Application of Spectroscopic Methods for the Identification of Superoxide Dismutases in Cyanobacteria
Abstract
:1. Introduction
2. Results
2.1. The 16S rRNA Tree and the Relationship between the Cyanobacteria Studied
2.2. Identification of SODs
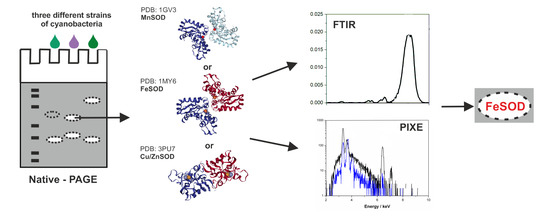
2.3. Metal Identification by the Spectroscopic Methods
2.3.1. Fourier Transformed Infrared (FTIR) Spectroscopy
2.3.2. Particle-Induced X-Ray Emission (PIXE) and Atomic Emission Spectroscopy (AES)
3. Discussion
4. Materials and Methods
4.1. Cyanobacteria
4.2. Databases Query and Phylogenetic Analysis
4.2.1. Sequencing of 16S rRNA
4.2.2. The Reconstruction of 16S rRNA Tree
4.3. Growth Conditions
4.4. Preparation of Soluble Proteins from Cyanobacteria
4.5. SOD Assay
4.6. Quantitative Analysis
4.7. Metal Identification in SOD Activity Bands by Spectroscopic Methods
4.8. Fourier Transformed Infrared (FTIR) Spectroscopy
4.9. Particle-Induced X-ray Emission (PIXE)
4.10. Atomic Emission Spectroscopy (AES)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2015; ISBN 9780198717478. [Google Scholar]
- Hayyan, M.; Hashim, M.A.; Alnashef, I.M. Superoxide ion: Generation and chemical implications. Chem. Rev. 2016, 116, 3029–3085. [Google Scholar] [CrossRef] [Green Version]
- Szymańska, R.; Ślesak, I.; Orzechowska, A.; Kruk, J. Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 2017, 139, 165–177. [Google Scholar] [CrossRef]
- Taverne, Y.J.; Merkus, D.; Bogers, A.J.; Halliwell, B.; Duncker, D.J.; Lyons, T.W. Reactive oxygen species: Radical factors in the evolution of animal life. BioEssays 2018, 40, 1700158. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Takahashi, M. Production and Scavenging of Active Oxygen in Photosynthesis; Kyle, C.D.J., Ed.; Elsevier: Amsterdam, The Netherlands, 1987. [Google Scholar]
- Miller, A.F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012, 586, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef] [PubMed]
- Case, A.J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 2017, 6, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ślesak, I.; Kula, M.; Ślesak, H.; Miszalski, Z.; Strzałka, K. How to define obligatory anaerobiosis? An evolutionary view on the antioxidant response system and the early stages of the evolution of life on Earth. Free Radic. Biol. Med. 2019, 140, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.D.; Huang, B.-W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [Green Version]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Priya, B.; Premanandh, J.; Dhanalakshmi, R.T.; Seethalakshmi, T.; Uma, L.; Prabaharan, D.; Subramanian, G. Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics 2007, 8, 435. [Google Scholar] [CrossRef]
- del Río, L.A.; Corpas, F.J.; López-Huertas, E.; Palma, J.M. Plant Superoxide Dismutases: Function under abiotic stress conditions. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–26. ISBN 978-3-319-75088-0. [Google Scholar]
- Fridovich, I. Superoxide dismutases. Adv. Enzymol. Relat. Areas Mol. Biol. 1986, 58, 61–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, H.P.; Hah, Y.C.; Roe, J.H. Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur. J. Biochem. 1996, 241, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Ślesak, I.; Ślesak, H.; Zimak-Piekarczyk, P.; Rozpądek, P. Enzymatic antioxidant systems in early anaerobes: Theoretical considerations. Astrobiology 2016, 16, 348–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, L.A.; Hug, L.A. Distribution of reactive oxygen species defense mechanisms across domain bacteria. Free Radic. Biol. Med. 2019, 140, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Jabłońska, J.; Tawfik, D.S. The evolution of oxygen-utilizing enzymes suggests early biosphere oxygenation. Nat. Ecol. Evol. 2021, 5, 442–448. [Google Scholar] [CrossRef]
- Boden, J.S.; Konhauser, K.O.; Robbins, L.J.; Sánchez-Baracaldo, P. Timing the evolution of antioxidant enzymes in cyanobacteria. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Schirrmeister, B.E.; Sanchez-Baracaldo, P.; Wacey, D. Cyanobacterial evolution during the Precambrian. Int. J. Astrobiol. 2016, 15, 187–204. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, T.L.; Bryant, D.A.; Macalady, J.L. The role of biology in planetary evolution: Cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ. Microbiol. 2016, 18, 325–340. [Google Scholar] [CrossRef]
- Dick, G.J.; Grim, S.L.; Klatt, J.M. Controls on O2 production in cyanobacterial mats and implications for Earth’s oxygenation. Annu. Rev. Earth Planet. Sci. 2018, 46, 123–147. [Google Scholar] [CrossRef]
- Sánchez-Baracaldo, P.; Cardona, T. On the origin of oxygenic photosynthesis and cyanobacteria. New Phytol. 2020, 225, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Ślesak, I.; Libik, M.; Karpinska, B.; Karpinski, S.; Miszalski, Z. The role of hydrogen peroxide in regulation of plant metabolism and cellular signalling in response to environmental stresses. Acta Biochim. Pol. 2007, 54, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lea-Smith, D.J.; Bombelli, P.; Vasudevan, R.; Howe, C.J. Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Kaneko, T.; Sato, S.; Mimuro, M.; Miyashita, H.; Tsuchiya, T.; Sasamoto, S.; Watanabe, A.; Kawashima, K.; Kishida, Y.; et al. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003, 10, 137–145. [Google Scholar] [CrossRef]
- Gupta, R.S. Protein signatures (molecular synapomorphies) that are distinctive characteristics of the major cyanobacterial clades. Int. J. Syst. Evol. Microbiol. 2009, 59, 2510–2526. [Google Scholar] [CrossRef] [PubMed]
- Mareš, J.; Strunecký, O.; Bučinská, L.; Wiedermannová, J. Evolutionary patterns of thylakoid architecture in cyanobacteria. Front. Microbiol. 2019, 10, 1–22. [Google Scholar] [CrossRef]
- Raven, J.A.; Sánchez-Baracaldo, P. Gloeobacter and the implications of a freshwater origin of Cyanobacteria. Phycologia 2021, 60, 402–418. [Google Scholar] [CrossRef]
- Mareš, J.; Hrouzek, P.; Kaňa, R.; Ventura, S.; Strunecký, O.; Komárek, J. The primitive thylakoid-less cyanobacterium Gloeobacter is a common rock-dwelling organism. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Suh, K.H. Light-dependent expression of superoxide dismutase from cyanobacterium Synechocystis sp. strain PCC 6803. Arch. Microbiol. 2005, 183, 218–223. [Google Scholar] [CrossRef]
- Ke, W.T.; Dai, G.Z.; Jiang, H.B.; Zhang, R.; Qiu, B.S. Essential roles of iron superoxide dismutase in photoautotrophic growth of Synechocystis sp. PCC 6803 and heterogeneous expression of marine Synechococcus sp. CC9311 copper/zinc superoxide dismutase within its sodB knockdown mutant. Microbiology 2014, 160, 228–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duygu, D.Y.; Udoh, A.U.; Ozer, T.B.; Akbulut, A.; Erkaya, I.A.; Yildiz, K.; Guler, D. Fourier transform infrared (FTIR) spectroscopy for identification of Chlorella vulgaris Beijerinck 1890 and Scenedesmus obliquus (Turpin) Kützing 1833. AFRICAN J. Biotechnol. 2012, 11, 3817–3824. [Google Scholar] [CrossRef]
- Wang, X.; Han, M.; Bao, J.; Tu, W.; Dai, Z. A superoxide anion biosensor based on direct electron transfer of superoxide dismutase on sodium alginate sol–gel film and its application to monitoring of living cells. Anal. Chim. Acta 2012, 717, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wu, J.; Ji, X.; Ma, Y.; Liu, L.; Zong, X.; Yang, H.; Dai, J.; Chen, X.; Xue, W. Advanced biomimetic nanoreactor for specifically killing tumor cells through multi-enzyme cascade. Theranostics 2020, 10, 6245–6260. [Google Scholar] [CrossRef]
- Ayala, I.; Perry, J.J.P.; Szczepanski, J.; Tainer, J.A.; Vala, M.T.; Nick, H.S.; Silverman, D.N. Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine. Biophys. J. 2005, 89, 4171–4179. [Google Scholar] [CrossRef] [Green Version]
- David, C.; D’Andrea, C.; Lancelot, E.; Bochterle, J.; Guillot, N.; Fazio, B.; Maragò, O.M.; Sutton, A.; Charnaux, N.; Neubrech, F.; et al. Raman and IR spectroscopy of manganese superoxide dismutase, a pathology biomarker. Vib. Spectrosc. 2012, 62, 50–58. [Google Scholar] [CrossRef]
- Cooper, E.A.; Knutson, K. Fourier Transform Infrared spectroscopy investigations of protein structure. Pharm. Biotechnol. 1995, 7, 101–143. [Google Scholar]
- Vedantham, G.; Sparks, H.G.; Sane, S.U.; Tzannis, S.; Przybycien, T.M. A holistic approach for protein secondary structure estimation from infrared spectra in H2O solutions. Anal. Biochem. 2000, 285, 33–49. [Google Scholar] [CrossRef]
- Sun, W.; Fang, J.; Cheng, M.; Xia, P.; Tang, W. Secondary structure dependent on metal ions of copper, zinc superoxide dismutase investigated by Fourier Transform IR spectroscopy. Biopolymers 1997, 42, 297–303. [Google Scholar] [CrossRef]
- Chatterjee, S.K.; Maji, R.C.; Barman, S.K.; Olmstead, M.M.; Patra, A.K. Hexacoordinate nickel(II)/(III) complexes that mimic the catalytic cycle of nickel superoxide dismutase. Angew. Chemie Int. Ed. 2014, 53, 10184–10189. [Google Scholar] [CrossRef]
- Harada, M.; Akiyama, A.; Furukawa, R.; Yokobori, S.I.; Tajika, E.; Yamagishi, A. Evolution of superoxide dismutases and catalases in cyanobacteria: Occurrence of the antioxidant enzyme genes before the rise of atmospheric oxygen. J. Mol. Evol. 2021, 89, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Canini, A.; Leonardi, D.; Caiola, M.G. Superoxide dismutase activity in the cyanobacterium Microcystis aeruginosa after surface bloom formation. New Phytol. 2001, 152, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Regelsberger, G.; Laaha, U.; Dietmann, D.; Rüker, F.; Canini, A.; Grilli-Caiola, M.; Furtmüller, P.G.; Jakopitsch, C.; Peschek, G.A.; Obinger, C. The iron superoxide dismutase from the filamentous cyanobacterium Nostoc PCC 7120: Localization, overexpression, and biochemical characterization. J. Biol. Chem. 2004, 279, 44384–44393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismaiel, M.M.S.; El-Ayouty, Y.M.; Loewen, P.C.; Piercey-Normore, M.D. Characterization of the iron-containing superoxide dismutase and its response to stress in cyanobacterium Spirulina (Arthrospira) platensis. J. Appl. Phycol. 2014, 26, 1649–1658. [Google Scholar] [CrossRef]
- Kesheri, M.; Kanchan, S.; Richa; Sinha, R.P. Isolation and in silico analysis of Fe-superoxide dismutase in the cyanobacterium Nostoc commune. Gene 2014, 553, 117–125. [Google Scholar] [CrossRef]
- Kesheri, M.; Kanchan, S.; Sinha, R.P. Isolation and in silico analysis of antioxidants in response to temporal variations in the cyanobacterium Oscillatoria sp. Gene Rep. 2021, 23, 101023. [Google Scholar] [CrossRef]
- Takao, M.; Yasui, A.; Oikawa, A. Unique characteristics of superoxide dismutase of a strictly anaerobic archaebacterium Methanobacterium thermoautotrophicum. J. Biol. Chem. 1991, 266, 14151–14154. [Google Scholar] [CrossRef]
- Meier, B.; Sehn, A.P.; Christa, M.; Manfred, S. Reactions of hydrogen peroxide with superoxide dismutase from Propionibacterium shermanii-an enzyme which is equally active with iron or manganese-are independent of the prosthetic metal. Arch. Biochem. Biophys. 1994, 313, 296–303. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Pudake, R.N.; Bae, H. Genome-wide identification of major protein families of cyanobacteria and genomic insight into the circadian rhythm. Eur. J. Phycol. 2017, 52, 149–165. [Google Scholar] [CrossRef]
- Mareš, J.; Komárek, J.; Compère, P.; Oren, A. Validation of the generic name Gloeobacter Rippka et al. 1974, Cyanophyceae. Cryptogam. Algol. 2013, 34, 255–262. [Google Scholar] [CrossRef]
- Liang, W.; Wang, L.; Shi, J.; Lei, X.; Yang, J.; Wu, S.; Chen, W. Differential expression of antioxidant proteins in the drought-tolerant cyanobacterium Nostoc flagelliforme under desiccation. Plant Omics 2014, 7, 205–212. [Google Scholar]
- Wang, H.; Ki, J.S. Molecular characterization and expression analysis of copper-zinc superoxide dismutases from the freshwater alga Closterium ehrenbergii under metal stress. Environ. Toxicol. 2020, 35, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Allen, J.; Foyer, C.H.; Matthijs, H.C.P. The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. B Biol. Sci. 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rippka, R.; Waterbury, J.; Cohen-Bazire, G. A cyanobacterium which lacks thylakoids. Arch. Microbiol. 1974, 100, 419–436. [Google Scholar] [CrossRef]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Rippka, R.; Herdman, M. Pasteur culture collection of cyanobacterial strains in axenic culture. In Catalogue of Strains; Institut Pasteur: Paris, France, 1992; p. 103. ISBN 9783440126349. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miszalski, Z.; Ślesak, I.; Niewiadomska, E.; Baczek-Kwinta, R.; Lüttge, U.; Ratajczak, R. Subcellular localization and stress responses of superoxide dismutase isoforms from leaves in the C3-CAM intermediate halophyte Mesembryanthemum crystallinum L. Plant Cell Environ. 1998, 21, 169–179. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Lekki, J.; Lebed, S.; Paszkowski, M.L.; Kusiak, M.; Vogt, J.; Hajduk, R.; Polak, W.; Potempa, A.; Stachura, Z.; Styczeń, J. Age determination of monazites using the new experimental chamber of the Cracow proton microprobe. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2003, 210, 472–477. [Google Scholar] [CrossRef]
- Campbell, J.L.; Boyd, N.I.; Grassi, N.; Bonnick, P.; Maxwell, J.A. The Guelph PIXE software package IV. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2010, 268, 3356–3363. [Google Scholar] [CrossRef]





| Band of SOD | Strain | Fe (µg g−1) | Mn (µg g−1) | Identified Form of SOD |
|---|---|---|---|---|
| 1A | Synechocystis sp. PCC 6803 | 5.0 ± 1.0 | LOQ a | FeSOD1 |
| 1B | G. violaceus CCALA 979 | 7.9 ± 2.7 | LOQ | FeSOD1 |
| 2B | 1.0 ± 0.4 | LOQ | FeSOD2 | |
| 1C | Geitlerinema sp. ZHR1A | 9.6 ± 1.8 | LOQ | FeSOD1 |
| 2C | 2.6 ± 1.0 | LOQ | FeSOD2 |
| Element | Wavelengths (nm) | Detection Limit (mg dm−3) | Content in Certificated Material (mg kg−1) | Measured (mg kg−1) | Recovery (%) |
|---|---|---|---|---|---|
| Fe | 238.2 | 0.005 | 185 | 201.2 | 108.8 |
| Mn | 257.6 | 0.001 | 47 | 48.6 | 103.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kula-Maximenko, M.; Zieliński, K.J.; Depciuch, J.; Lekki, J.; Niemiec, M.; Ślesak, I. Application of Spectroscopic Methods for the Identification of Superoxide Dismutases in Cyanobacteria. Int. J. Mol. Sci. 2022, 23, 13819. https://doi.org/10.3390/ijms232213819
Kula-Maximenko M, Zieliński KJ, Depciuch J, Lekki J, Niemiec M, Ślesak I. Application of Spectroscopic Methods for the Identification of Superoxide Dismutases in Cyanobacteria. International Journal of Molecular Sciences. 2022; 23(22):13819. https://doi.org/10.3390/ijms232213819
Chicago/Turabian StyleKula-Maximenko, Monika, Kamil Jan Zieliński, Joanna Depciuch, Janusz Lekki, Marcin Niemiec, and Ireneusz Ślesak. 2022. "Application of Spectroscopic Methods for the Identification of Superoxide Dismutases in Cyanobacteria" International Journal of Molecular Sciences 23, no. 22: 13819. https://doi.org/10.3390/ijms232213819