Systematic Identification and Expression Analysis of the Sorghum Pht1 Gene Family Reveals Several New Members Encoding High-Affinity Phosphate Transporters
Abstract
:1. Introduction
2. Results
2.1. Identification of Putative SbPht1 Genes
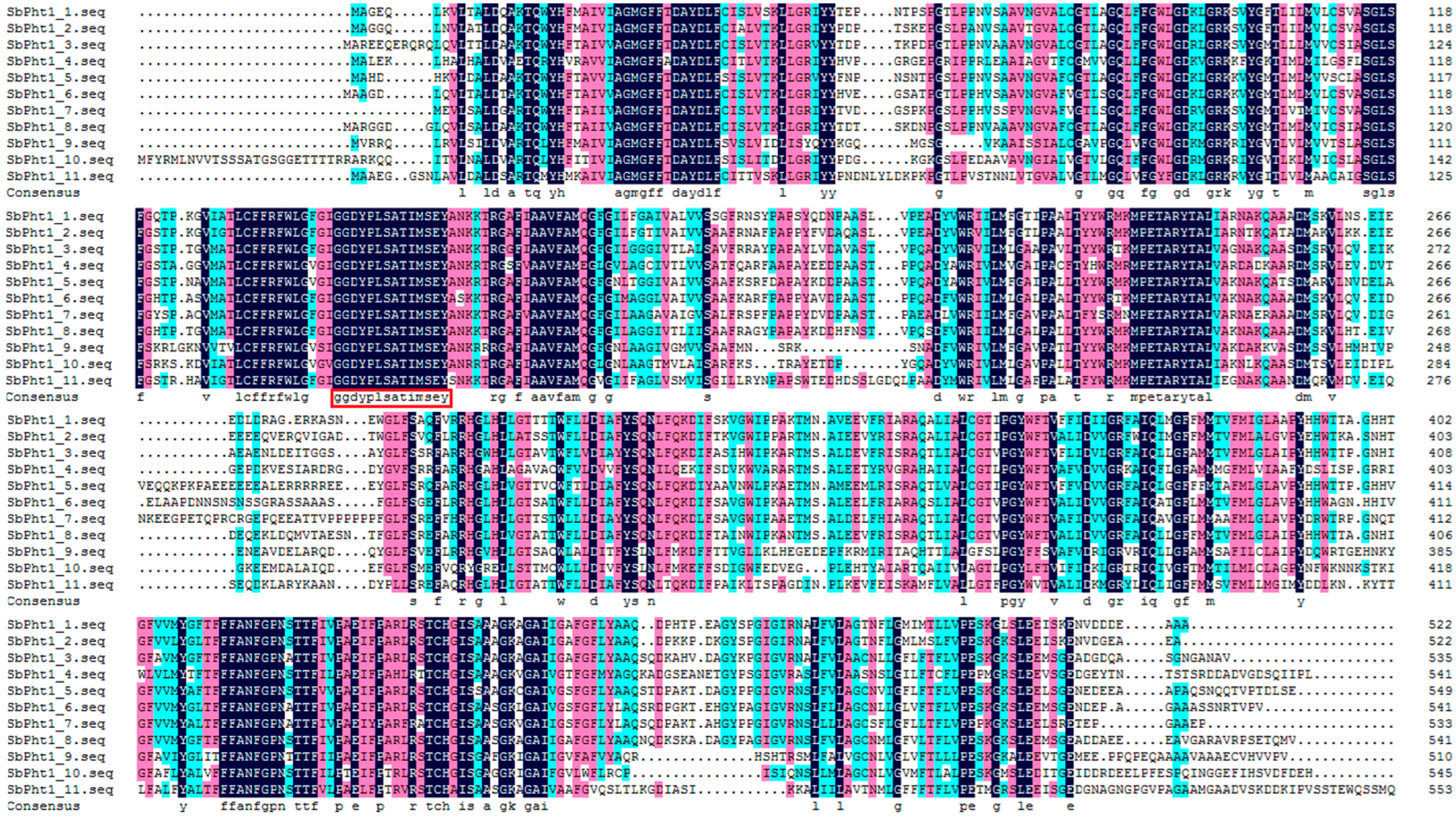
2.2. Bioinformatics Analysis of Pht1 Transporters from Sorghum
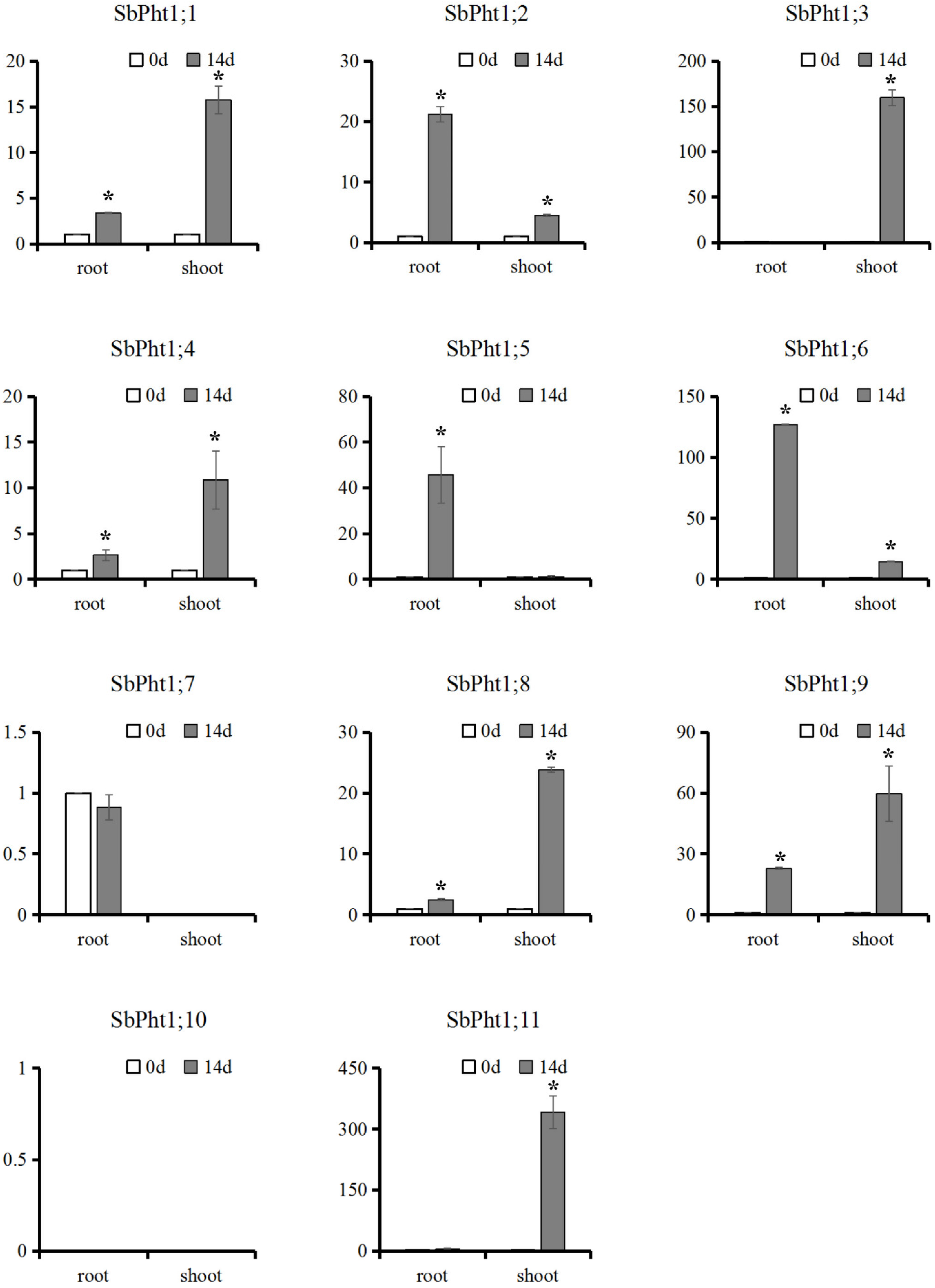
2.3. Expression Analysis of SbPht1s in Responss to Low-P Stress
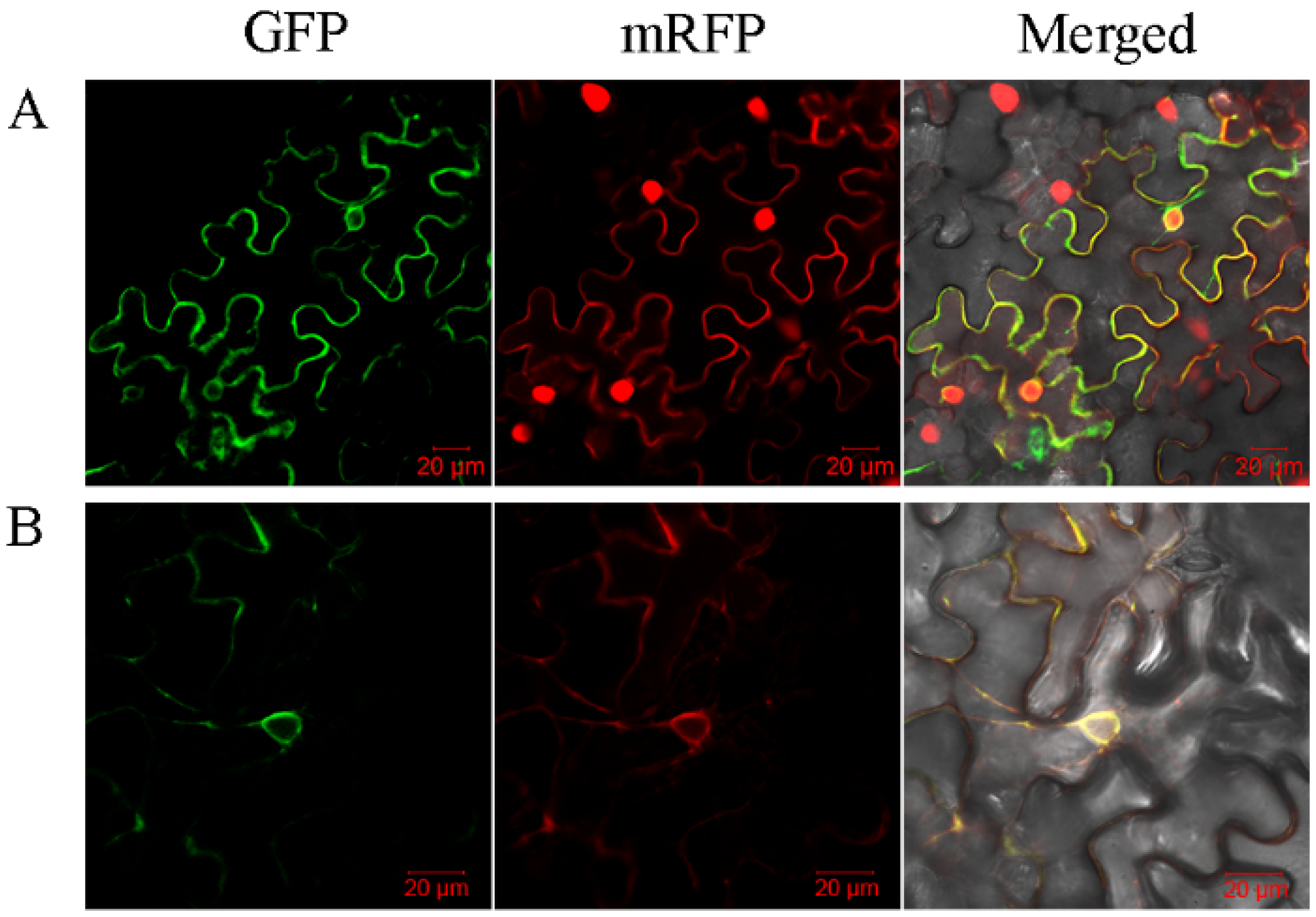
2.4. Subcellular Localization of SbPht1s
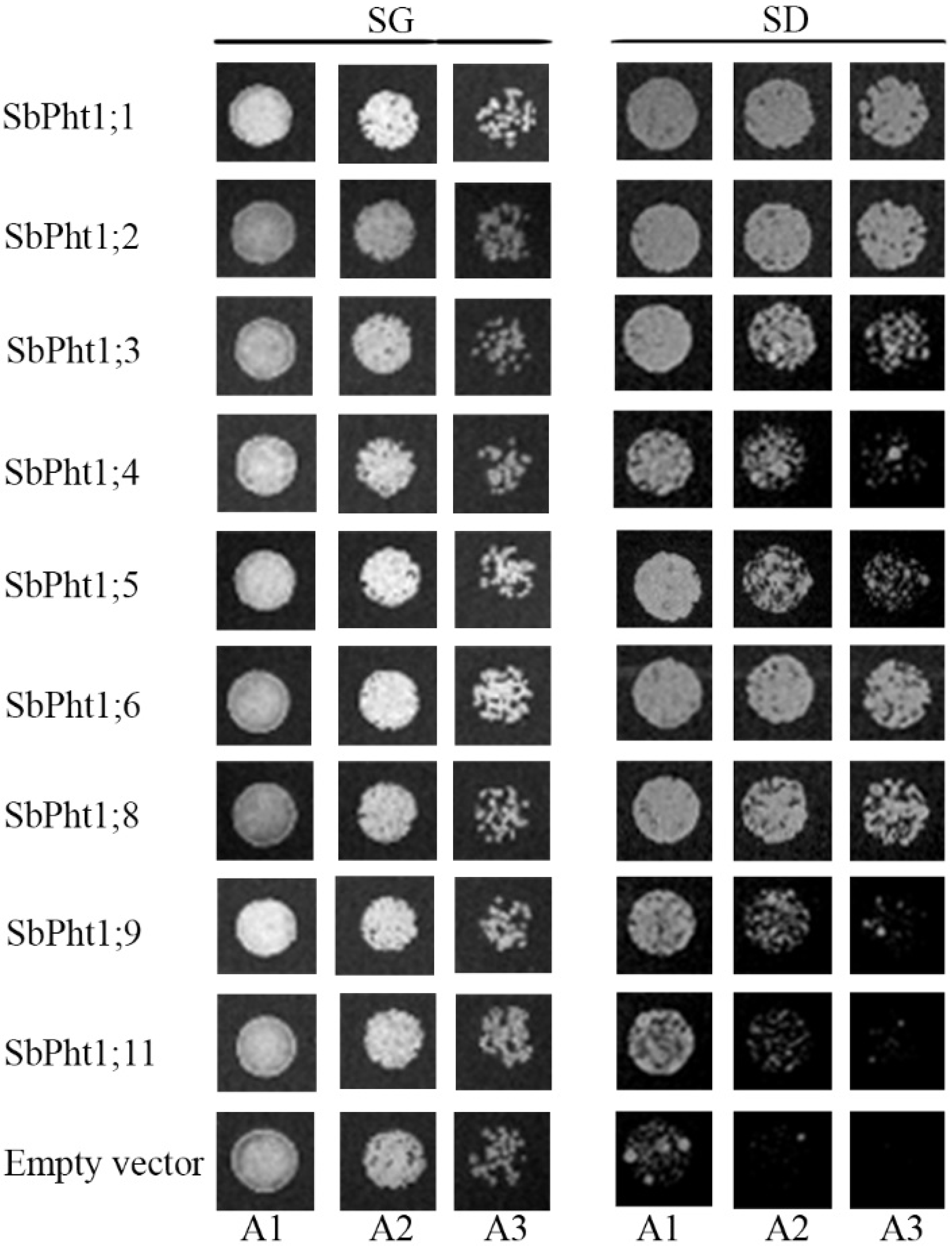
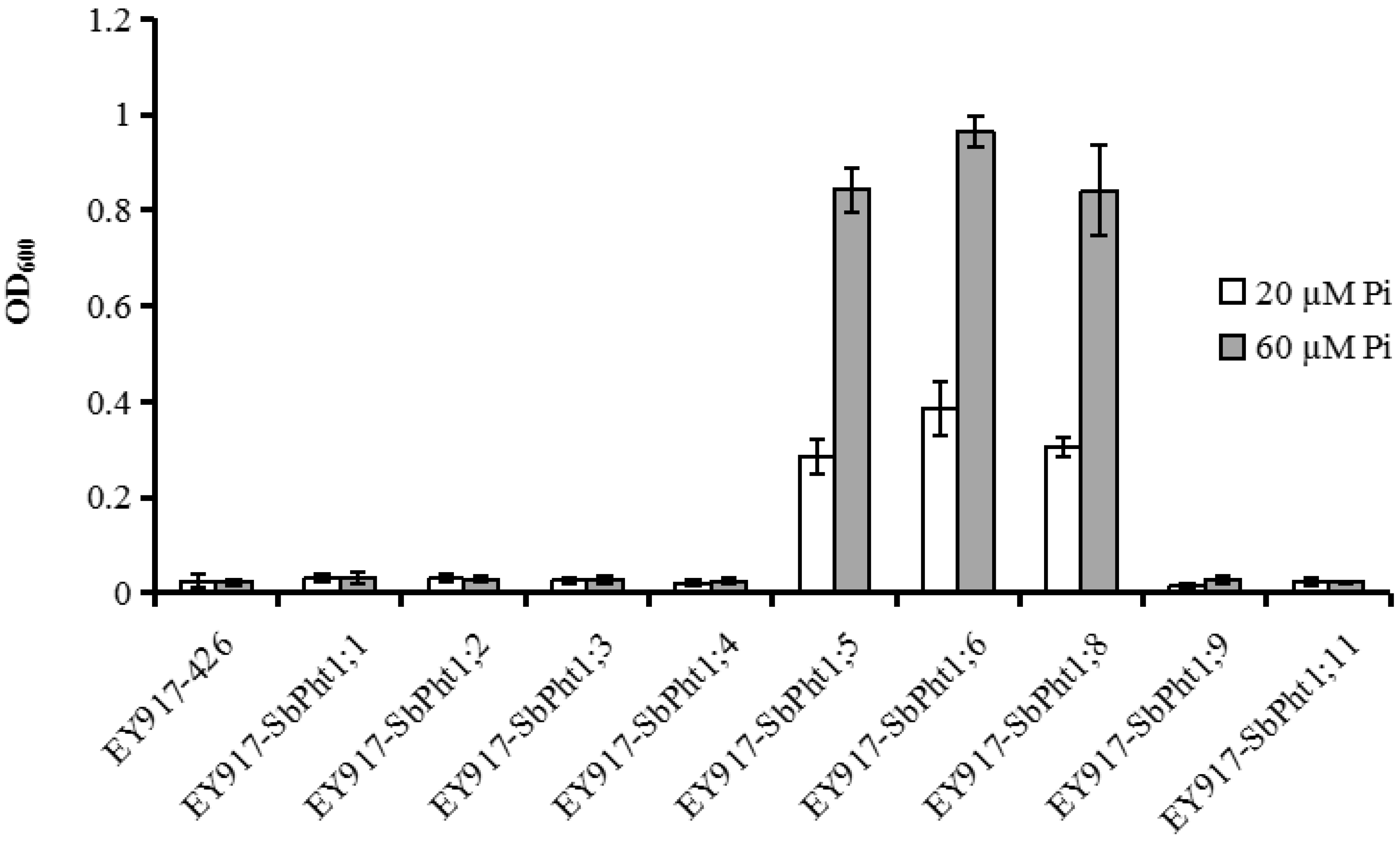
2.5. Functional Analysis of SbPht1s in Yeast
3. Discussion
3.1. Identification of SbPht1 Family Genes
3.2. SbPht1;5, SbPht1;6, and SbPht1;8 Play Important Roles in Sorghum in Response to Low-P Stress
3.3. SbPht1;7 and SbPht1;10 May Have Divergent Functions
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Isolation of Pht1s from Sorghum
4.3. Bioinformatics Analyses
4.4. Gene Expression Analysis
4.5. Subcellular Location
4.6. Yeast Mutant Complement Growth Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bucher, M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 2007, 173, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, P.A. Properties and Management of Soils in the Tropics. Soil Sci. 1977, 124, 187. [Google Scholar] [CrossRef]
- Bieleski, R.L. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [Green Version]
- Marschner, H. Marschner’s Mineral Nutrition of Higher Plants; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Casieri, L.; Ait Lahmidi, N.; Doidy, J.; Veneault-Fourrey, C.; Migeon, A.; Bonneau, L.; Courty, P.E.; Garcia, K.; Charbonnier, M.; Delteil, A.; et al. Biotrophic transportome in mutualistic plant–fungal interactions. Mycorrhiza 2013, 23, 597–625. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef]
- Lopez-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, L.; Luan, M.; Luan, S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E6571–E6578. [Google Scholar] [CrossRef] [Green Version]
- Ullrich-Eberius, C.I.; Novacky, A.; Van Bel, A.J.E. Phosphate uptake in Lemna gibba G1: Energetics and kinetics. Planta 1984, 161, 46–52. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef]
- Bucher, M.; Rausch, C.; Daram, P. Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant Nutr. Soil Sci. 2001, 164, 209–217. [Google Scholar] [CrossRef]
- Muchhal, U.S.; Pardo, J.M.; Raghothama, K.G. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 10519–10523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Lapis-Gaza, H.R.; Jost, R.; Finnegan, P.M. Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1; 8 and PHT1; 9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 2014, 14, 334. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Wang, T.; Zhang, Y.; Li, L.; Wang, C.; Guo, S.; Zhang, T.; Wang, C. GTPase ROP6 negatively modulates phosphate deficiency through inhibition of PHT1; 1 and PHT1; 4 in Arabidopsis thaliana. J. Integr. Plant Biol. 2021, 63, 1775–1786. [Google Scholar] [CrossRef]
- Liu, C.; Muchhal, U.S.; Uthappa, M.; Kononowicz, A.K.; Raghothama, K.G. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998, 116, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Chen, X.; Wang, H.; Liao, D.; Gu, M.; Qu, H.; Sun, S.; Xu, G. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 2014, 14, 61. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; You, Q.; Lei, M. A CRISPR/Cas9 deletion into the phosphate transporter SlPHO1; 1 reveals its role in phosphate nutrition of tomato seedlings. Physiol. Plant. 2019, 167, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Liu, J.; Li, Y.; Zhang, J.; Li, S.; An, Y.; Hu, T.; Yang, P. Functional analysis of the phosphate transporter gene MtPT6 from Medicago truncatula. Front. Plant Sci. 2021, 11, 620377. [Google Scholar] [CrossRef]
- Leggewie, G.; Willmitzer, L.; Riesmeier, J.W. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: Identification of phosphate transporters from higher plants. Plant Cell 1997, 9, 381–392. [Google Scholar] [PubMed] [Green Version]
- Cao, M.; Liu, H.; Zhang, C.; Wang, D.; Liu, X.; Chen, Q. Functional analysis of StPHT1; 7, a Solanum tuberosum L. phosphate transporter gene, in growth and drought tolerance. Plants 2020, 9, 1384. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef] [Green Version]
- Teng, W.; Zhao, Y.Y.; Zhao, X.Q.; He, X.; Ma, W.Y.; Deng, Y.; Chen, X.P.; Tong, Y.P. Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat. Front. Plant Sci. 2017, 8, 543. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hu, L.; Yu, D.; Xu, K.; Zhang, J.; Li, X.; Wang, P.; Chen, G.; Liu, Z.; Peng, C.; et al. Integrative analysis of the wheat PHT1 gene family reveals a novel member involved in Arbuscular mycorrhizal phosphate transport and immunity. Cells 2019, 8, 490. [Google Scholar] [CrossRef] [Green Version]
- Roch, G.V.; Maharajan, T.; Krishna, T.P.; Ignacimuthu, S.; Ceasar, S.A. Expression of PHT1 family transporter genes contributes for low phosphate stress tolerance in foxtail millet (Setaria italica) genotypes. Planta 2020, 252, 98. [Google Scholar] [CrossRef]
- Bulgarelli, R.G.; De Oliveira, V.H.; de Andrade, S.A.L. Arbuscular mycorrhizal symbiosis alters the expression of PHT1 phosphate transporters in roots and nodules of P-starved soybean plants. Theor. Exp. Plant Phys. 2020, 32, 243–253. [Google Scholar] [CrossRef]
- Hufnagel, B.; de Sousa, S.M.; Assis, L.; Guimaraes, C.T.; Leiser, W.; Azevedo, G.C.; Negri, B.; Larson, B.G.; Shaff, J.E.; Pastina, M.M.; et al. Duplicate and conquer: Multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 2014, 166, 659–677. [Google Scholar] [CrossRef]
- Khoddami, A.; Messina, V.; Vadabalija Venkata, K.; Farahnaky, A.; Blanchard, C.L.; Roberts, T.H. Sorghum in foods: Functionality and potential in innovative products. Crit. Rev. Food Sci. 2021, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Liu, X.B.; Dong, C.F.; Lu, X.Y.; Shen, Y.X. Field performance and nutritive value of sweet sorghum in eastern China. Field Crop. Res. 2014, 157, 84–88. [Google Scholar] [CrossRef]
- Otani, T.; Ae, N. Phosphorus (P) uptake mechanisms of crops grown in soils with low P status: I. Screening of crops for efficient P uptake. Soil Sci. Plant Nutr. 1996, 42, 155–163. [Google Scholar] [CrossRef]
- Walder, F.; Brulé, D.; Koegel, S.; Wiemken, A.; Boller, T.; Courty, P.E. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Glassop, D.; Godwin, R.M.; Smith, S.E.; Smith, F.W. Rice phosphate transporters associated with phosphate uptake in rice roots colonised with arbuscular mycorrhizal fungi. Botany 2007, 85, 644–651. [Google Scholar] [CrossRef]
- Smith, S.E.; Dickson, S.; Smith, F.A. Nutrient transfer in arbuscular mycorrhizas: How are fungal and plant processes integrated? Funct. Plant Biol. 2001, 28, 685–696. [Google Scholar] [CrossRef]
- Smith, F.W.; Rae, A.L.; Hawkesford, M.J. Molecular mechanisms of phosphate and sulphate transport in plants. BBA-Biomembranes 2000, 1465, 236–245. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Huang, W.; Liu, F.; Tang, N.; Liu, Y.; Lin, H.; Zhao, B. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef]
- Karthikeyan, A.S.; Varadarajan, D.K.; Mukatira, U.T.; D’Urzo, M.P.; Damsz, B.; Raghothama, K.G. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002, 130, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Lian, X.M. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate from source to sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, L.; Guo, Y.; Chen, L.; Liang, R.; Gu, M.; Xu, G.; Zhao, J.; Walk, T.; Liao, H. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 2012, 7, e47726. [Google Scholar] [CrossRef]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, J.; Zhang, X.; Fan, H.; Gu, M.; Qu, H.; Xu, G. Phosphate transporter OsPht1; 8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 2015, 230, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-pcr data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Jiang, F.; Shen, Y.; Zhan, Q.; Bai, B.; Chen, W.; Chi, Y. Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol. 2019, 19, 306. [Google Scholar] [CrossRef]
- Wang, X.; Fan, C.; Zhang, X.; Zhu, J.; Fu, Y.F. BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol. 2013, 13, 198. [Google Scholar] [CrossRef] [Green Version]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Darbani, B.; Motawia, M.S.; Olsen, C.E.; Nour-Eldin, H.H.; Møller, B.L.; Rook, F. The biosynthetic gene cluster for the cyanogenic glucoside dhurrin in Sorghum bicolor contains its co-expressed vacuolar MATE transporter. Sci. Rep. 2016, 6, 37079. [Google Scholar] [CrossRef]
- Wang, C.; Yue, W.; Ying, Y.; Wang, S.; Secco, D.; Liu, Y.; Whelan, J.; Tyerman, S.D.; Shou, H. Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 2015, 169, 2822–2831. [Google Scholar] [PubMed] [Green Version]
- Wykoff, D.D.; O’Shea, E.K. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 2001, 159, 1491–1499. [Google Scholar] [CrossRef] [PubMed]


| Gene | Alias | Accession Number | CDS Size (bp) | Amino acid Length (AA) | Protein Molecular Mass (Da) | Isoelectric Point of Protein |
|---|---|---|---|---|---|---|
| SbPht1;1 | Sb01g020580 | XM_002467113.2 | 1566 | 522 | 56,952.80 | 7.85 |
| SbPht1;2 | Sb01g046890 | XM_002465800.2 | 1566 | 522 | 57,129.30 | 8.69 |
| SbPht1;3 | Sb01g046900 | XM_002468450.2 | 1605 | 535 | 57,871.40 | 8.74 |
| SbPht1;4 | Sb02g009880 | XM_002461845.1 | 1623 | 541 | 58,903.60 | 7.95 |
| SbPht1;5 | Sb06g002800 | XM_002447480.2 | 1647 | 549 | 60,197.30 | 7.04 |
| SbPht1;6 | Sb07g023780 | XM_002445623.2 | 1623 | 541 | 57,785.30 | 8.88 |
| SbPht1;7 | Sb01g047910 | DQ459071.1 | 1599 | 533 | 57,492.10 | 6.92 |
| SbPht1;8 | Sb01g020570 | XM_002464513.2 | 1623 | 541 | 58,768.40 | 7.82 |
| SbPht1;9 | Sb06g002560 | XM_002446106.2 | 1530 | 510 | 56,245.90 | 9.52 |
| SbPht1;10 | Sb06g002540 | XM_002447472.2 | 1635 | 545 | 60,421.60 | 6.67 |
| SbPht1;11 | Sb03g029970 | XM_002458208.2 | 1662 | 554 | 60,245.10 | 7.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Shen, Y.; Chen, W.; Bai, B.; Ji, X.; Chi, Y. Systematic Identification and Expression Analysis of the Sorghum Pht1 Gene Family Reveals Several New Members Encoding High-Affinity Phosphate Transporters. Int. J. Mol. Sci. 2022, 23, 13855. https://doi.org/10.3390/ijms232213855
Zhang J, Shen Y, Chen W, Bai B, Ji X, Chi Y. Systematic Identification and Expression Analysis of the Sorghum Pht1 Gene Family Reveals Several New Members Encoding High-Affinity Phosphate Transporters. International Journal of Molecular Sciences. 2022; 23(22):13855. https://doi.org/10.3390/ijms232213855
Chicago/Turabian StyleZhang, Jinglong, Yixin Shen, Wei Chen, Binqiang Bai, Xiaomin Ji, and Yingjun Chi. 2022. "Systematic Identification and Expression Analysis of the Sorghum Pht1 Gene Family Reveals Several New Members Encoding High-Affinity Phosphate Transporters" International Journal of Molecular Sciences 23, no. 22: 13855. https://doi.org/10.3390/ijms232213855




