Immune Response Gaps Linked to SARS-CoV-2 Infection: Cellular Exhaustion, Senescence, or Both?
Abstract
:1. Introduction
2. Results
3. Discussion
3.1. Demographic and Histopathological Findings
3.2. Cellular Response
3.3. Perforin Polymorphism
3.4. Study Limitations
4. Materials and Methods
4.1. Ethical Approval
4.2. Samples
4.3. Immunohistochemistry Analysis
4.4. Genetic Alleles Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gustine, J.N.; Jones, D. Immunopathology of Hyperinflammation in COVID-19. Am. J. Pathol. 2021, 191, 4–17. Available online: http://ajp.amjpathol.org/article/S0002944020304090/fulltext (accessed on 2 April 2022). [CrossRef] [PubMed]
- Viasus, D.; Oteo Revuelta, J.A.; Martínez-Montauti, J.; Carratalà, J. Influenza A(H1N1)pdm09-related pneumonia and other complications. Enferm. Infecc. Microbiol. Clin. 2012, 30 (Suppl. S4), 43–48. Available online: https://pubmed.ncbi.nlm.nih.gov/23116792/ (accessed on 2 April 2022). [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; The Northwell COVID-19 Research Consortium. Presenting Characteristics, Comorbidities, and Outcomes among 5700 Patients Hospitalized with COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. Available online: https://pubmed.ncbi.nlm.nih.gov/32320003/ (accessed on 2 April 2022). [CrossRef] [PubMed]
- Chen, Z.; John Wherry, E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020, 20, 529–536. Available online: https://www.nature.com/articles/s41577-020-0402-6 (accessed on 30 March 2022). [CrossRef] [PubMed]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Imunologia Celular e Molecular, 8th ed.; Elsevier: Rio de Janeiro, Brazil, 2012; Available online: http://library1.nida.ac.th/termpaper6/sd/2554/19755.pdf (accessed on 9 April 2022).
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7803150/ (accessed on 2 April 2022). [CrossRef]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2020, 93, 250–256. [Google Scholar] [CrossRef]
- Acharya, D.; Liu, G.Q.; Gack, M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. Available online: https://www.nature.com/articles/s41577-020-0346-x (accessed on 3 April 2022). [CrossRef]
- Varchetta, S.; Mele, D.; Oliviero, B.; Mantovani, S.; Ludovisi, S.; Cerino, A.; Bruno, R.; Castelli, A.; Mosconi, M.; Vecchia, M.; et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2020, 18, 604–612. Available online: https://www.nature.com/articles/s41423-020-00557-9 (accessed on 16 April 2022). [CrossRef]
- Miyamoto, T. [Interferon and bone]. Clin. Calcium 2015, 25, 1653–1658. Available online: https://pubmed.ncbi.nlm.nih.gov/26503870/ (accessed on 16 April 2022).
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 12, 2952. [Google Scholar] [CrossRef]
- de Paula, C.B.V.; de Azevedo, M.L.V.; Nagashima, S.; Martins, A.P.C.; Malaquias, M.A.S.; Miggiolaro, A.F.R.D.S.; da Silva Motta Júnior, J.; Avelino, G.; do Carmo, L.A.P.; Carstens, L.B.; et al. IL-4/IL-13 remodeling pathway of COVID-19 lung injury. Sci. Rep. 2020, 10, 4–11. [Google Scholar] [CrossRef]
- Lee, J.-W.; Chun, W.; Lee, H.; Min, J.-H.; Kim, S.-M.; Seo, J.-Y.; Ahn, K.-S.; Oh, S.-R. The Role of Macrophages in the Development of Acute and Chronic Inflammatory Lung Diseases. Cells 2021, 10, 897. Available online: https://pubmed.ncbi.nlm.nih.gov/33919784/ (accessed on 16 April 2022). [CrossRef]
- Li, M.; Guo, W.; Dong, Y.; Wang, X.; Dai, D.; Liu, X.; Wu, Y.; Li, M.; Zhang, W.; Zhou, H.; et al. Elevated Exhaustion Levels of NK and CD8+ T Cells as Indicators for Progression and Prognosis of COVID-19 Disease. Front. Immunol. 2020, 11, 2681. [Google Scholar] [CrossRef]
- Westmeier, J.; Paniskaki, K.; Karaköse, Z.; Werner, T.; Sutter, K.; Dolff, S.; Overbeck, M.; Limmer, A.; Liu, J.; Zheng, X.; et al. Impaired cytotoxic CD8+ T cell response in elderly COVID-19 patients. mBio 2020, 11, e02243-20. Available online: https://journals.asm.org/doi/full/10.1128/mBio.02243-20 (accessed on 30 March 2022). [CrossRef]
- de Paula, C.B.V.; Nagashima, S.; Liberalesso, V.; Collete, M.; da Silva, F.P.G.; Oricil, A.G.G.; Barbosa, G.S.; da Silva, G.V.C.; Wiedmer, D.B.; Dezidério, F.D.S.; et al. COVID-19: Immunohistochemical Analysis of TGF-β Signaling Pathways in Pulmonary Fibrosis. Int. J. Mol. Sci. 2021, 23, 168. [Google Scholar] [CrossRef]
- Paramsothy, A.; Lartey Jalloh, S.; Davies, R.A.; Guttormsen, A.B.; Cox, R.J.; Mohn, K.G.I. Humoral and cellular immune responses in critically ill influenza A/H1N1-infected patients. Scand. J. Immunol. 2021, 94, e13045. Available online: https://pubmed.ncbi.nlm.nih.gov/33891354/ (accessed on 22 April 2022). [CrossRef]
- Hariri, L.P.; North, C.M.; Shih, A.R.; Israel, R.A.; Maley, J.H.; Villalba, J.A.; Vinarsky, V.; Rubin, J.; Okin, D.A.; Sclafani, A.; et al. Lung Histopathology in Coronavirus Disease 2019 as Compared With Severe Acute Respiratory Sydrome and H1N1 Influenza: A Systematic Review. Chest 2021, 159, 73–84. Available online: https://pubmed.ncbi.nlm.nih.gov/33038391/ (accessed on 22 April 2022). [CrossRef]
- Zhou, S.; Wang, Y.; Zhu, T.; Xia, L. CT Features of Coronavirus Disease 2019 (COVID-19) Pneumonia in 62 Patients in Wuhan, China. Am. J. Roentgenol. 2020, 214, 1287–1294. Available online: https://www.ajronline.org (accessed on 30 August 2021). [CrossRef]
- Nagashima, S.; Mendes, M.C.; Martins, A.P.C.; Borges, N.H.; Godoy, T.M.; Dos Santos Miggiolaro, A.F.R.; Dos Santos Dezidério, F.; Machado-Souza, C.; De Noronha, L. Endothelial Dysfunction and Thrombosis in Patients with COVID-19—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2404. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7505138/ (accessed on 22 April 2022). [CrossRef]
- Jia, L.; Xie, J.; Zhao, J.; Cao, D.; Liang, Y.; Hou, X.; Wang, L.; Li, Z. Mechanisms of severe mortality-associated bacterial co-infections following influenza virus infection. Front. Cell. Infect. Microbiol. 2017, 7, 338. [Google Scholar] [CrossRef] [Green Version]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. Available online: https://pubmed.ncbi.nlm.nih.gov/33980688/ (accessed on 24 April 2022). [CrossRef] [PubMed]
- Doglioni, C.; Ravaglia, C.; Chilosi, M.; Rossi, G.; Dubini, A.; Pedica, F.; Piciucchi, S.; Vizzuso, A.; Stella, F.; Maitan, S.; et al. COVID-19 Interstitial Pneumonia: Histological and Immunohistochemical Features on Cryobiopsies. Respiration 2021, 100, 488–498. Available online: https://www.karger.com/Article/FullText/514822 (accessed on 24 April 2022). [CrossRef] [PubMed]
- Haller, O.; Kochs, G.; Weber, F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007, 18, 425–433. Available online: https://pubmed.ncbi.nlm.nih.gov/17683972/ (accessed on 26 April 2022). [CrossRef] [PubMed]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature 2021, 600, 295–301. Available online: https://www.nature.com/articles/s41586-021-04142-6 (accessed on 24 April 2022). [CrossRef] [PubMed]
- Kaneko, N.; Boucau, J.; Kuo, H.-H.; Perugino, C.; Mahajan, V.S.; Farmer, J.R.; Liu, H.; Diefenbach, T.J.; Piechocka-Trocha, A.; Lefteri, K.; et al. Temporal changes in T cell subsets and expansion of cytotoxic CD4+ T cells in the lungs in severe COVID-19. Clin. Immunol. 2022, 237, 108991. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8961941/ (accessed on 24 April 2022). [CrossRef]
- García, L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020, 11, 1441. [Google Scholar] [CrossRef]
- Gadotti, A.C.; de Castro Deus, M.; Telles, J.P.; Wind, R.; Goes, M.; Ossoski, R.G.C.; de Padua, A.M.; de Noronha, L.; Moreno-Amaral, A.; Baena, C.P.; et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020, 289, 198171. [Google Scholar] [CrossRef]
- Schmidt, M.E.; Varga, S.M. The CD8 T cell response to respiratory virus infections. Front. Immunol. 2018, 9, 678. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Larbi, A. Markers of T Cell Senescence in Humans. Int. J. Mol. Sci. 2017, 18, 1742. Available online: https://www.mdpi.com/1422-0067/18/8/1742/htm (accessed on 1 April 2022). [CrossRef] [Green Version]
- Virgin, H.W.; Wherry, E.J.; Ahmed, R. Redefining chronic viral infection. Cell 2009, 138, 30–50. Available online: https://pubmed.ncbi.nlm.nih.gov/19596234/ (accessed on 6 May 2022). [CrossRef] [Green Version]
- Arcanjo, A.; Guimarães Pinto, K.; Logullo, J.; Leite, P.E.C.; Menezes, C.C.B.; Freire-de-Lima, L.; Diniz-Lima, I.; Decoté-Ricardo, D.; Nunes Rodrigues-da-Silva, R.; Geraldo Freire-de-Lima, C.; et al. Critically Ill Coronavirus Disease 2019 Patients Exhibit Hyperactive Cytokine Responses Associated with Effector Exhausted Senescent T Cells in Acute Infection. J. Infect. Dis. 2021, 224, 1672–1683. Available online: https://academic.oup.com/jid/article/224/10/1672/6357075 (accessed on 6 May 2022). [CrossRef]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. Available online: https://pubmed.ncbi.nlm.nih.gov/21739672/ (accessed on 2 April 2022). [CrossRef]
- Fujita, J.; Higa, H.; Azuma, M.; Kin, M.; Kakazu, K.; Toume, M.; Nakazato, I.; Cash, H.L.; Higa, F.; Tateyama, M.; et al. Immunohistochemical findings of an autopsied lung specimen from a patient with pandemic influenza (A/H1N1pdm) virus infection. Intern Med. 2012, 51, 507–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- House, I.G.; Thia, K.; Brennan, A.J.; Tothill, R.; Dobrovic, A.; Yeh, W.Z.; Saffery, R.; Chatterton, Z.; Trapani, J.A.; Voskoboinik, I. Heterozygosity for the common perforin mutation, p.A91V, impairs the cytotoxicity of primary natural killer cells from healthy individuals. Immunol. Cell Biol. 2015, 93, 575–580. Available online: https://pubmed.ncbi.nlm.nih.gov/25776844/ (accessed on 6 May 2022). [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. Available online: https://pubmed.ncbi.nlm.nih.gov/25998963/ (accessed on 6 May 2022). [CrossRef]
- Zanchettin, A.C.; Barbosa, L.V.; Dutra, A.A.; Prá, D.M.M.; Pereira, M.R.C.; Stocco, R.B.; Martins, A.P.C.; Vaz de Paula, C.B.; Nagashima, S.; de Noronha, L.; et al. Role of Genetic Polymorphism Present in Macrophage Activation Syndrome Pathway in Post Mortem Biopsies of Patients with COVID-19. Viruses 2022, 14, 1699. [Google Scholar] [CrossRef]
- Jaworowska, A.; Pastorczak, A.; Trelinska, J.; Wypyszczak, K.; Borowiec, M.; Fendler, W.; Sedek, L.; Szczepanski, T.; Ploski, R.; Młynarski, W. Perforin gene variation influences survival in childhood acute lymphoblastic leukemia. Leuk. Res. 2018, 65, 29–33. [Google Scholar] [CrossRef]
- dbSNP. NCBI Reference SNP Report. Available online: https://www.ncbi.nlm.nih.gov/snp/?term=rs885822 (accessed on 16 October 2021).

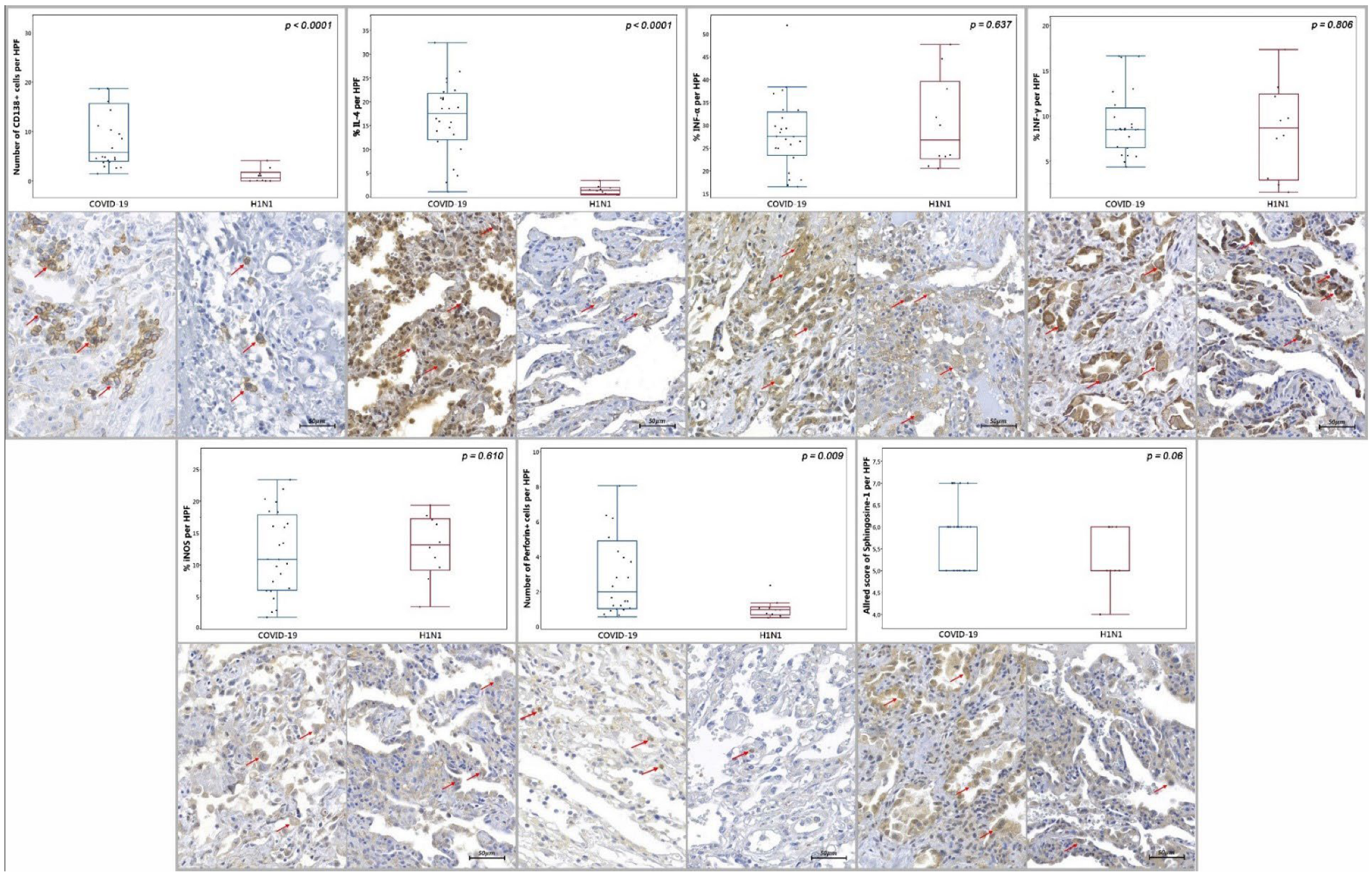
| Data | COVID-19 (n = 24) | H1N1 (n = 10) | p-Value |
|---|---|---|---|
| Gender a | Male 15 (62.5%) Female 9 (37.5%) | Male 8 (80%) Female 2 (20%) | 0.437 * |
| Age (years) b | 71.96 ± 12.5 | 43.5 ± 14 | <0.001 ** |
| Time from hospitalization to death (days) b | 15.87 ± 10.2 | 4.70 ± 6.13 | 0.003 ** |
| Mechanical ventilation b | 12 ± 9.2 | 4.7 ± 6.13 | 0.028 ** |
| Previous pulmonary diseases | Bronchial Asthma (4/24) Interstitial Pulmonary Fibrosis (1/24) | ----- | ----- |
| Histopathological findings | Interstitial pneumonitis with scarce septal neutrophils, hyaline membrane, type II pneumocyte hyperplasia, fibrosis, and micro thrombosis | Interstitial pneumonitis with high septal neutrophils infiltration and no micro thrombosis | ----- |
| Tissue Arginase-1 c | 14.98/9.36 (5.32–37) | 3.81/ 8.60 (1.1–24) | 0.002 *** |
| Tissue CCR4 c | 4.77/10.34 (0.19–18.18) | 6.14/8.06 (1.19–11.86) | 0.845 *** |
| Number of CD3+ d | 45/52.8 (9.3–168) | 48.55/38.92 (29–112.45) | 0.533 *** |
| Number of CD4+ d | 5.9/6.14 (1.45–56.4) | 2.8/2.32 (1.55–5.65) | 0.009 *** |
| Number of CD8+ d | 18.8/20.77 (1.9–52.65) | 35.7/82.84 (7.5–81.05) | 0.03 *** |
| Number of CD20+ d | 4.02/4.4 (0.6–24.95) | 6.3/9.51 (1.85–17.05) | 0.416 *** |
| Number of CD57+ d | 1.08/1.15 (0.3–4.3) | 2.58/1.92 (1.35–5.45) | 0.001 *** |
| Number of CD68+ d | 71.52/42.53 (25.75–118.05) | 46.7/35.54 (17.8–67.15) | 0.013 *** |
| Number of CD138+ d | 5.78/12.75 (1.45–57.85) | 0.58/1.75 (0–4.1) | <0.0001 *** |
| Tissue IL-4 c | 17.5/9.72 (1.02–32.4) | 1.36/1.41 (0.36–3.41) | <0.0001 *** |
| Tissue INF-α c | 10.14/14.5 (0.52–32.75) | 10.35/7.54 (1.36–20) | 0.637 *** |
| Tissue INF-γ c | 8.5/4.39 (4.36–16.62) | 8.66/9.5 (1.57–17.34) | 0.806 *** |
| Tissue iNOS c | 10.88/11.81 (1.78–23.35) | 13.14/8.08 (3.43–19.36) | 0.610 *** |
| Number of PD-1+ a,d | 24 (72.7 %) | 9 (27.3 %) | 0.07 * |
| Number of Perforin-1+ d | 1.98/3.89 (0.55–12) | 0.95/0.44 (0.5–2.35) | 0.009 *** |
| Allred of Sphingosine-1 e | 6/1 (5–7) | 5/1 (4–6) | 0.06 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, L.V.; Prá, D.M.M.; Nagashima, S.; Pereira, M.R.C.; Stocco, R.B.; da Silva, F.d.L.F.; Cruz, M.R.; Dallagassa, D.; Stupak, T.J.; da Rosa Götz, G.W.X.; et al. Immune Response Gaps Linked to SARS-CoV-2 Infection: Cellular Exhaustion, Senescence, or Both? Int. J. Mol. Sci. 2022, 23, 13734. https://doi.org/10.3390/ijms232213734
Barbosa LV, Prá DMM, Nagashima S, Pereira MRC, Stocco RB, da Silva FdLF, Cruz MR, Dallagassa D, Stupak TJ, da Rosa Götz GWX, et al. Immune Response Gaps Linked to SARS-CoV-2 Infection: Cellular Exhaustion, Senescence, or Both? International Journal of Molecular Sciences. 2022; 23(22):13734. https://doi.org/10.3390/ijms232213734
Chicago/Turabian StyleBarbosa, Leonardo Vinicius, Daniele Margarita Marani Prá, Seigo Nagashima, Marcos Roberto Curcio Pereira, Rebecca Benicio Stocco, Francys de Luca Fernandes da Silva, Milena Rueda Cruz, Djessyka Dallagassa, Thiago João Stupak, George Willian Xavier da Rosa Götz, and et al. 2022. "Immune Response Gaps Linked to SARS-CoV-2 Infection: Cellular Exhaustion, Senescence, or Both?" International Journal of Molecular Sciences 23, no. 22: 13734. https://doi.org/10.3390/ijms232213734
APA StyleBarbosa, L. V., Prá, D. M. M., Nagashima, S., Pereira, M. R. C., Stocco, R. B., da Silva, F. d. L. F., Cruz, M. R., Dallagassa, D., Stupak, T. J., da Rosa Götz, G. W. X., Nasimoto, G. G., Cracco, L. A. F., Silva, I. B., de Moura, K. F., Deus, M. d. C., Martins, A. P. C., Spitzenbergen, B. A. K. V., Amaral, A. N. M., de Paula, C. B. V., ... de Noronha, L. (2022). Immune Response Gaps Linked to SARS-CoV-2 Infection: Cellular Exhaustion, Senescence, or Both? International Journal of Molecular Sciences, 23(22), 13734. https://doi.org/10.3390/ijms232213734






