A. The long non-coding road to atherosclerosis. Curr. Atheroscler. Rep. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Leung, A.; Trac, C.; Jin, W.; Lanting, L.; Akbany, A.; Saetrom, P.; Schones, D.E.; Natarajan, R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ. Res. 2013, 113, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Yuan, Y.; Tan, M.; Zhang, L.; Xue, X.; Yan, Y.; Han, L.; Xu, Z. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur. J. Cardiothorac. Surg. 2015, 47, 439–446. [Google Scholar] [CrossRef]
- He, Q.; Tan, J.; Yu, B.; Shi, W.; Liang, K. Long noncoding RNA HIF1A-AS1A reduces apoptosis of vascular smooth muscle cells: Implications for the pathogenesis of thoracoabdominal aorta aneurysm. Pharmazie 2015, 70, 310–315. [Google Scholar]
- Ma, Y.; Huang, D.; Yang, F.; Tian, M.; Wang, Y.; Shen, D.; Wang, Q.; Chen, Q.; Zhang, L. Long Noncoding RNA Highly Upregulated in Liver Cancer Regulates the Tumor Necrosis Factor-alpha-Induced Apoptosis in Human Vascular Endothelial Cells. DNA Cell Biol. 2016, 35, 296–300. [Google Scholar] [CrossRef]
- Wu, G.; Cai, J.; Han, Y.; Chen, J.; Huang, Z.P.; Chen, C.; Cai, Y.; Huang, H.; Yang, Y.; Liu, Y.; et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014, 130, 1452–1465. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Chen, C.; Cheng, G.; Yang, X.; Li, C.; Shi, R.; Zhao, N. Tanshinol suppresses endothelial cells apoptosis in mice with atherosclerosis via lncRNA TUG1 up-regulating the expression of miR-26a. Am. J. Transl. Res. 2016, 8, 2981–2991. [Google Scholar] [PubMed]
- Li, F.P.; Lin, D.Q.; Gao, L.Y. LncRNA TUG1 promotes proliferation of vascular smooth muscle cell and atherosclerosis through regulating miRNA-21/PTEN axis. Eur. Rev. Med. Pharm. Sci. 2018, 22, 7439–7447. [Google Scholar] [CrossRef]
- Tang, Y.; Jin, X.; Xiang, Y.; Chen, Y.; Shen, C.X.; Zhang, Y.C.; Li, Y.G. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015, 589, 3189–3196. [Google Scholar] [CrossRef] [PubMed]
- Sallam, T.; Jones, M.; Thomas, B.J.; Wu, X.; Gilliland, T.; Qian, K.; Eskin, A.; Casero, D.; Zhang, Z.; Sandhu, J.; et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018, 24, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, H.; Yue, Y.; Li, S.; Zhang, D.; He, R. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL -stimulated vascular smooth muscle cells. J. Biomed. Sci. 2018, 25, 11. [Google Scholar] [CrossRef]
- Li, K.; Chowdhury, T.; Vakeel, P.; Koceja, C.; Sampath, V.; Ramchandran, R. Delta-like 4 mRNA is regulated by adjacent natural antisense transcripts. Vasc. Cell 2015, 7, 3. [Google Scholar] [CrossRef]
- Shen, Z.; She, Q. Association Between the Deletion Allele of Ins/Del Polymorphism (Rs145204276) in the Promoter Region of GAS5 with the Risk of Atherosclerosis. Cell Physiol. Biochem. 2018, 49, 1431–1443. [Google Scholar] [CrossRef]
- Zhong, X.; Ma, X.; Zhang, L.; Li, Y.; Li, Y.; He, R. MIAT promotes proliferation and hinders apoptosis by modulating miR-181b/STAT3 axis in ox-LDL-induced atherosclerosis cell models. Biomed. Pharm. 2018, 97, 1078–1085. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Lyu, Q.; Xu, S.; Lyu, Y.; Choi, M.; Christie, C.K.; Slivano, O.J.; Rahman, A.; Jin, Z.G.; Long, X.; Xu, Y.; et al. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc. Natl. Acad. Sci. USA 2019, 116, 546–555. [Google Scholar] [CrossRef]
- Bell, R.D.; Long, X.; Lin, M.; Bergmann, J.H.; Nanda, V.; Cowan, S.L.; Zhou, Q.; Han, Y.; Spector, D.L.; Zheng, D.; et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arter. Thromb. Vasc. Biol. 2014, 34, 1249–1259. [Google Scholar] [CrossRef]
- Xu, X.; Ma, C.; Liu, C.; Duan, Z.; Zhang, L. Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem. Biophys. Res. Commun. 2018, 503, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Robb, G.B.; Carson, A.R.; Tai, S.C.; Fish, J.E.; Singh, S.; Yamada, T.; Scherer, S.W.; Nakabayashi, K.; Marsden, P.A. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J. Biol. Chem. 2004, 279, 37982–37996. [Google Scholar] [CrossRef] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zornig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Wu, Z.; He, Y.; Li, D.; Fang, X.; Shang, T.; Zhang, H.; Zheng, X. Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am. J. Transl. Res. 2017, 9, 3326–3335. [Google Scholar] [PubMed]
- Zhang, Y.; Liu, X.; Bai, X.; Lin, Y.; Li, Z.; Fu, J.; Li, M.; Zhao, T.; Yang, H.; Xu, R.; et al. Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J. Pineal. Res. 2018, 64, e12449. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Kawai, T.; Kusunoki, H.; Yamamoto, H.; Takeya, Y.; et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012, 220, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.F.; Wu, K.; Hu, K.; Chen, Y.; Xiao, J. NAMPT regulates senescence, proliferation, and migration of endothelial progenitor cells through the SIRT1 AS lncRNA/miR-22/SIRT1 pathway. Biochem. Biophys. Res. Commun. 2016, 478, 1382–1388. [Google Scholar] [CrossRef]
- Wang, J.; Su, Z.; Lu, S.; Fu, W.; Liu, Z.; Jiang, X.; Tai, S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta 2018, 485, 229–233. [Google Scholar] [CrossRef]
- Wu, M.; Feng, Y.; Shi, X. Advances with Long Non-Coding RNAs in Diabetic Peripheral Neuropathy. Diabetes Metab. Syndr. Obes. 2020, 13, 1429–1434. [Google Scholar] [CrossRef]
- Hüttenhofer, A.; Schattner, P.; Polacek, N. Non-coding RNAs: Hope or hype? TRENDS Genet. 2005, 21, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.A.; Fraczek, M.G.; Parker, S.; Delneri, D.; O’Keefe, R.T. Non-coding RNAs and disease: The classical ncRNAs make a comeback. Biochem. Soc. Trans. 2016, 44, 1073–1078. [Google Scholar] [CrossRef]
- Bhartiya, D.; Scaria, V. Genomic variations in non-coding RNAs: Structure, function and regulation. Genomics 2016, 107, 59–68. [Google Scholar] [CrossRef] [PubMed]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef]
- Ann, S.J.; Bang, H.; Lee, C.J.; Oh, J.; Park, S.; Kang, S.M.; Choi, J.K.; Lee, S.H. LncRNA HSPA7 in human atherosclerotic plaques sponges miR-223 and promotes the proinflammatory vascular smooth muscle cell transition. Exp. Mol. Med. 2021, 53, 1842–1849. [Google Scholar] [CrossRef]
- Hu, Y.W.; Guo, F.X.; Xu, Y.J.; Li, P.; Lu, Z.F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.H.; Kang, C.M.; et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J. Clin. Investig. 2019, 129, 1115–1128. [Google Scholar] [CrossRef]
- Fitzwalter, B.E.; Thorburn, A. Recent insights into cell death and autophagy. FEBS J. 2015, 282, 4279–4288. [Google Scholar] [CrossRef]
- Guo, F.X.; Wu, Q.; Li, P.; Zheng, L.; Ye, S.; Dai, X.Y.; Kang, C.M.; Lu, J.B.; Xu, B.M.; Xu, Y.J.; et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. 2019, 26, 1670–1687. [Google Scholar] [CrossRef]
- Vacante, F.; Rodor, J.; Lalwani, M.K.; Mahmoud, A.D.; Bennett, M.; De Pace, A.L.; Miller, E.; Van Kuijk, K.; de Bruijn, J.; Gijbels, M.; et al. CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice. Circ. Res. 2021, 128, 1258–1275. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Zhang, T.; Jia, S.; Ma, X.; Zhang, M.; Wang, L.; Ma, A. LncRNA SNHG16 accelerates atherosclerosis and promotes ox-LDL-induced VSMC growth via the miRNA-22-3p/HMGB2 axis. Eur. J. Pharm. 2022, 915, 174601. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yan, J.H.; Ge, Z.W.; Fei, A.H.; Zhang, Y.C. LncRNA Gaplinc promotes the pyroptosis of vascular endothelial cells through SP1 binding to enhance NLRP3 transcription in atherosclerosis. Cell Signal. 2022, 99, 110420. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Haemmig, S.; Deng, Y.; Chen, J.; Simion, V.; Yang, D.; Sukhova, G.; Shvartz, E.; Wara, A.; Cheng, H.S.; et al. A Smooth Muscle Cell-Enriched Long Noncoding RNA Regulates Cell Plasticity and Atherosclerosis by Interacting With Serum Response Factor. Arter. Thromb. Vasc. Biol. 2021, 41, 2399–2416. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, J.; Zhang, J.; Guo, X.; Liu, H.; Li, P.; Zhang, Y.; Lin, C.; Fan, Z. LncRNA PVT1 knockdown alleviated ox-LDL-induced vascular endothelial cell injury and atherosclerosis by miR-153-3p/GRB2 axis via ERK/p38 pathway. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 3508–3521. [Google Scholar] [CrossRef]
- Khyzha, N.; Khor, M.; DiStefano, P.V.; Wang, L.; Matic, L.; Hedin, U.; Wilson, M.D.; Maegdefessel, L.; Fish, J.E. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 16410–16419. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Bibli, S.I.; Siasos, G.; Oikonomou, E.; Perrea, D.N.; Filis, K.; Tousoulis, D.; Sigala, F. Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis. Biomolecules 2021, 11, 623. [Google Scholar] [CrossRef]
- Ulrich, V.; Rotllan, N.; Araldi, E.; Luciano, A.; Skroblin, P.; Abonnenc, M.; Perrotta, P.; Yin, X.; Bauer, A.; Leslie, K.L.; et al. Chronic miR-29 antagonism promotes favorable plaque remodeling in atherosclerotic mice. EMBO Mol. Med. 2016, 8, 643–653. [Google Scholar] [CrossRef]
- Marsh, E.E.; Steinberg, M.L.; Parker, J.B.; Wu, J.; Chakravarti, D.; Bulun, S.E. Decreased expression of microRNA-29 family in leiomyoma contributes to increased major fibrillar collagen production. Fertil. Steril. 2016, 106, 766–772. [Google Scholar] [CrossRef]
- Lopes, J.; Adiguzel, E.; Gu, S.; Liu, S.L.; Hou, G.; Heximer, S.; Assoian, R.K.; Bendeck, M.P. Type VIII collagen mediates vessel wall remodeling after arterial injury and fibrous cap formation in atherosclerosis. Am. J. Pathol. 2013, 182, 2241–2253. [Google Scholar] [CrossRef]
- Kothapalli, D.; Liu, S.L.; Bae, Y.H.; Monslow, J.; Xu, T.; Hawthorne, E.A.; Byfield, F.J.; Castagnino, P.; Rao, S.; Rader, D.J.; et al. Cardiovascular protection by ApoE and ApoE-HDL linked to suppression of ECM gene expression and arterial stiffening. Cell Rep. 2012, 2, 1259–1271. [Google Scholar] [CrossRef]
- Zampetaki, A.; Attia, R.; Mayr, U.; Gomes, R.S.; Phinikaridou, A.; Yin, X.; Langley, S.R.; Willeit, P.; Lu, R.; Fanshawe, B.; et al. Role of miR-195 in aortic aneurysmal disease. Circ. Res. 2014, 115, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Di Gregoli, K.; Mohamad Anuar, N.N.; Bianco, R.; White, S.J.; Newby, A.C.; George, S.J.; Johnson, J.L. MicroRNA-181b Controls Atherosclerosis and Aneurysms Through Regulation of TIMP-3 and Elastin. Circ. Res. 2017, 120, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Kwan, P.; Ding, J.; Tredget, E.E. MicroRNA 181b regulates decorin production by dermal fibroblasts and may be a potential therapy for hypertrophic scar. PLoS ONE 2015, 10, e0123054. [Google Scholar] [CrossRef] [PubMed]
- Merline, R.; Moreth, K.; Beckmann, J.; Nastase, M.V.; Zeng-Brouwers, J.; Tralhao, J.G.; Lemarchand, P.; Pfeilschifter, J.; Schaefer, R.M.; Iozzo, R.V.; et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci. Signal. 2011, 4, ra75. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Wong, G.; Shiina, M. Up-regulation of Histone Methyltransferase, DOT1L, by Matrix Hyaluronan Promotes MicroRNA-10 Expression Leading to Tumor Cell Invasion and Chemoresistance in Cancer Stem Cells from Head and Neck Squamous Cell Carcinoma. J. Biol. Chem. 2016, 291, 10571–10585. [Google Scholar] [CrossRef]
- Wang, X.; Ling, C.C.; Li, L.; Qin, Y.; Qi, J.; Liu, X.; You, B.; Shi, Y.; Zhang, J.; Jiang, Q.; et al. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016, 110, 140–150. [Google Scholar] [CrossRef]
- Chen, C.H.; Cheng, C.Y.; Chen, Y.C.; Sue, Y.M.; Liu, C.T.; Cheng, T.H.; Hsu, Y.H.; Chen, T.H. MicroRNA-328 inhibits renal tubular cell epithelial-to-mesenchymal transition by targeting the CD44 in pressure-induced renal fibrosis. PLoS ONE 2014, 9, e99802. [Google Scholar] [CrossRef]
- Xie, B.; Zhang, C.; Kang, K.; Jiang, S. miR-599 Inhibits Vascular Smooth Muscle Cells Proliferation and Migration by Targeting TGFB2. PLoS ONE 2015, 10, e0141512. [Google Scholar] [CrossRef]
- Al-U’datt, D.G.; Allen, B.G.; Hiram, R.; Alrabadi, N. Current knowledge into the role of the peptidylarginine deiminase (PAD) enzyme family in cardiovascular disease. Eur. J. Pharm. 2021, 891, 173765. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Mao, C.; Jia, Y.; Fu, Y.; Kong, W. Extracellular matrix dynamics in vascular remodeling. Am. J. Physiol. Cell Physiol. 2020, 319, C481–C499. [Google Scholar] [CrossRef]
- Boon, L.; Ugarte-Berzal, E.; Martens, E.; Fiten, P.; Vandooren, J.; Janssens, R.; Blanter, M.; Yu, K.; Boon, M.; Struyf, S.; et al. Citrullination as a novel posttranslational modification of matrix metalloproteinases. Matrix. Biol. 2021, 95, 68–83. [Google Scholar] [CrossRef]
- Budatha, M.; Zhang, J.; Schwartz, M.A. Fibronectin-Mediated Inflammatory Signaling Through Integrin alpha5 in Vascular Remodeling. J. Am. Heart Assoc. 2021, 10, e021160. [Google Scholar] [CrossRef] [PubMed]
- Stefanelli, V.L.; Choudhury, S.; Hu, P.; Liu, Y.; Schwenzer, A.; Yeh, C.R.; Chambers, D.M.; Pesson, K.; Li, W.; Segura, T.; et al. Citrullination of fibronectin alters integrin clustering and focal adhesion stability promoting stromal cell invasion. Matrix. Biol. 2019, 82, 86–104. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Mostafa, R.; Ibili, E.; Fert-Bober, J. Role of protein deimination in cardiovascular diseases: Potential new avenues for diagnostic and prognostic biomarkers. Expert Rev. Proteom. 2021, 18, 1059–1071. [Google Scholar] [CrossRef]
- Cai, Z.; Gong, Z.; Li, Z.; Li, L.; Kong, W. Vascular Extracellular Matrix Remodeling and Hypertension. Antioxid. Redox Signal. 2021, 34, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Song, D.; Wu, J.; Wang, J. Long Non-Coding RNAs Link Oxidized Low-Density Lipoprotein with the Inflammatory Response of Macrophages in Atherogenesis. Front. Immunol. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Pu, L.; Luo, X.; Wang, B.; Li, F.; Liu, B. Regulatory Roles of Related Long Non-coding RNAs in the Process of Atherosclerosis. Front. Physiol. 2020, 11, 564604. [Google Scholar] [CrossRef]
- Li, D.; Ma, Y.; Deng, W.; Feng, J. Construction and Analysis of lncRNA-Associated ceRNA Network in Atherosclerotic Plaque Formation. Biomed. Res. Int. 2022, 2022, 4895611. [Google Scholar] [CrossRef]
- Kraczkowska, W.; Jagodzinski, P.P. The Long Non-Coding RNA Landscape of Atherosclerotic Plaques. Mol. Diagn. Ther. 2019, 23, 735–749. [Google Scholar] [CrossRef]
- Newby, A.C. Matrix metalloproteinases regulate migration, proliferation, and death of vascular smooth muscle cells by degrading matrix and non-matrix substrates. Cardiovasc. Res. 2006, 69, 614–624. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Cao, C.; Yan, H.; Zhou, B.; Gao, Y.; Li, Q.; Li, J. The Long Non-Coding RNA LncRNA8975-1 is Upregulated in Hypertrophic Scar Fibroblasts and Controls Collagen Expression. Cell Physiol. Biochem. 2016, 40, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Li, Q.; Li, X.; Gao, Y.; Hua, X.; Zhou, B.; Li, J. Overexpression of LncRNA AC067945.2 Down-Regulates Collagen Expression in Skin Fibroblasts and Possibly Correlates with the VEGF and Wnt Signalling Pathways. Cell Physiol. Biochem. 2018, 45, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Guo, Y.; Chen, T.; Jin, T.; Ma, L.; Ai, L.; Guo, J.; Niu, Z.; Yang, R.; Wang, Q.; et al. Systematic analyses identify the anti-fibrotic role of lncRNA TP53TG1 in IPF. Cell Death Dis. 2022, 13, 525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, Y.; Hu, Z.; Zhang, Z.; Shao, S.; Yao, Q.; Zheng, L.; Wang, J.; Han, X.; Zhang, Y.; et al. SCARNA10, a nuclear-retained long non-coding RNA, promotes liver fibrosis and serves as a potential biomarker. Theranostics 2019, 9, 3622–3638. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, F.A.; Wang, L.; Wang, Y.F.; Wu, C.F. Long noncoding RNA NEAT1 promotes pulmonary fibrosis by regulating the microRNA4553p/SMAD3 axis. Mol. Med. Rep. 2021, 23, 218. [Google Scholar] [CrossRef]
- Cao, F.; Li, Z.; Ding, W.M.; Yan, L.; Zhao, Q.Y. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-beta1-Smad axis in atrial fibrillation. Mol. Med. 2019, 25, 7. [Google Scholar] [CrossRef]
- Wei, Z.; Chen, L.; Meng, L.; Han, W.; Huang, L.; Xu, A. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric. Cancer 2020, 23, 1018–1032. [Google Scholar] [CrossRef]
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell-Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272. [Google Scholar] [CrossRef]
- Zhong, Q.; Wang, L.; Qi, Z.; Cao, J.; Liang, K.; Zhang, C.; Duan, J. Long Non-coding RNA TUG1 Modulates Expression of Elastin to Relieve Bronchopulmonary Dysplasia via Sponging miR-29a-3p. Front. Pediatr. 2020, 8, 573099. [Google Scholar] [CrossRef]
- Huang, Z.P.; Ding, Y.; Chen, J.; Wu, G.; Kataoka, M.; Hu, Y.; Yang, J.H.; Liu, J.; Drakos, S.G.; Selzman, C.H.; et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc. Res. 2016, 112, 543–554. [Google Scholar] [CrossRef]
- Neumann, P.; Jae, N.; Knau, A.; Glaser, S.F.; Fouani, Y.; Rossbach, O.; Kruger, M.; John, D.; Bindereif, A.; Grote, P.; et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, S.; Li, Q.; Ma, Y.; Sun, P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed. Pharm. 2018, 105, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.J.; Zhang, X.; Ding, N.; Zhang, J.Y.; Chen, Y.P. LncRNA NEAT1 regulates MMP-16 by targeting miR-200a/b to aggravate inflammation in asthma. Autoimmunity 2021, 54, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Chen, C.H.; Sun, Y.J. Overexpression of lncRNA GAS5 attenuates cardiac fibrosis through regulating PTEN/MMP-2 signal pathway in mice. Eur. Rev. Med. Pharm. Sci. 2019, 23, 4414–4418. [Google Scholar] [CrossRef]
- Li, W.D.; Zhou, D.M.; Sun, L.L.; Xiao, L.; Liu, Z.; Zhou, M.; Wang, W.B.; Li, X.Q. LncRNA WTAPP1 Promotes Migration and Angiogenesis of Endothelial Progenitor Cells via MMP1 Through MicroRNA 3120 and Akt/PI3K/Autophagy Pathways. Stem Cells 2018, 36, 1863–1874. [Google Scholar] [CrossRef]
- Zhou, L.; Ren, M.; Zeng, T.; Wang, W.; Wang, X.; Hu, M.; Su, S.; Sun, K.; Wang, C.; Liu, J.; et al. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019, 10, 813. [Google Scholar] [CrossRef]
- Huang, C.K.; Kafert-Kasting, S.; Thum, T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ. Res. 2020, 126, 663–678. [Google Scholar] [CrossRef]
- Libby, P.; Sasiela, W. Plaque stabilization: Can we turn theory into evidence? Am. J. Cardiol. 2006, 98, 26P–33P. [Google Scholar] [CrossRef]
- Prashar, Y.; Bais, S.; Gill, N.S. Emerging role of various signaling pathways in the pathogenesis and therapeutics of atherosclerosis. Rev. Vasc. Med. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Kowara, M.; Cudnoch-Jedrzejewska, A. Different Approaches in Therapy Aiming to Stabilize an Unstable Atherosclerotic Plaque. Int. J. Mol. Sci. 2021, 22, 4354. [Google Scholar] [CrossRef]
- Migdalski, A.; Jawien, A. New insight into biology, molecular diagnostics and treatment options of unstable carotid atherosclerotic plaque: A narrative review. Ann. Transl. Med. 2021, 9, 1207. [Google Scholar] [CrossRef] [PubMed]
- Ormrod, D.J.; Holmes, C.C.; Miller, T.E. Dietary chitosan inhibits hypercholesterolaemia and atherogenesis in the apolipoprotein E-deficient mouse model of atherosclerosis. Atherosclerosis 1998, 138, 329–334. [Google Scholar] [CrossRef]
- Belalcazar, L.M.; Merched, A.; Carr, B.; Oka, K.; Chen, K.H.; Pastore, L.; Beaudet, A.; Chan, L. Long-term stable expression of human apolipoprotein A-I mediated by helper-dependent adenovirus gene transfer inhibits atherosclerosis progression and remodels atherosclerotic plaques in a mouse model of familial hypercholesterolemia. Circulation 2003, 107, 2726–2732. [Google Scholar] [CrossRef]
- Veseli, B.E.; Perrotta, P.; De Meyer, G.R.; Roth, L.; Van der Donckt, C.; Martinet, W.; De Meyer, G.R. Animal models of atherosclerosis. Eur. J. Pharm. 2017, 816, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.M.; Giannarelli, C. Immune cell profiling in atherosclerosis: Role in research and precision medicine. Nat. Rev. Cardiol. 2022, 19, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Bui, A.V.; Diesch, J.; Manasseh, R.; Hausding, C.; Rivera, J.; Haviv, I.; Agrotis, A.; Htun, N.M.; Jowett, J.; et al. A novel mouse model of atherosclerotic plaque instability for drug testing and mechanistic/therapeutic discoveries using gene and microRNA expression profiling. Circ. Res. 2013, 113, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Allahverdian, S.; Pannu, P.S.; Francis, G.A. Contribution of monocyte-derived macrophages and smooth muscle cells to arterial foam cell formation. Cardiovasc. Res. 2012, 95, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Pryma, C.S.; Ortega, C.; Dubland, J.A.; Francis, G.A. Pathways of smooth muscle foam cell formation in atherosclerosis. Curr. Opin. Lipidol. 2019, 30, 117–124. [Google Scholar] [CrossRef]
- Wang, Y.; Dubland, J.A.; Allahverdian, S.; Asonye, E.; Sahin, B.; Jaw, J.E.; Sin, D.D.; Seidman, M.A.; Leeper, N.J.; Francis, G.A. Smooth Muscle Cells Contribute the Majority of Foam Cells in ApoE (Apolipoprotein E)-Deficient Mouse Atherosclerosis. Arter. Thromb. Vasc. Biol. 2019, 39, 876–887. [Google Scholar] [CrossRef]
- Ridker, P.M.; Everett, B.M.; Thuren, T.; MacFadyen, J.G.; Chang, W.H.; Ballantyne, C.; Fonseca, F.; Nicolau, J.; Koenig, W.; Anker, S.D.; et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N. Engl. J. Med. 2017, 377, 1119–1131. [Google Scholar] [CrossRef]
- Tardif, J.C.; Kouz, S.; Waters, D.D.; Bertrand, O.F.; Diaz, R.; Maggioni, A.P.; Pinto, F.J.; Ibrahim, R.; Gamra, H.; Kiwan, G.S.; et al. Efficacy and Safety of Low-Dose Colchicine after Myocardial Infarction. N. Engl. J. Med. 2019, 381, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Investigators, S.; White, H.D.; Held, C.; Stewart, R.; Tarka, E.; Brown, R.; Davies, R.Y.; Budaj, A.; Harrington, R.A.; Steg, P.G.; et al. Darapladib for preventing ischemic events in stable coronary heart disease. N. Engl. J. Med. 2014, 370, 1702–1711. [Google Scholar] [CrossRef] [PubMed]
- Tall, A.R.; Rader, D.J. Trials and Tribulations of CETP Inhibitors. Circ. Res. 2018, 122, 106–112. [Google Scholar] [CrossRef]
- Kitahara, S.; Kataoka, Y.; Sugane, H.; Otsuka, F.; Asaumi, Y.; Noguchi, T.; Yasuda, S. In vivo imaging of vulnerable plaque with intravascular modalities: Its advantages and limitations. Cardiovasc. Diagn. Ther. 2020, 10, 1461–1479. [Google Scholar] [CrossRef]
- Homorodean, C.; Leucuta, D.C.; Ober, M.; Homorodean, R.; Spinu, M.; Olinic, M.; Tataru, D.; Olinic, D.M. Intravascular ultrasound insights into the unstable features of the coronary atherosclerotic plaques: A systematic review and meta-analysis. Eur. J. Clin. Investig. 2022, 52, e13671. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, M.; Li, X.; Zhao, Y.; Shao, J.; Liu, Z.; Lin, L.; Xu, Q.; Wang, L.; Lu, X.; et al. Anti-inflammatory Therapies for Coronary Heart Disease: A Systematic Review and Meta-Analysis. Front. Cardiovasc. Med. 2021, 8, 726341. [Google Scholar] [CrossRef] [PubMed]
- Tardif, J.C.; McMurray, J.J.; Klug, E.; Small, R.; Schumi, J.; Choi, J.; Cooper, J.; Scott, R.; Lewis, E.F.; L’Allier, P.L.; et al. Effects of succinobucol (AGI-1067) after an acute coronary syndrome: A randomised, double-blind, placebo-controlled trial. Lancet 2008, 371, 1761–1768. [Google Scholar] [CrossRef]
- Wudexi, I.; Shokri, E.; Abo-Aly, M.; Shindo, K.; Abdel-Latif, A. Comparative Effectiveness of Anti-Inflammatory Drug Treatments in Coronary Heart Disease Patients: A Systematic Review and Network Meta-Analysis. Mediat. Inflamm. 2021, 2021, 5160728. [Google Scholar] [CrossRef] [PubMed]
- Kuhnast, S.; van der Tuin, S.J.; van der Hoorn, J.W.; van Klinken, J.B.; Simic, B.; Pieterman, E.; Havekes, L.M.; Landmesser, U.; Luscher, T.F.; Willems van Dijk, K.; et al. Anacetrapib reduces progression of atherosclerosis, mainly by reducing non-HDL-cholesterol, improves lesion stability and adds to the beneficial effects of atorvastatin. Eur. Heart J. 2015, 36, 39–48. [Google Scholar] [CrossRef]
- Kaikita, K.; Yasuda, S.; Akao, M.; Ako, J.; Matoba, T.; Nakamura, M.; Miyauchi, K.; Hagiwara, N.; Kimura, K.; Hirayama, A.; et al. Bleeding and Subsequent Cardiovascular Events and Death in Atrial Fibrillation With Stable Coronary Artery Disease: Insights From the AFIRE Trial. Circ. Cardiovasc. Interv. 2021, 14, e010476. [Google Scholar] [CrossRef]
- Kitayama, K.; Koga, T.; Maeda, N.; Inaba, T.; Fujioka, T. Pactimibe stabilizes atherosclerotic plaque through macrophage acyl-CoA:cholesterol acyltransferase inhibition in WHHL rabbits. Eur. J. Pharm. 2006, 539, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, D.B.; Lehmann, R.A.; Schneck, L.; Keenan, R.T.; Shah, B.; Greenberg, J.D.; Cronstein, B.N.; Sedlis, S.P.; Pillinger, M.H. Colchicine use is associated with decreased prevalence of myocardial infarction in patients with gout. J. Rheumatol. 2012, 39, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Toth, P.P.; Barter, P.J.; Rosenson, R.S.; Boden, W.E.; Chapman, M.J.; Cuchel, M.; D’Agostino, R.B., Sr.; Davidson, M.H.; Davidson, W.S.; Heinecke, J.W.; et al. High-density lipoproteins: A consensus statement from the National Lipid Association. J. Clin. Lipidol. 2013, 7, 484–525. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.H.; Liu, C.C.; Kuo, I.H.; Zak, A.; Kim, S.C. Effects of colchicine on risk of cardiovascular events and mortality among patients with gout: A cohort study using electronic medical records linked with Medicare claims. Ann. Rheum. Dis. 2016, 75, 1674–1679. [Google Scholar] [CrossRef]
- White, H.; Held, C.; Stewart, R.; Watson, D.; Harrington, R.; Budaj, A.; Steg, P.G.; Cannon, C.P.; Krug-Gourley, S.; Wittes, J.; et al. Study design and rationale for the clinical outcomes of the STABILITY Trial (STabilization of Atherosclerotic plaque By Initiation of darapLadIb TherapY) comparing darapladib versus placebo in patients with coronary heart disease. Am. Heart J. 2010, 160, 655–661. [Google Scholar] [CrossRef]
- Nissen, S.E.; Wolski, K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N. Engl. J. Med. 2007, 356, 2457–2471. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. The changing landscape of atherosclerosis. Nature 2021, 592, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, M.W.; Moore, K.J. MicroRNA regulation of atherosclerosis. Circ. Res. 2016, 118, 703–720. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).