Genome-Wide Identification of the Highly Conserved INDETERMINATE DOMAIN (IDD) Zinc Finger Gene Family in Moso Bamboo (Phyllostachys edulis)
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of PheIDD Family Genes in Moso Bamboo
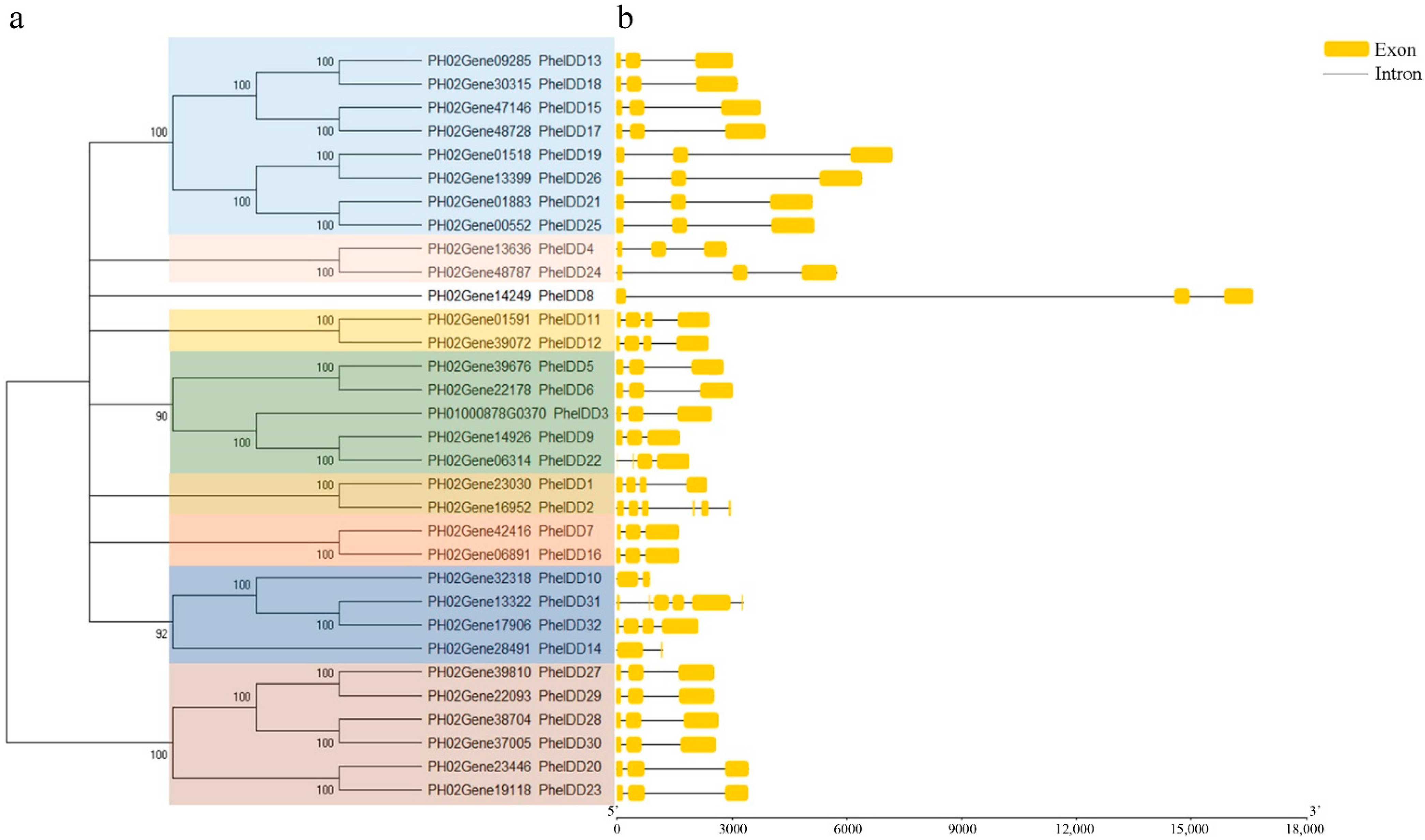
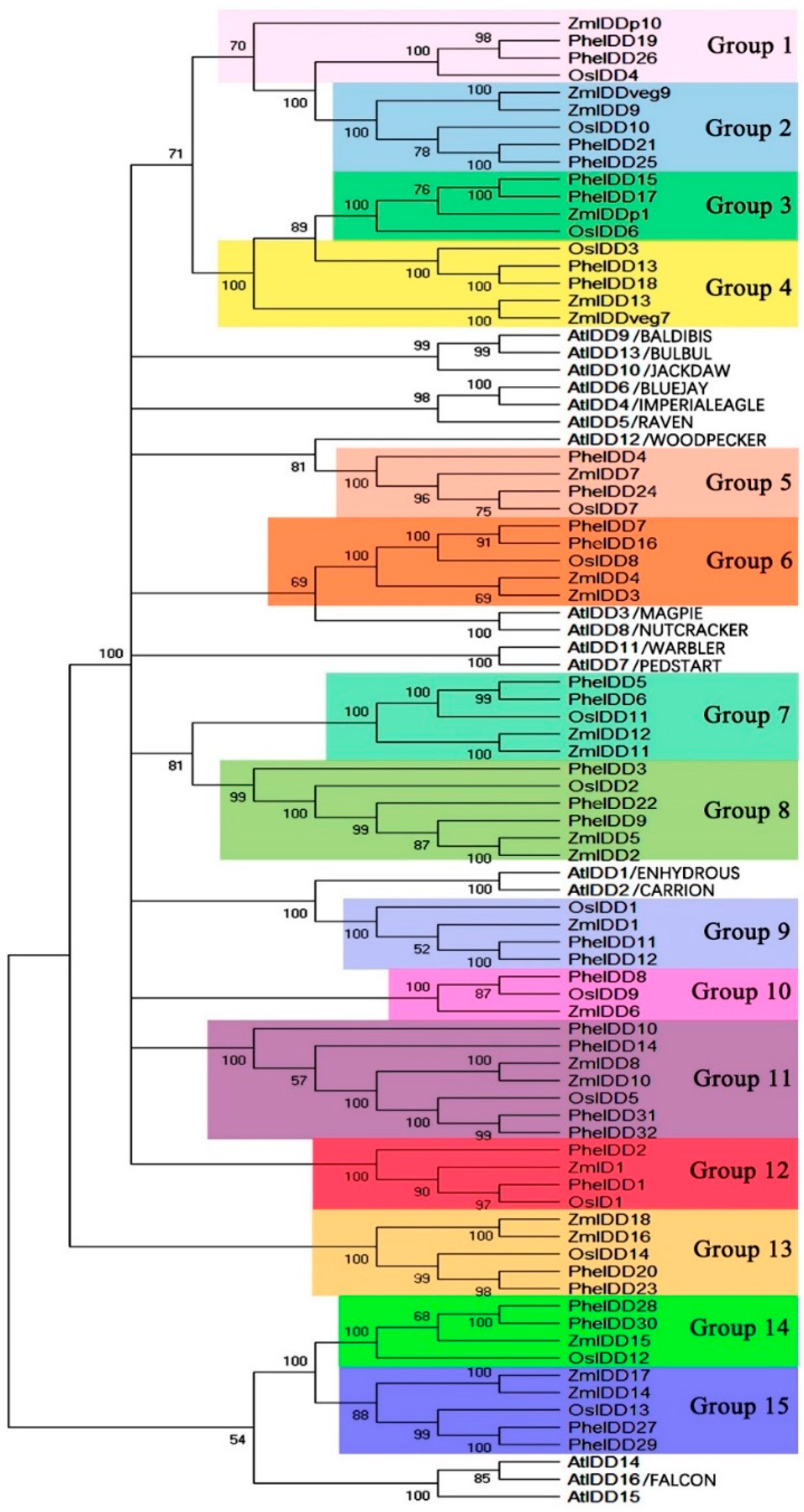
2.2. Phylogenetic Analyses, Synteny, and Duplication of the PheIDDFamily Genes
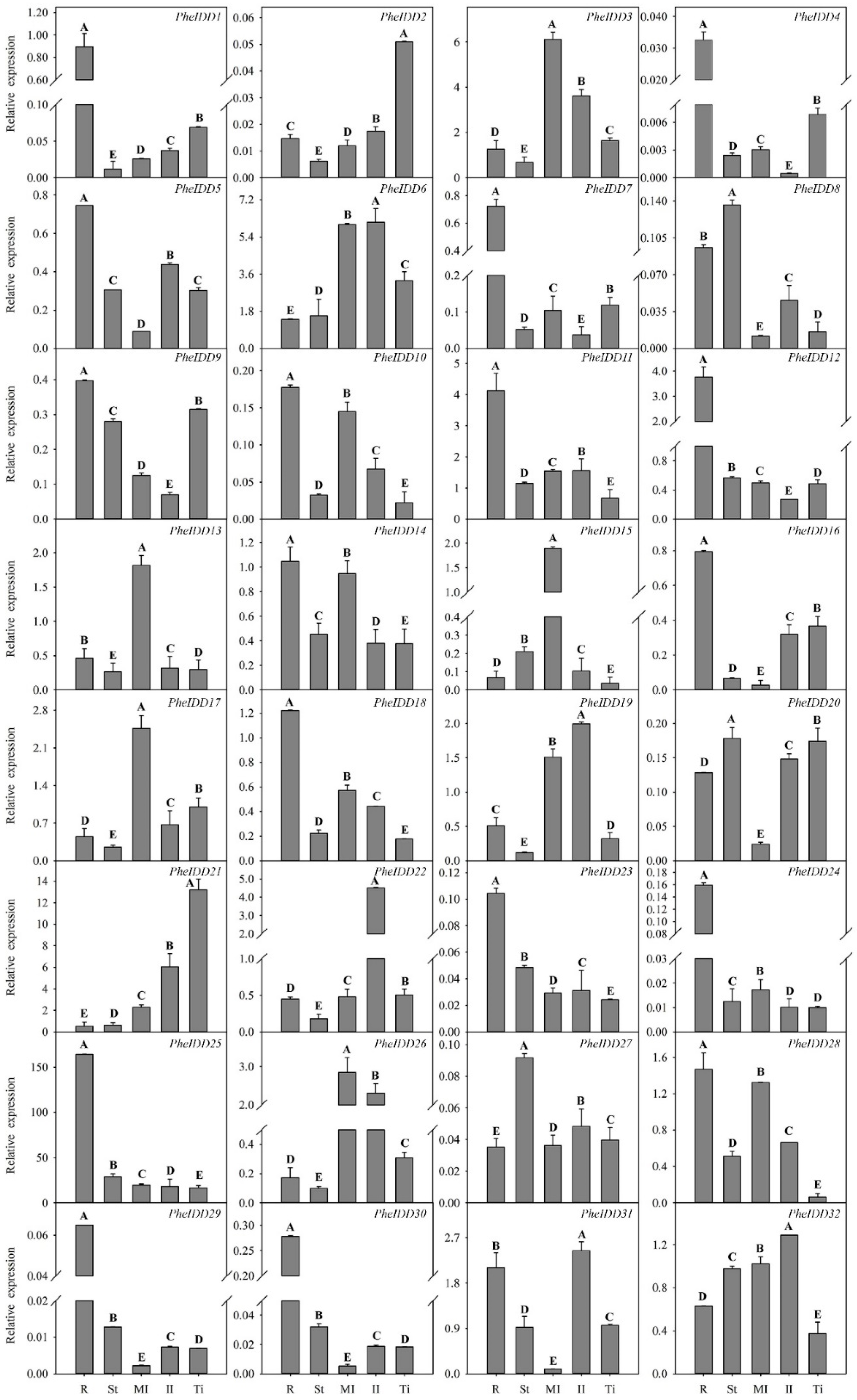
2.3. Expression Profiles of PheIDDs in Different Tissues
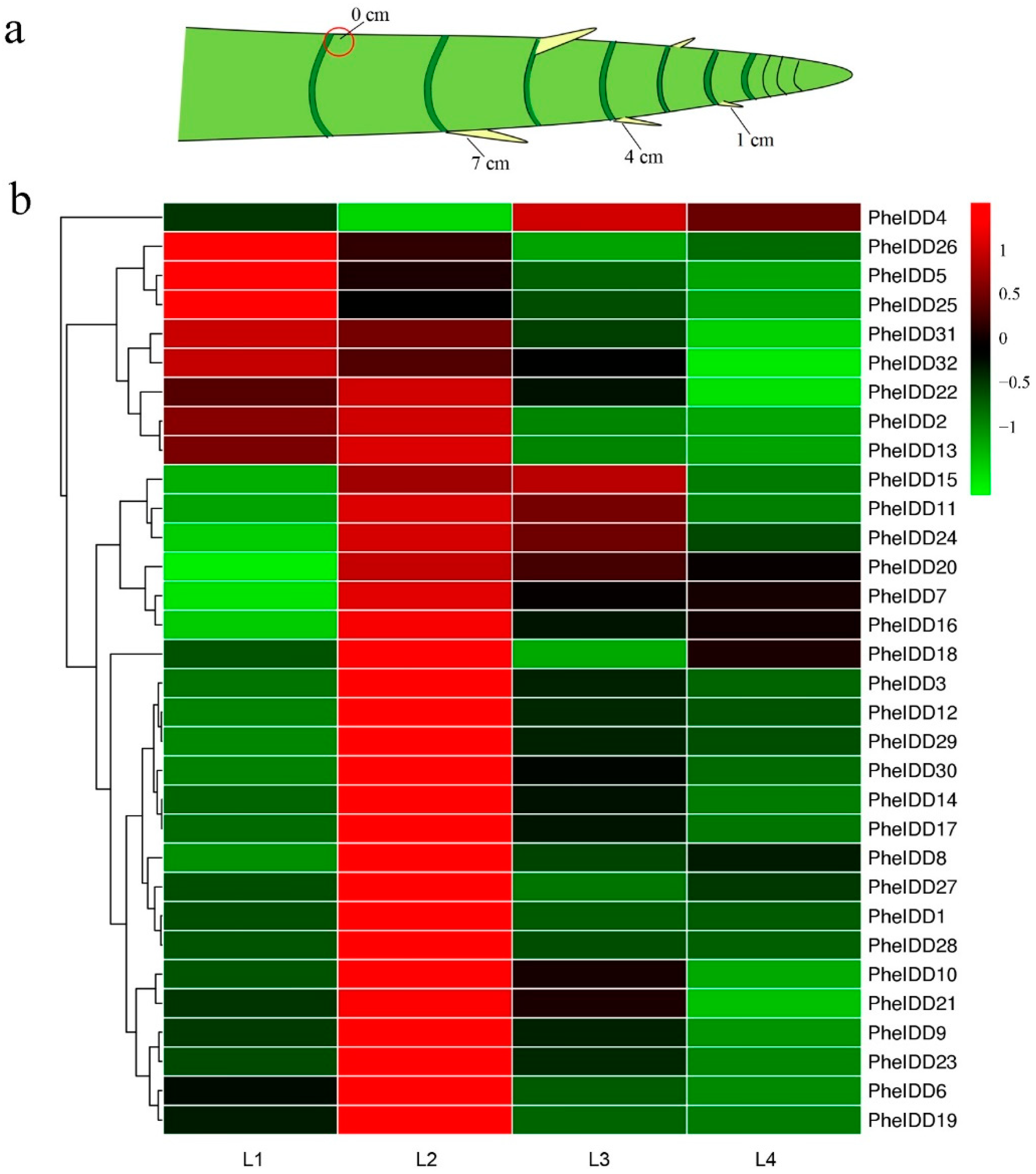
2.4. Dynamic Expression Profiles of PheIDDs in Underground Lateral Buds/Shoots at Different Developmental Stages
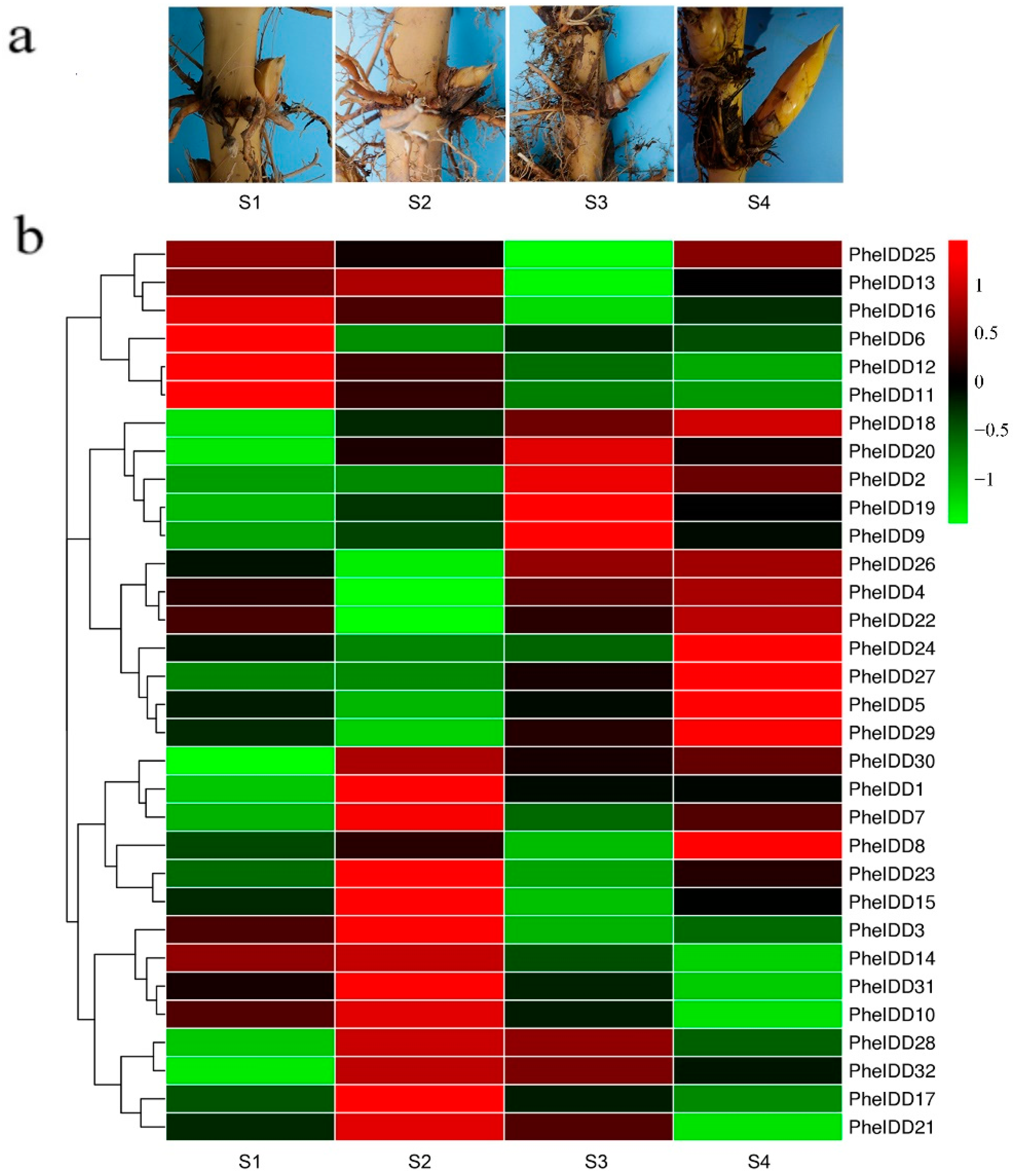
2.5. Dynamic Expression Profiles of PheIDDs in Aboveground Branches of Different Sizes
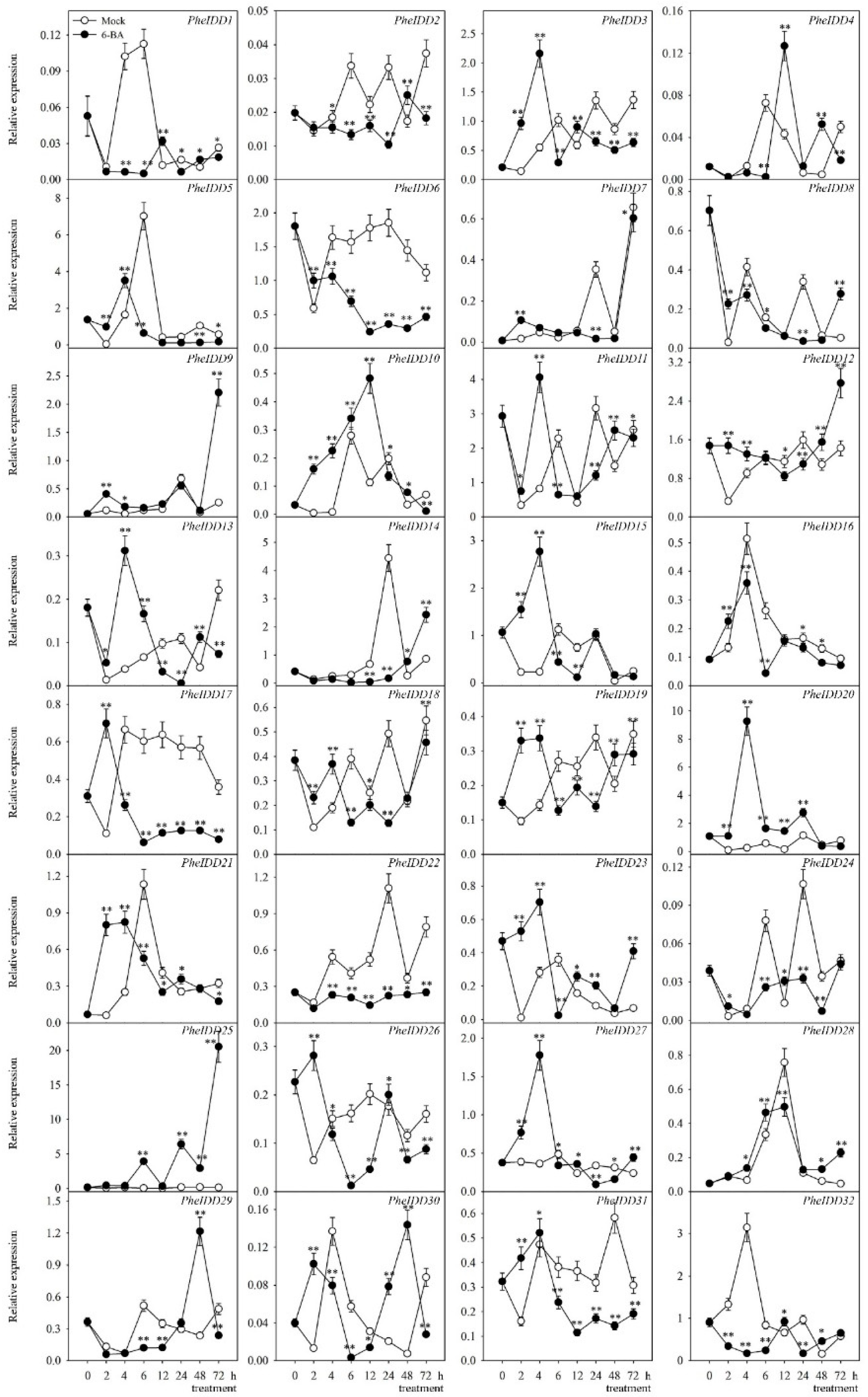
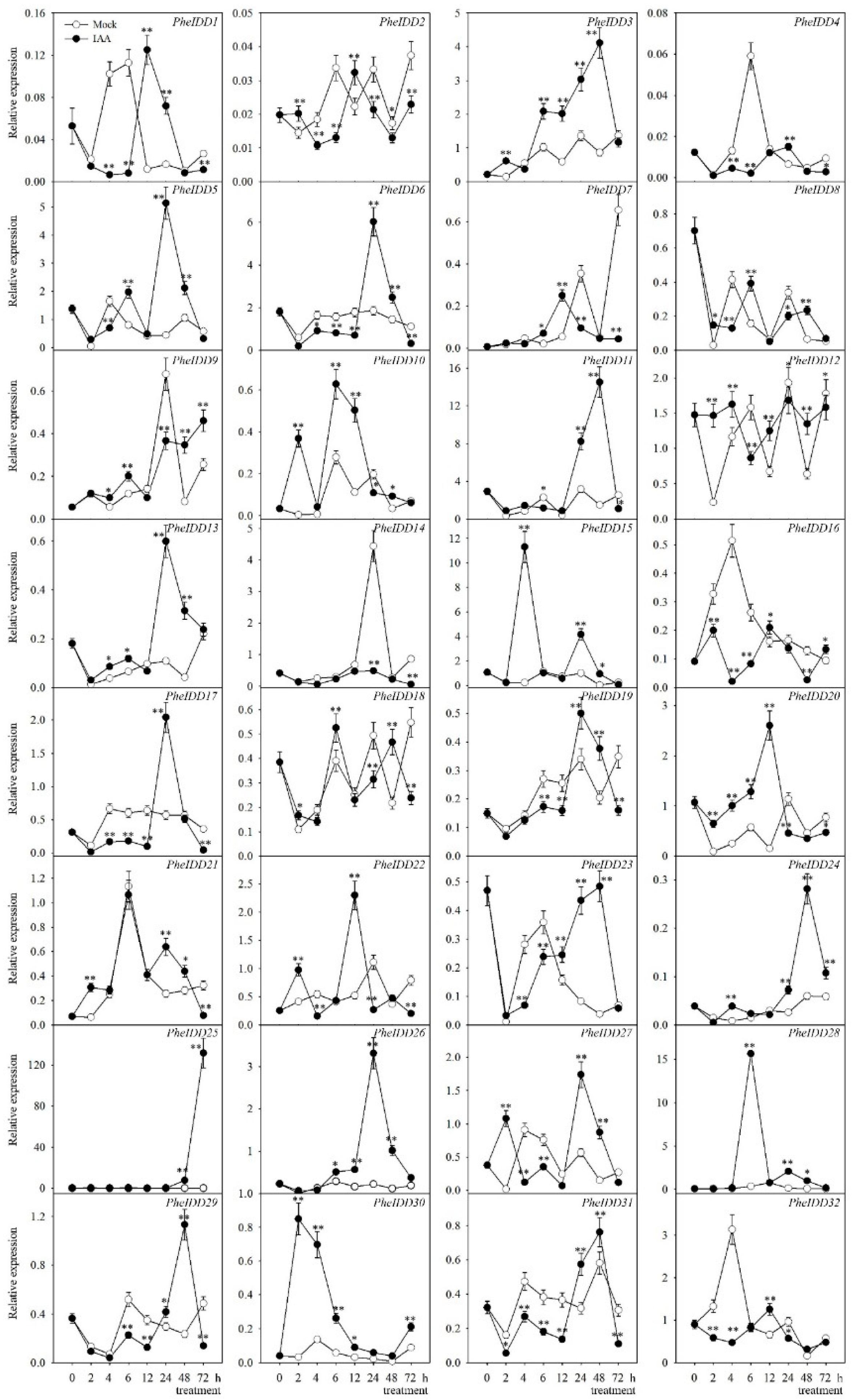
2.6. Transcript Profiling of PheIDDs under IAA and 6-BA Treatments
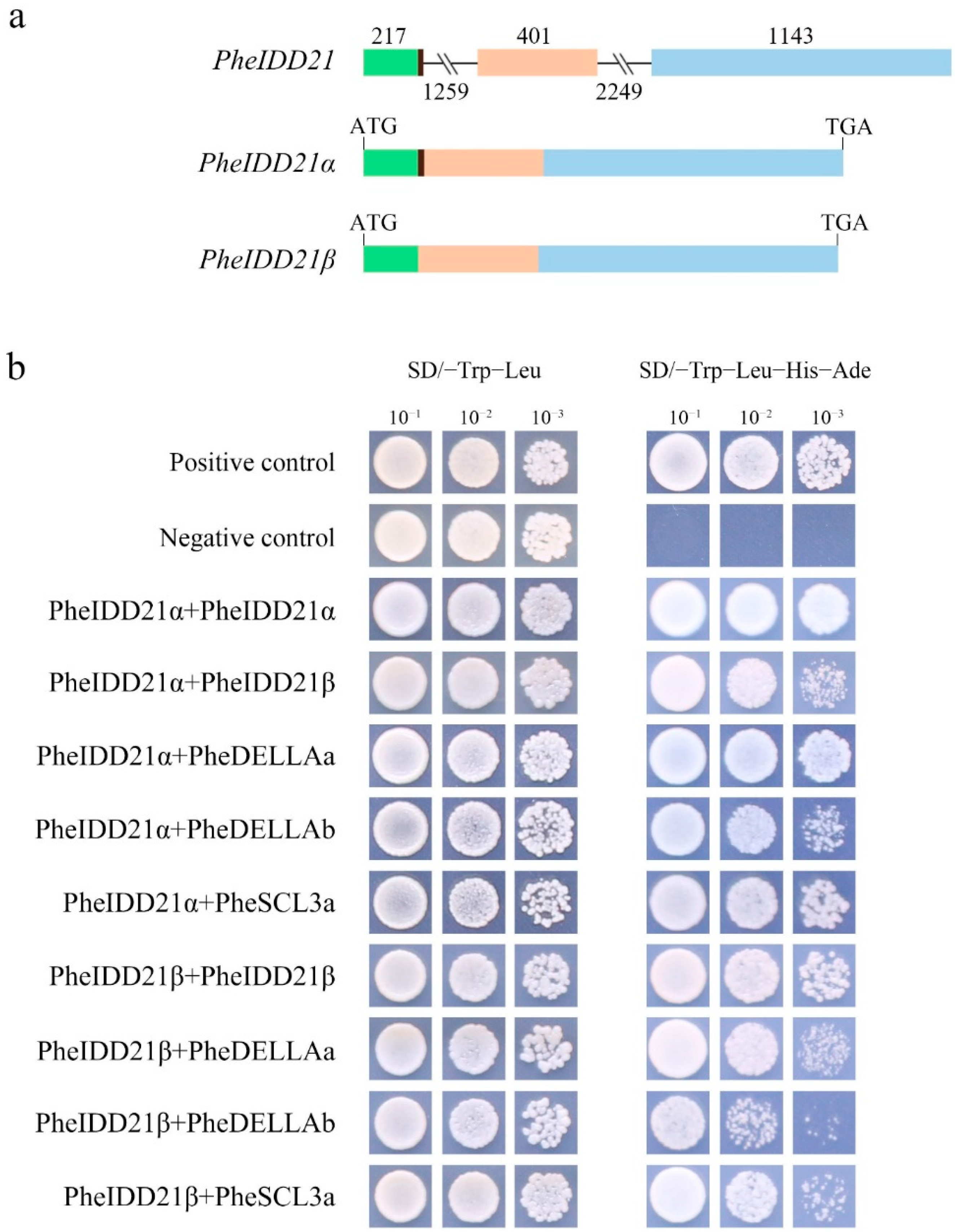
2.7. PheIDD21Generated Two Transcripts by Alternative Splicing
2.8. Interaction of PheIDD21 with RGAS Proteins and Its Binding to Target Genes
3. Discussion
3.1. The IDDGene Family in Moso Bamboo
3.2. PheIDDs Respond to Exogenous Hormone Treatments
3.3. Expression Profile Analysis of PheIDDGenes in the Development of Aboveground Branches, and Underground Buds/Shoots
3.4. IDD Interacting Proteins and Target Genes
4. Methods and Materials
4.1. Plant Materials and Treatments
4.2. Identification of PheIDDFamilyGenes in the Moso Bamboo Genome
4.3. Bioinformatic Analyses
4.3.1. Analysis of Chemical Characteristics
4.3.2. Gene structures and Scaffold Locations
4.3.3. Screening of Gene Pairs and Calculation of Ka/Ks Ratios
4.3.4. Synteny Analysis and Gene Duplication
4.3.5. Prediction of Phosphorylation Sites in the PheIDDs
4.3.6. Prediction of Protein–Protein Interactions
4.3.7. Multiple Sequence Alignment and Phylogenetic Analyses
4.4. Quantitative Real-Time PCR (qRT–PCR)
4.5. Yeast Two-Hybrid Assay (Y2H)
4.6. Yeast One-Hybrid Assay (Y1H)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colasanti, J.; Yuan, Z.; Sundaresan, V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Curr. Opin. Plant Biol. 1998, 1, 276. [Google Scholar] [CrossRef]
- Prochetto, S.; Reinheimer, R. Step by step evolution of Indeterminate Domain (IDD) transcriptional regulators: From algae to angiosperms. Ann. Bot. 2020, 126, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Le, D.T.; Hwang, S.; Seo, P.J.; Kim, H.U. Role of the INDETERMINATE DOMAIN genes in plants. Int. J. Mol. Sci. 2019, 20, 2286. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.P.; Huang, P.; Lee, D.Y.; Brutnell, T.P. Making roots, shoots, and seeds: IDD gene family diversification in plants. Trends Plant Sci. 2018, 23, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef]
- Hirano, Y.; Nakagawa, M.; Suyama, T.; Murase, K.; Shirakawa, M.; Takayama, S.; Sun, T.; Hakoshima, T. Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat. Plants 2017, 3, 17010. [Google Scholar] [CrossRef]
- Feurtado, J.A.; Huang, D.; Wicki-Stordeur, L.; Hemstock, L.E.; Potentier, M.S.; Tsang, E.W.T.; Cutler, A.J. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 2011, 23, 1772–1794. [Google Scholar] [CrossRef]
- Cui, H.; Levesque, M.P.; Vernoux, T.; Jung, J.W.; Paquette, A.J.; Gallagher, K.L.; Wang, J.Y.; Blilou, I.; Scheres, B.; Benfey, P.N. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 2007, 316, 421–425. [Google Scholar] [CrossRef]
- Welch, D.; Hassan, H.; Blilou, I.; Immink, R.; Heidstra, R.; Scheres, B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007, 21, 2196–2204. [Google Scholar] [CrossRef]
- Hassan, H.; Scheres, B.; Blilou, I. JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 2010, 137, 1523–1529. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Liu, T.; Newell, N.R.; Magnani, E.; Huang, T.; Kerstetter, R.; Michaels, S.; Barton, M.K. Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 2013, 25, 3228–3249. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Zhao, J.; Jing, Y.; Fan, M.; Liu, J.; Wang, Z.; Xin, W.; Hu, Y. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 2013, 9, e1003759. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Teramura, H.; Murakoshi, S.; Nasuno, K.; Nishida, N.; Ito, T.; Yoshida, M.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 2014, 26, 2920–2938. [Google Scholar] [CrossRef]
- Zentella, R.; Zhang, Z.-L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Ryu, J.; Kang, S.K.; Park, C.M. Modulation of sugar metabolism by an INDETER MINATEDOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011, 65, 418–429. [Google Scholar] [CrossRef]
- Colasanti, J.; Tremblay, R.; Wong, A.Y.; Coneva, V.; Kozaki, A.; Mable, B.K. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genom. 2006, 7, 158. [Google Scholar] [CrossRef]
- Wong, A.Y.M.; Colasanti, J. Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J Exp Bot. 2007, 58, 403–414. [Google Scholar] [CrossRef]
- Matsubara, K.; Yamanouchi, U.; Wang, Z.X.; Minobe, Y.; Izawa, T.; Yano, M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 2008, 148, 1425–1435. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, S.L.; Je, B.I.; Piao, H.L.; Park, S.H.; Kim, C.M.; Ryu, C.H.; Park, S.H.; Xuan, Y.H.; Joseph, C.; et al. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008, 56, 1018–1029. [Google Scholar] [CrossRef]
- Wu, C.; You, C.; Li, C.; Long, T.; Chen, G.; Byrne, M.E.; Zhang, Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12915–12920. [Google Scholar] [CrossRef]
- Hu, S.; Dong, G.; Xu, J.; Su, Y.; Shi, Z.; Ye, W.; Li, Y.; Li, G.; Zhang, B.; Hu, J.; et al. A point mutation in the zinc finger motif of RID1/EHD2/OsID1 protein leads to outstanding yield-related traits in japonica rice variety Wuyunjing 7. Rice 2013, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Cheng, S.; Zhao, B.; Xuan, Y.; Shao, M. The indeterminate domain protein ROC1 regulates chilling tolerance via activation of DREB1B/CBF1 in rice. Int. J. Mol. Sci. 2016, 17, 233. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Neelakandan, A.K.; Gontarek, B.C.; Vollbrecht, E.; Becraft, P.W. The naked endosperm genes encode duplicate INDETERMINATE domain transcription factors required for maize endosperm cell patterning and differentiation. Plant Physiol. 2015, 167, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Jöst, M.; Hensel, G.; Kappel, C.; Druka, A.; Sicard, A.; Hohmann, U.; Beier, S.; Himmelbach, A.; Waugh, R.; Kumlehn, J.; et al. The INDETERMINATE DOMAIN protein BROAD LEAF1 limits barley leaf width by restricting lateral proliferation. Curr. Biol. 2016, 26, 903–909. [Google Scholar] [CrossRef]
- Kozaki, A.; Hake, S.; Colasanti, J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004, 32, 1710–1720. [Google Scholar] [CrossRef]
- Xuan, Y.; Priatama, R.A.; Huang, J.; Je, B.I.; Liu, J.; Park, S.J.; Piao, H.L.; Son, D.Y.; Lee, J.J.; Park, S.H.; et al. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol. 2013, 197, 791–804. [Google Scholar] [CrossRef]
- Sun, Q.; Li, D.; Chu, J.; Yuan, D.; Li, C.; Zhong, L.; Han, X.; Xuan, Y. Indeterminate domain proteins regulate rice defense to sheath blight disease. Rice 2020, 13, 1–12. [Google Scholar] [CrossRef]
- Morita, M.T.; Sakaguchi, K.; Kiyose, S.; Taira, K.; Kato, T.; Nakamura, M.; Tasaka, M. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 2006, 47, 619–628. [Google Scholar] [CrossRef]
- Tanimoto, M.; Tremblay, R.; Colasanti, J. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol. Biol. 2008, 67, 57–69. [Google Scholar] [CrossRef]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Zhang, Y.; Cheng, Z.; Hou, D.; Li, X.; Gao, J. Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol. J. 2017, 15, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, H.; Cai, D.; Gao, Y.; Zhang, H.; Wang, Y.; Lin, C.; Ma, L.; Gu, L. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J. 2017, 91, 684–699. [Google Scholar] [CrossRef]
- Wei, Q.; Jiao, C.; Ding, Y.; Gao, S.; Guo, L.; Chen, M.; Hu, P.; Xia, S.; Ren, G.; Fei, Z. Cellular and molecular characterizations of a slow-growth variant provide insights into the fast growth of bamboo. Tree Physiol. 2018, 38, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Guo, L.; Jiao, C.; Fei, Z.; Chen, M.; Cao, J.; Ding, Y.; Yuan, Q. Characterization of the developmental dynamics of the elongation of a bamboo internode during the fast growth stage. Tree Physiol. 2019, 39, 1201–1214. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H.; et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). GigaScience 2018, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Li, L.; Zhang, S.; Shen, J.; Li, S.; Hu, S.; Peng, Q.; Xiao, J.; Wu, C. Suppressor of rid1 (SID1) shares common targets with RID1 on florigen genes to initiate floral transition in rice. PLoS Genet. 2017, 13, e1006642. [Google Scholar] [CrossRef]
- Jeong, E.Y.; Seo, P.J.; Woo, J.C.; Park, C.M. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 2015, 15, 110. [Google Scholar] [CrossRef]
- Gontarek, B.C.; Neelakandan, A.K.; Wu, H.; Becraft, P.W. NKD Transcription factors are central regulators of maize endosperm development. Plant Cell 2016, 28, 2916–2936. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Xing, L.; Qi, S.; Du, L.; Wu, H.; Shao, H.; Li, Y.; Ma, J.; Han, M. Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol. Genet. Genom. 2017, 292, 755–771. [Google Scholar] [CrossRef]
- Su, X.; Meng, T.; Zhao, Y.; Li, G.; Cheng, X.; Abdullah, M.; Sun, X.; Cai, Y.; Lin, Y. Comparative genomic analysis of the IDD genes in five Rosaceae species and expression analysis in Chinese white pear (Pyrus bretschneideri). Peer J. 2019, 7, e6628. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A.; Bennetzen, J.L. The evolution of nuclear genome structure in seed plants. Am. J. Bot. 2004, 91, 1709–1725. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. Progress and challenges in studies of the evolution of development. J. Exp. Bot. 2006, 57, 3505–3516. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Völz, R.; Rayapuram, N.; Hirt, H. Phosphorylation regulates the activity of INDETERMINATE-DOMAIN (IDD/BIRD) proteins in response to diverse environmental conditions. Plant Signal. Behav. 2019, 14, e1642037. [Google Scholar] [CrossRef]
- Halford, N.G.; Hey, S.J. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 2009, 419, 247–259. [Google Scholar] [CrossRef]
- Gamuyao, R.; Nagai, K.; Ayano, M.; Mori, Y.; Minami, A.; Kojima, M.; Suzuki, T.; Sakakibara, H.; Higashiyama, T.; Ashikari, M.; et al. Hormone distribution and transcriptome profiles in bamboo shoots provide insights on bamboo stem emergence and growth. Plant Cell Physiol. 2017, 58, 702–716. [Google Scholar] [CrossRef]
- Long, Y.; Smet, W.; Cruz-Ramírez, A.; Castelijns, B.; Jonge, W.D.; Mähönen, A.P.; Bouchet, B.P.; Perez, G.S.; Akhmanova, A.; Scheres, B.; et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 2015, 27, 1185–1199. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.A.; Sozzani, R.; Yardımcı, G.G.; Petricka, J.J.; Vernoux, T.; Blilou, I.; Jose, A.; Cara, M.W.; Uwe, O.; Ben, S.; et al. Transcriptional control of tissue formation throughout root development. Science 2015, 350, 426–430. [Google Scholar] [CrossRef]
- Zhao, J.; Gao, P.; Li, C.; Lin, X.; Guo, X.; Liu, S. PhePEBP family genes regulated by plant hormones and drought are associated with the activation of lateral buds and seedling growth in Phyllostachys edulis. Tree Physiol. 2019, 39, 1387–1404. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, Z.; Fei, B.; Li, L.; Hu, T.; Gao, Z.; Jiang, Z. BambooGDB: A bamboo genome database with functional annotation and an analysis platform. Database 2014, 2014, bau006. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Rui, X. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Pan, F.; Wang, Y.; Liu, H.; Wu, M.; Chu, W.; Chen, D.; Xiang, Y. Genome-wide identification and expression analysis of SBP-like transcription factor genes in moso bamboo (Phyllostachys edulis). BMC Genom. 2017, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Coulter, J.A.; Li, Y.; Zhang, X.; Meng, J.; Zhang, J.; Liu, Y. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 153, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2017, 8, e56573. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Phe-Phe | Phe-Os |
|---|---|
| PheIDD1/PheIDD2 | PheIDD1/OsID1 |
| PheIDD5/PheIDD6 | PheIDD2/OsID1 |
| PheIDD7/PheIDD16 | PheIDD4/OsIDD7 |
| PheIDD9/PheIDD22 | PheIDD5/OsIDD11 |
| PheIDD10/PheIDD14 | PheIDD6/OsIDD11 |
| PheIDD11/PheIDD12 | PheIDD7/OsIDD8 |
| PheIDD13/PheIDD18 | PheIDD8/OsIDD9 |
| PheIDD15/PheIDD17 | PheIDD9/OsIDD2 |
| PheIDD19/PheIDD26 | PheIDD11/OsIDD1 |
| PheIDD20/PheIDD23 | PheIDD12/OsIDD1 |
| PheIDD21/PheIDD25 | PheIDD13/OsIDD3 |
| PheIDD27/PheIDD29 | PheIDD15/OsIDD6 |
| PheIDD28/PheIDD30 | PheIDD16/OsIDD8 |
| PheIDD31/PheIDD32 | PheIDD17/OsIDD6 |
| PheIDD18/OsIDD3 | |
| PheIDD19OsIDD4 | |
| PheIDD20/OsIDD14 | |
| PheIDD21/OsIDD10 | |
| PheIDD22/OsIDD2 | |
| PheIDD23/OsIDD14 | |
| PheIDD24/OsIDD7 | |
| PheIDD25/OsIDD10 | |
| PheIDD26/OsIDD4 | |
| PheIDD27/OsIDD13 | |
| PheIDD28/OsIDD12 | |
| PheIDD29/OsIDD13 | |
| PheIDD30/OsIDD12 | |
| PheIDD31/OsIDD5 | |
| PheIDD32//OsIDD5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Zhou, M.; Chen, J.; Shao, M.; Zou, L.; Ying, Y.; Liu, S. Genome-Wide Identification of the Highly Conserved INDETERMINATE DOMAIN (IDD) Zinc Finger Gene Family in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2022, 23, 13952. https://doi.org/10.3390/ijms232213952
Guo X, Zhou M, Chen J, Shao M, Zou L, Ying Y, Liu S. Genome-Wide Identification of the Highly Conserved INDETERMINATE DOMAIN (IDD) Zinc Finger Gene Family in Moso Bamboo (Phyllostachys edulis). International Journal of Molecular Sciences. 2022; 23(22):13952. https://doi.org/10.3390/ijms232213952
Chicago/Turabian StyleGuo, Xiaoqin, Minshu Zhou, Jiaoyu Chen, Mingxia Shao, Longhai Zou, Yeqing Ying, and Shenkui Liu. 2022. "Genome-Wide Identification of the Highly Conserved INDETERMINATE DOMAIN (IDD) Zinc Finger Gene Family in Moso Bamboo (Phyllostachys edulis)" International Journal of Molecular Sciences 23, no. 22: 13952. https://doi.org/10.3390/ijms232213952





