Novel Insights into Anthocyanin Synthesis in the Calyx of Roselle Using Integrated Transcriptomic and Metabolomic Analyses
Abstract
1. Introduction
2. Results
2.1. Anthocyanin Content in Roselle Calyxes
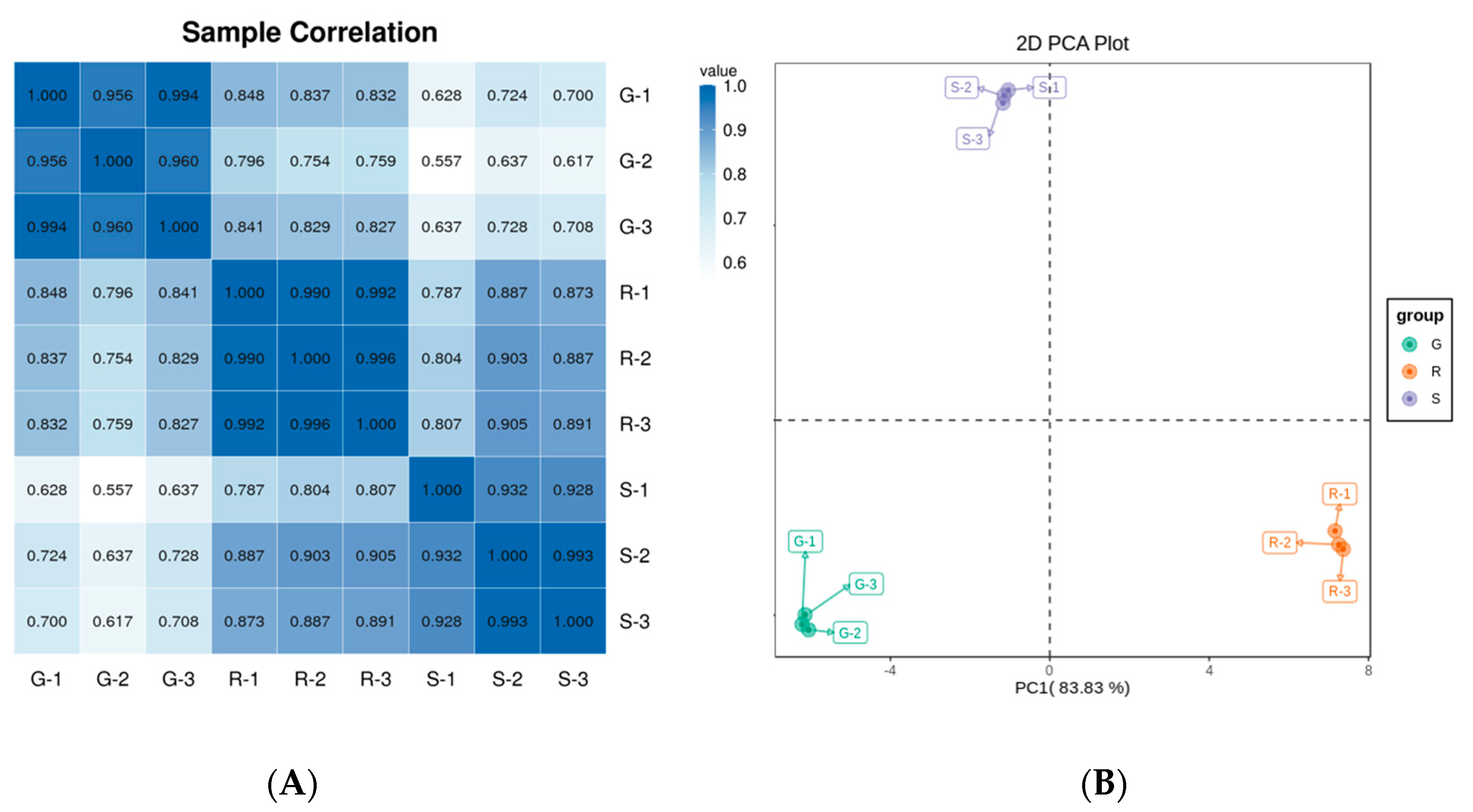
2.2. Sample Correlation Analysis of Roselle Calyxes and Principal Component Analysis
2.3. Differential Metabolite of Roselle
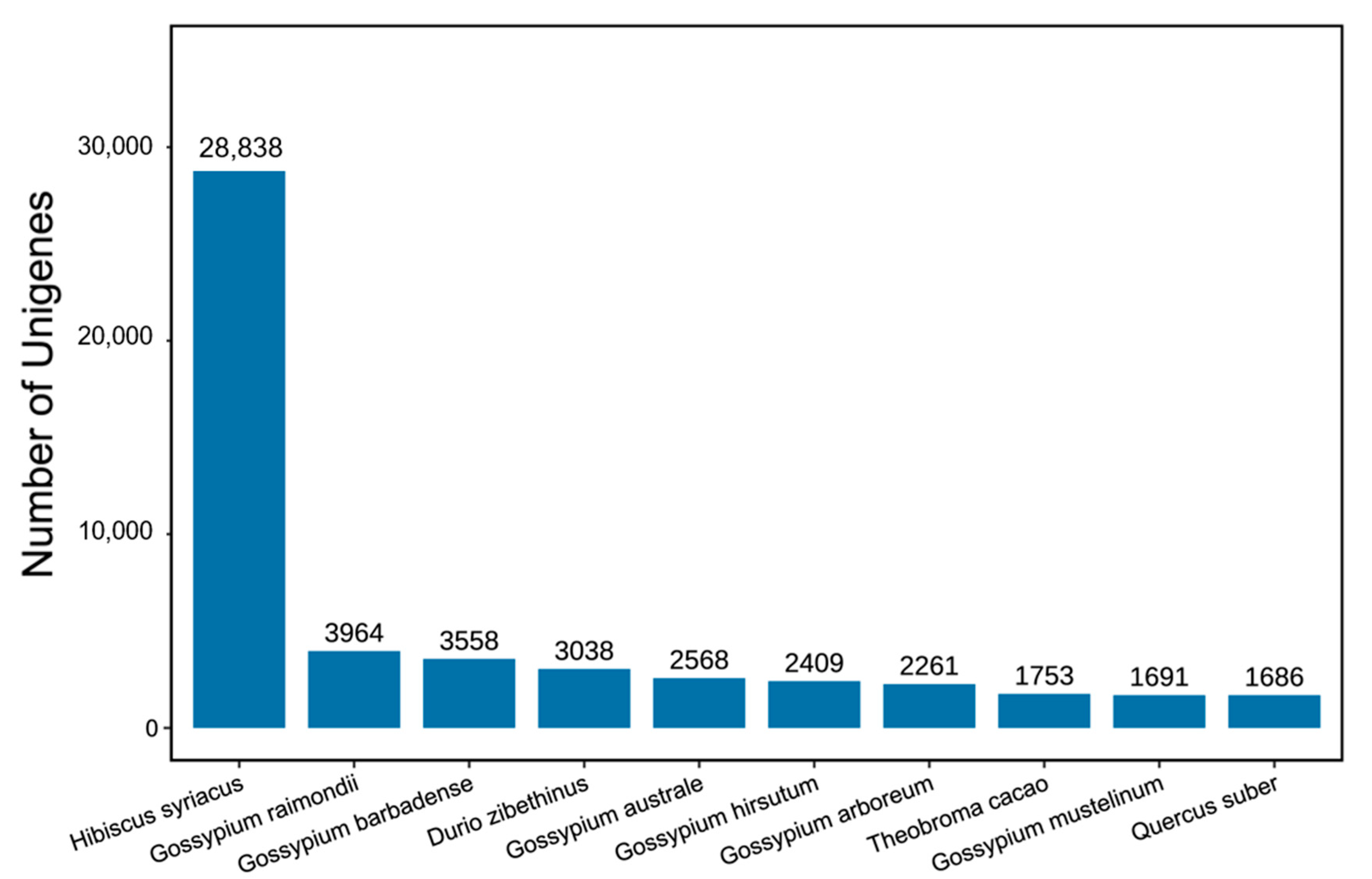
2.4. Roselle Transcriptome and De Novo Assembly
2.5. Differentially Expressed Genes (DEGs)
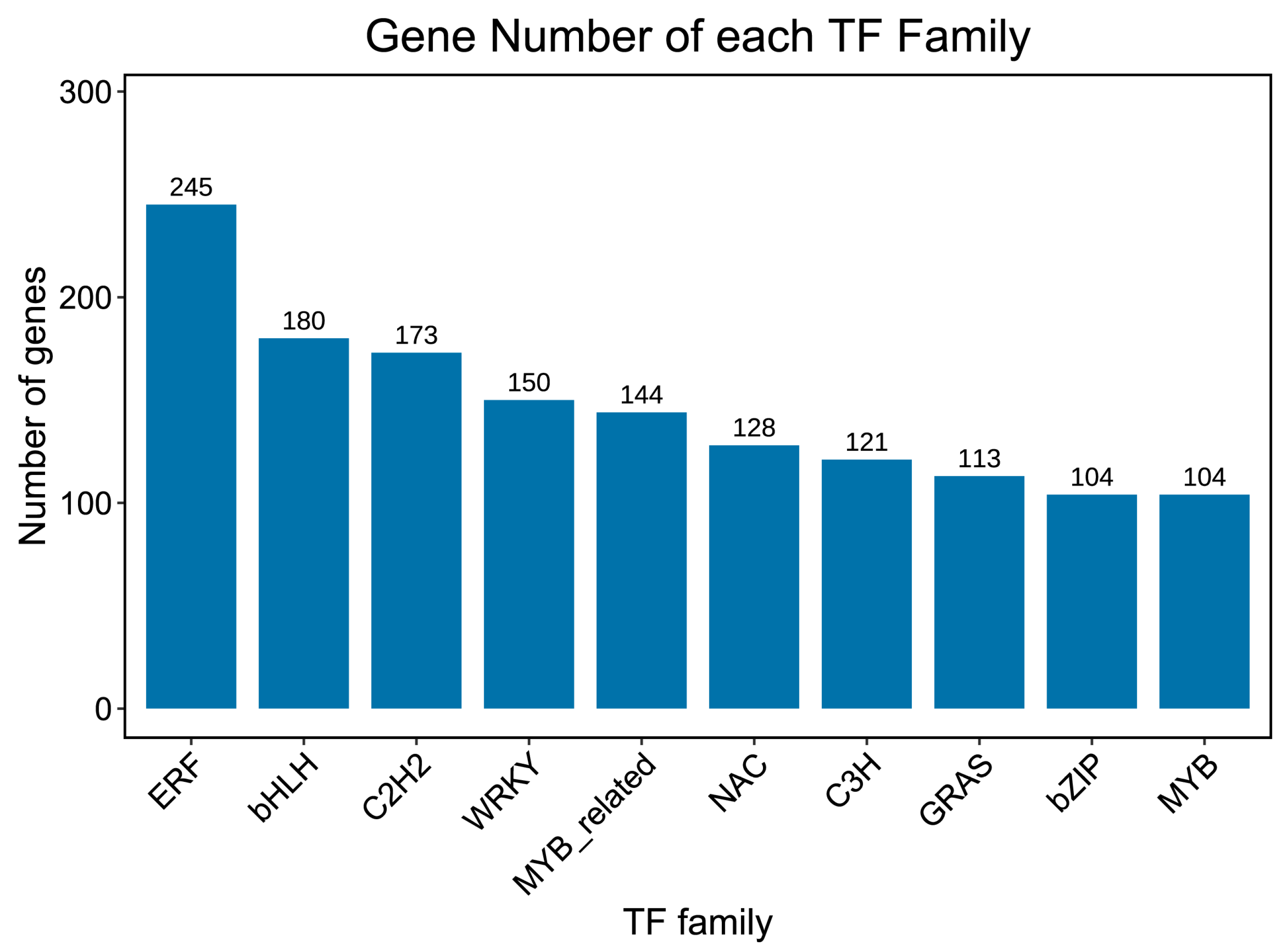
2.6. Transcription Factor
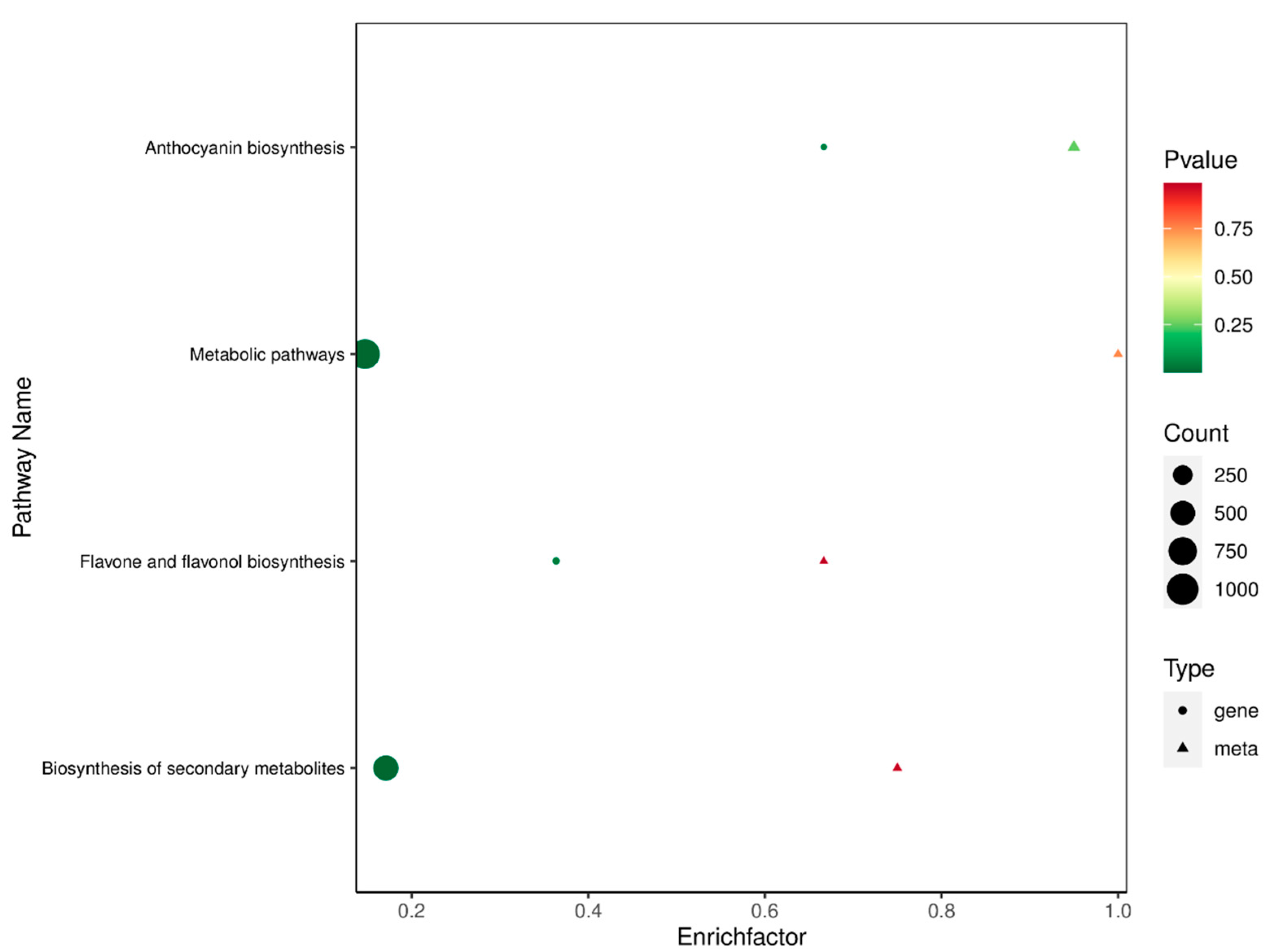
2.7. Conjoint Analysis
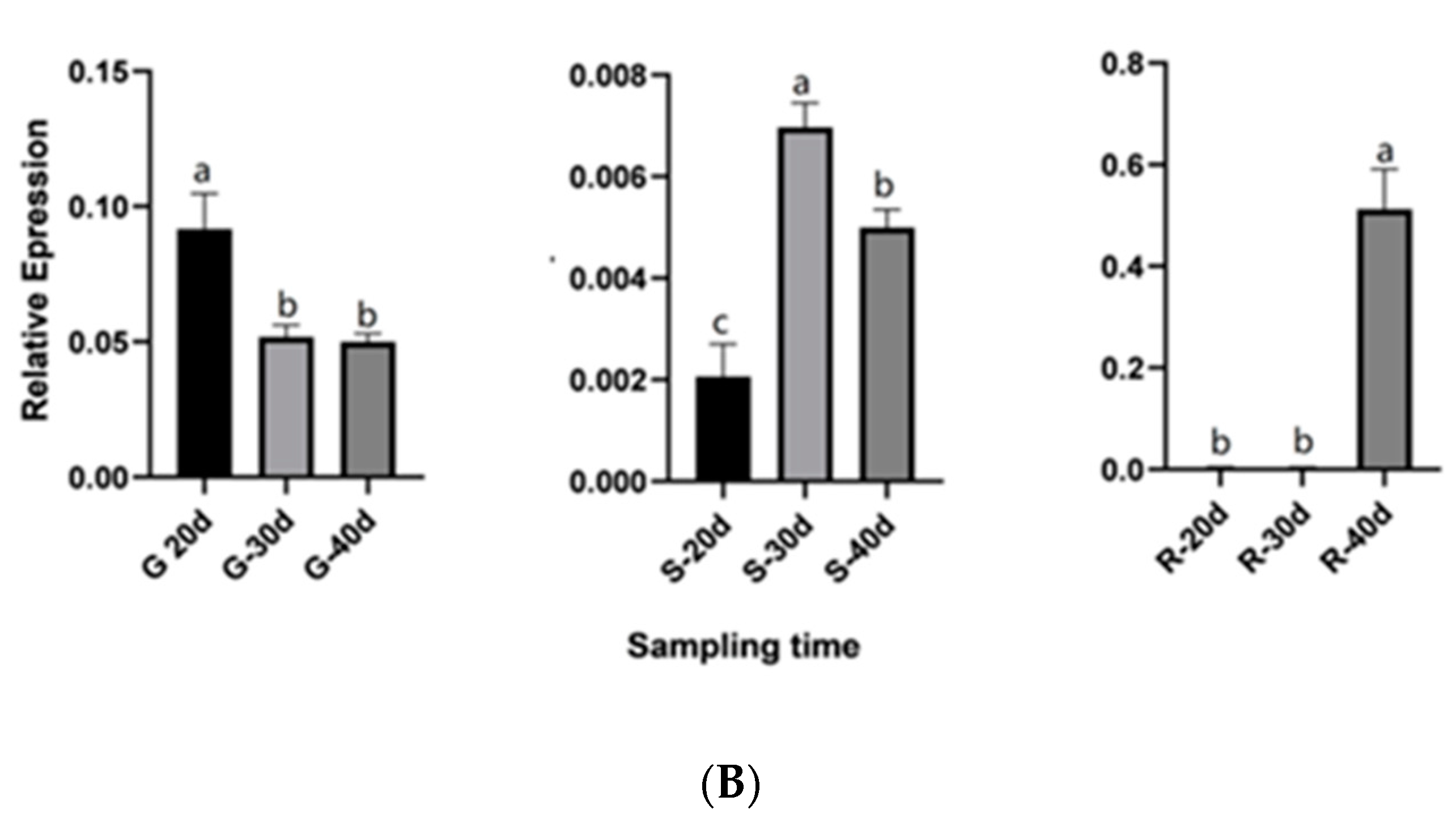
2.8. QRT-PCR Validation for Candidate Genes
2.9. Essential Genes Responsible for Anthocyanin Synthesis in Roselle Calyxes
3. Discussion
3.1. Anthocyanin Types and Content Identified in the Calyxes of Roselle
3.2. There Were Several Essential Genes Controlling Anthocyanin Synthesis in Roselle Calyx
3.3. Integrated Analysis of the Transcriptome and Metabolome
4. Materials and Methods
4.1. Plant Materials
4.2. Analysis of Anthocyanins in Roselle Using UPLC-Q-TOF/MS
4.3. Metabolite Analysis
4.4. Screening of Differential Metabolites
4.5. RNA Extraction and Illumina Sequencing
4.6. RNA-Seq Analysis of Roselle Calyxes from Three Cultivars
4.7. Conjoint Analysis
4.8. Protein Domain Prediction
4.9. Transcription Factor (TF) Identification
4.10. Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibrahim, E.B.; Abdalla, A.W.H.; Ibrahim, E.H.A.; El Naim, A.M. Variability in Some Roselle (Hibiscus sabdariffa L.) Genotypes for Yield and its Attributes. Int. J. Agric. For. 2013, 3, 261–266. [Google Scholar]
- Salami, S.O.; Afolayan, A.J. Evaluation of roselle’s nutritional and elemental compositions of green and red cultivars: Hibiscus Sabrina L. Sci. Rep. 2021, 11, 1030. [Google Scholar] [CrossRef] [PubMed]
- Othman, S.N.; Yong, S.Y.C.; Karjiban, R.A.; Shakri, A. Cloning and comparative protein modeling of two MADS-box genes, HsMADS1 and HsMADS2 isolated from Hibiscus sabdariffa L. var. UML (roselle). Aust. J. Crop Sci. 2016, 10, 207–215. [Google Scholar]
- Hashemi, A.; Shahani, A. Effects of salt stress on the morphological characteristics, total phenol, and total anthocyanin contents of Roselle (Hibiscus sabdariffa L.). Plant Physiol. Rep. 2019, 24, 210–214. [Google Scholar] [CrossRef]
- Riaz, G.; Chopra, R. A review on phytochemistry and therapeutic uses of Hibiscus sabdariffa L. Biomed. Pharmacother. 2018, 102, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Pattanittum, P.; Ngamjarus, C.; Buttramee, F.; Somboonporn, C. Roselle for hypertension in adults. Cochrane Database Syst. Rev. 2021, 11, CD007894. [Google Scholar]
- Ashaye, O. Studies on moisture sorption isotherm and nutritional properties of dried Roselle calyces. Int. Food Res. J. 2013, 20, 509–513. [Google Scholar]
- Chauhan, A.; Kaith, B. Evaluation of dynamic materials procured from waste biomass. Int. Sch. Res. Netw. 2011, 7, 393489. [Google Scholar] [CrossRef][Green Version]
- Yao, Y. Study on the Antioxidant Activity and Application of Hibiscus sabdariffa L.; Xihua University: Chengdu, China, 2020; pp. 1–2. [Google Scholar]
- Ni, Y.; Chen, H.M.; Liu, D.; Zeng, L.H.; Chen, P.H.; Liu, C. Discovery of genes involved in anthocyanin biosynthesis from the rind and pith of three sugarcane cultivars using integrated metabolic profiling and RNA-seq analysis. BMC Plant Biol. 2021, 21, 214. [Google Scholar] [CrossRef]
- Fan, R.H.; Huang, M.L. Progress in regulation of anthocyanins. Chin. J. Cell Biol. 2013, 35, 741–746. [Google Scholar]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Yola, M.L.; Eren, T.; Atar, N. Molecular imprinted nanosensor based on surface plasmon resonance: Application to the sensitive determination of amoxicillin. Sens. Actuators B. Chem. 2014, 195, 28–35. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A Review. Front. Chemistry. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, M.D.; Wen, C.X.; Xie, X.L.; Tian, W.; Wen, S.Q.; Lu, R.K.; Liu, L.D. Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. Flowers. BMC Plant Biol. 2020, 20, 349. [Google Scholar] [CrossRef]
- Zhang, K.K.; Su, H.F.; Lin, Y.G.; Zhang, L.Y. Research progress on the mechanism of color generation and anthocyanin accumulation in anthocyanin-rich tea. J. Food Saf. Qual. 2022, 13, 3585–3592. [Google Scholar]
- Zhang, H.; Wu, Y.L.; Jin, S.W.; Ding, M.Q. The Function Analysis of Cotton GaHD1 Gene in the Regulation of the Development of Anthocyanin Synthesis. Mol. Plant Breed. Available online: http://kns.cnki.net/kcms/detail/46.1068.S.20220328.1618.018.html (accessed on 6 September 2022).
- Sun, C.L.; Deng, L.; Du, M.M.; Zhao, J.H.; Chen, Q.; Huang, T.T.; Jiang, H.L.; Li, C.B.; Li, C.Y. A Transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant. 2020, 13, 42–58. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.J.; Wu, Y.; Zheng, L.M.; Zhang, G.F. Regulatory mechanisms of anthocyanin biosynthesis in apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Lyu, J.; Choi, H.; Ryu, J.; Kwon, S.J.; Jo, Y.D.; Hong, M.J.; Kim, J.B.; Ahn, J.W.; Kang, S.Y. Transcriptome analysis and identification of genes related to the biosynthesis of anthocyanins and kaempferitrin in Kenaf (Hibiscus cannabinus L.). J. Plant Biol. 2020, 63, 51–62. [Google Scholar] [CrossRef]
- Yu, T.T.; Ni, X.Z.; Gao, L.H.; Han, G.J.; Zhu, C.F.; Shen, Y.M. Advances in study of Dihydroflavonol 4-Reductase (DFR) genes of higher plants. Bull. Bot. Res. 2018, 38, 632–640. [Google Scholar]
- Mizukami, H.; Tomita, K.; Ohashi, H. Anthocyanin accumulation and changes in activities of phenylalanine ammonia-lyase and chalcone synthase in roselle (Hibiscus sabdariffa L.) callus cultures. Plant Cell Rep. 1989, 8, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.S. Functional Analysis of Genes Related to Proanthocyanidin Synthesis and Regulation in Colored Cotton; Zhejiang Sci-Tech University: Hangzhou, China, 2021; pp. 1–2. [Google Scholar]
- Sukkhaeng, S.; Promdang, S.; Doungngern, U. Fruit characters and physicochemical properties of roselle (Hibiscus sabdariffa L.) in Thailand—A screening of 13 new genotypes. J. Appl. Res. Med. Aromat. Plants. 2018, 11, 47–53. [Google Scholar]
- Zhao, J.; Liu, R.; Yang, F.; Li, X.; Liu, H.S.; Yan, Q.; Xiao, Y.H. Cloning and expression analyses of R2R3-MYB genes related to anthocyanin biosynthesis in Rose. Sci. Agric. Sin. 2015, 48, 1392–1404. [Google Scholar]
- Hinojosa-Gómez, J.; San Martín-Hernández, C.; Heredia, J.B.; León-Félix, J.; Osuna-Enciso, T.; Muy-Rangel, M.D. Anthocyanin induction by drought stress in the calyx of Roselle Cultivars. Molecules. 2020, 25, 1555. [Google Scholar] [CrossRef] [PubMed]
- Trivellini, A.; Gordillo, B.; Rodríguez-Pulido, F.J.; Borghesi, E.; Ferrante, A.; Vernieri, P.; Quijada-Morín, N.; González-Miret, M.L.; Heredia, F.J. Effect of salt stress in the regulation of anthocyanins and color of Hibiscus flowers by digital image analysis. J. Agric. Food Chem. 2014, 62, 6966–6974. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lei, T.; Jin, X.H.; Fu, D.X. Flavonoid related compounds and their biosynthesis pathways in Linum penne. Guinea. 2017, 37, 1368–1377. [Google Scholar]
- Muhammed Amin, M.I.; Kanimarani, S.M. The influence of plant growth regulators on phytochemical components in the leaves and calyxes of Roselle (Hibiscus sabdariffa L.). Sci. Lett. 2020, 32, 193–199. [Google Scholar]
- Zhang, Q.; Wang, L.L.; Liu, Z.G.; Zhao, Z.H.; Zhao, J.; Wang, Z.T.; Zhou, G.F.; Liu, P.; Liu, M.J. Transcriptome and metabolome profiling unveils the Ziziphus jujuba Mill mechanisms. Peel coloration. Food Chem. 2020, 312, 125903. [Google Scholar] [CrossRef]
- Liu, M.M.; Wang, M.Z.; Lu, S.F. Cloning and Bioinformatics Analysis of homogentisate solanesyltransferase gene of salvia miltiorrhiza. Genom. Appl. Biol. 2019, 38, 3614–3621. [Google Scholar]
- Yu, J.W.; Han, R.; Qiu, T.; Li, Z.Y.; Pan, S.Y.; Dong, T.T. Research progress on microRNA in plant growth and development. Mol. Plant Breed. 2020, 18, 1495–1504. [Google Scholar]
- Liu, X.; Li, F.; Song, Q.F.; Niu, S.Z.; Li, L.T. Analysis of gene expression related to anthocyanin regulation of ‘P113’ purple strain of camellia tachangensis F. C. Zhang in different seasons. J. Tea Sci. 2021, 41, 789–801. [Google Scholar]
- Song, Y.Y. Activity Regulation of Serine Acetyltransferase, a Key Enzyme Involved in B-ODAP Biosynthesis of Lathyrus Sativus; Northwest A&F University: Xianyang, China, 2021; pp. 1–2. [Google Scholar]
- Roshani, R.M.; Nulit, R.; Yong, C.S.Y. Molecular dissection of novel anthocyanin transcripts HsCHS and HsF3H from the calyx tissue of Hibiscus sabdariffa L. Int. J. Environ. Sci. Technol. 2018, 16, 1909–1920. [Google Scholar] [CrossRef]
- Ogata, J.; Kanno, Y.; Itoh, Y.; Tsugawa, H.; Suzuki, M. Anthocyanin biosynthesis in roses. Nature 2005, 435, 757–758. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.F.; Zhu, Y.L.; Fan, J.Y.; Wang, D.J.; Gong, X.H.; Ouyang, Z. Accumulation of flavonoid glycosides and UFGT gene expression in mulberry leaves (Morus alba L.) before and after frost. Chem. Biodivers. 2017, 14, 201600496. [Google Scholar] [CrossRef]
- Wang, W.S. Identification and Function Analysis of Glycosyltransferase Genes Related to Flavonoids and Anthocyanins Metabolism in Rice; Shandong University: Jinan, China, 2020; pp. 65–68. [Google Scholar]
- Wang, J.; Hou, B.K. Glycosyltransferases: Key players involved in the modification of plant secondary metabolites. Front. Biol. 2009, 4, 39–46. [Google Scholar] [CrossRef]
- Poppenberger, B.; Fujioka, S.; Soeno, K.; George, G.L.; Vaistij, F.E.; Hiranuma, S.; Seto, H.; Takatsuto, S.; Adam, G.; Yoshida, S.; et al. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 2005, 102, 15253–15258. [Google Scholar] [CrossRef]
- Zhang, Q. Identification and Functional Analysis of R2R3-MYB Regulators in the Anthocyanin Biosynthetic Pathway from Prunus Mume; Beijing Forestry University: Beijing, China, 2017; pp. 31–48. [Google Scholar]
- Wang, B.; Zhang, T.X.; Zhao, Q.; Meng, X.C.; Li, Y.F.; Zhao, D.W. Aplication progress of plant metabonomics in medicinal plants. Chin. Arch. Tradit. Chin. Med. 2021, 39, 28–31. [Google Scholar]
- Paola-Naranjo, R.D.D.; Sánchez-Sánche, J.; González-Paramás, A.M.; Rivas-Gonzalo, J.C. Liquid chromatographic-mass spectrometric analysis of anthocyanin composition of dark blue bee pollen from Echium plantagineum. J. Chromatogr. A 2004, 1054, 205–210. [Google Scholar] [CrossRef]
- Cruz, A.D.; Hilber, G.; Rivière, C.; Mengin, V.; Ollat, N.; Bordenave, L.; Decroocq, S.; Delaunay, J.C.; Delrot, S.; Mérillon, J.M.; et al. Anthocyanin identification and composition of wild Vitis spp. accessions by using LC-MS and LC-NMR. Anal. Chim. Acta. 2012, 732, 145–152. [Google Scholar] [CrossRef]
- Ferrars, R.M.; Czank, C.; Saha, S.; Needs, P.W.; Zhang, Q.Z.; Raheem, K.S.; Botting, N.P.; Kroon, P.A.; Kay, C.D. Methods for isolating, identifying, and quantifying anthocyanin metabolites in clinical samples. Anal. Chem. 2014, 86, 10052–10058. [Google Scholar] [CrossRef]
- Zhu, Q.L.; Yu, S.Z.; Zeng, D.C.; Liu, H.M.; Wang, H.C.; Yang, Z.F.; Xie, X.R.; Shen, R.X.; Tan, J.T.; Li, H.Y.; et al. Development of “Purple endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant. 2017, 10, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Zhang, J.Q.; Zheng, S.; Wang, Z.Q.; Xu, C.M.; Zhang, Q.X.; Wu, J.S.; Lou, H.Q. Metabolite profiling and transcriptome analyses reveal novel regulatory mechanisms of melatonin biosynthesis in hickory. Hortic. Res. 2021, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fast: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 1884–1890. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, G.M.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, R.S.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families’ database: Towards a more sustainable future. Bioinformatics 2016, 44, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Jin, J.P.; Zhang, H.; Kong, L.; Gao, G.; Lou, J.C. Plant TFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Chen, H.; Liu, W.; Liu, H.; Gong, J.; Wang, H.L.; Guo, A.Y. Animal TFDB: A comprehensive animal transcription factor database. Nucleic Acids Res. 2011, 40, 144–149. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.H.; Yang, Y.Y.; Li, W.; Xu, Y.; Xie, Y.T.; Liu, Y. KLHL22 activates amino-acid-dependent mTORC1 signaling to promote tumorigenesis and aging. Nature 2018, 557, 585–589. [Google Scholar] [CrossRef] [PubMed]







| Compounds | Mean(Group G) (μg/g) | Mean(Group S) (μg/g) | Mean(Group R) (μg/g) | RT (min) |
|---|---|---|---|---|
| Cyanidin-3-O-glucoside | 0.16 | 12.23 | 40.58 | 7.48 |
| Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | ND | ND | 0.01 | 10.26 |
| Cyanidin-3-O-sambubioside-5-O-glucoside | ND | 1.27 | 5.72 | 5.66 |
| Cyanidin-3-O-xyloside | ND | 0.01 | 0.06 | 9.69 |
| Cyanidin-3-O-rutinoside-5-O-glucoside | ND | 0.01 | 0.01 | 5.51 |
| Cyanidin-3-O-galactoside | ND | 0.27 | 1.30 | 6.94 |
| Cyanidin-3-O-arabinoside | ND | 0.04 | 0.31 | 7.91 |
| Cyanidin-3-O-sophoroside | ND | 2.15 | 11.23 | 6.92 |
| Cyanidin-3-O-rutinoside | ND | 0.20 | 0.46 | 7.98 |
| Cyanidin-3,5,3’-O-triglucoside | ND | 0.03 | 0.11 | 5.18 |
| Cyanidin-3-O-sambubioside | 0.11 | 330.06 | 531.37 | 7.59 |
| Cyanidin-3,5-O-diglucoside | ND | 0.05 | 0.14 | 5.45 |
| Delphinidin-3-O-rutinoside | 0.03 | 2.19 | 3.53 | 7.08 |
| Delphinidin-3-O-(6-O-malonyl-beta-D-glucoside) | 0.08 | ND | ND | 9.56 |
| Delphinidin-3-O-rutinoside-5-O-glucoside | 0.16 | 0.06 | ND | 7.10 |
| Delphinidin-3-O-sambubioside-5-O-glucoside | ND | 1.36 | 2.32 | 4.30 |
| Delphinidin-3-O-rhamnoside | ND | 0.03 | 0.08 | 8.05 |
| Delphinidin-3-O-glucoside | ND | 35.37 | 70.47 | 6.48 |
| Delphinidin-3-O-arabinoside | ND | 0.07 | 0.17 | 7.07 |
| Delphinidin-3-O-sambubioside | 0.26 | 851.97 | 955.11 | 6.37 |
| Delphinidin-3-O-sophoroside | ND | 44.60 | 85.08 | 6.06 |
| Malvidin-3-O-glucoside | 0.01 | ND | 0.01 | 9.37 |
| Pelargonidin-3-O-galactoside | ND | ND | 0.15 | 7.77 |
| Pelargonidin-3-O-sambubioside | ND | 3.65 | 16.18 | 8.76 |
| Pelargonidin-3-O-glucoside | ND | 0.24 | 4.17 | 8.42 |
| Pelargonidin-3-O-sophoroside | ND | 0.01 | 0.06 | 7.71 |
| Peonidin-3-O-sambubioside-5-O-glucoside | ND | ND | 0.06 | 6.69 |
| Peonidin-3,5-O-diglucoside | ND | ND | 0.01 | 6.76 |
| Peonidin-3-O-sambubioside | ND | 0.36 | 1.38 | 9.00 |
| Peonidin-3-O-glucoside | ND | 0.08 | 0.11 | 9.02 |
| Peonidin | 0.02 | 0.07 | 0.03 | 9.76 |
| Petunidin-3-O-galactoside | ND | 2.84 | 7.90 | 8.10 |
| Petunidin-3-O-glucoside | ND | 0.58 | 1.62 | 8.11 |
| Petunidin-3-O-arabinoside | ND | 0.03 | 0.04 | 8.53 |
| Petunidin-3,5-O-diglucoside | ND | ND | 0.03 | 6.17 |
| Procyanidin B3 | 0.07 | 0.26 | 0.23 | 3.45 |
| Procyanidin B2 | 6.62 | 8.28 | 3.57 | 5.57 |
| Procyanidin B1 | 0.64 | 0.82 | 1.17 | 3.86 |
| Rutin | 615.25 | 308.06 | 61.94 | 11.31 |
| Quercetin-3-O-glucoside | 312.22 | 120.67 | 49.38 | 11.30 |
| Kaempferol-3-O-rutinoside | 17.90 | 43.03 | 12.72 | 12.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, Y.; Li, M.; Lin, L.; Qi, J.; Xu, J.; Zhang, L.; Fang, P.; Tao, A. Novel Insights into Anthocyanin Synthesis in the Calyx of Roselle Using Integrated Transcriptomic and Metabolomic Analyses. Int. J. Mol. Sci. 2022, 23, 13908. https://doi.org/10.3390/ijms232213908
Li J, Li Y, Li M, Lin L, Qi J, Xu J, Zhang L, Fang P, Tao A. Novel Insights into Anthocyanin Synthesis in the Calyx of Roselle Using Integrated Transcriptomic and Metabolomic Analyses. International Journal of Molecular Sciences. 2022; 23(22):13908. https://doi.org/10.3390/ijms232213908
Chicago/Turabian StyleLi, Jing, Yunqing Li, Mei Li, Lihui Lin, Jianmin Qi, Jiantang Xu, Liwu Zhang, Pingping Fang, and Aifen Tao. 2022. "Novel Insights into Anthocyanin Synthesis in the Calyx of Roselle Using Integrated Transcriptomic and Metabolomic Analyses" International Journal of Molecular Sciences 23, no. 22: 13908. https://doi.org/10.3390/ijms232213908
APA StyleLi, J., Li, Y., Li, M., Lin, L., Qi, J., Xu, J., Zhang, L., Fang, P., & Tao, A. (2022). Novel Insights into Anthocyanin Synthesis in the Calyx of Roselle Using Integrated Transcriptomic and Metabolomic Analyses. International Journal of Molecular Sciences, 23(22), 13908. https://doi.org/10.3390/ijms232213908





