Chromosomally Unstable Gastric Cancers Overexpressing Claudin-6 Disclose Cross-Talk between HNF1A and HNF4A, and Upregulated Cholesterol Metabolism
Abstract
:1. Introduction
2. Results
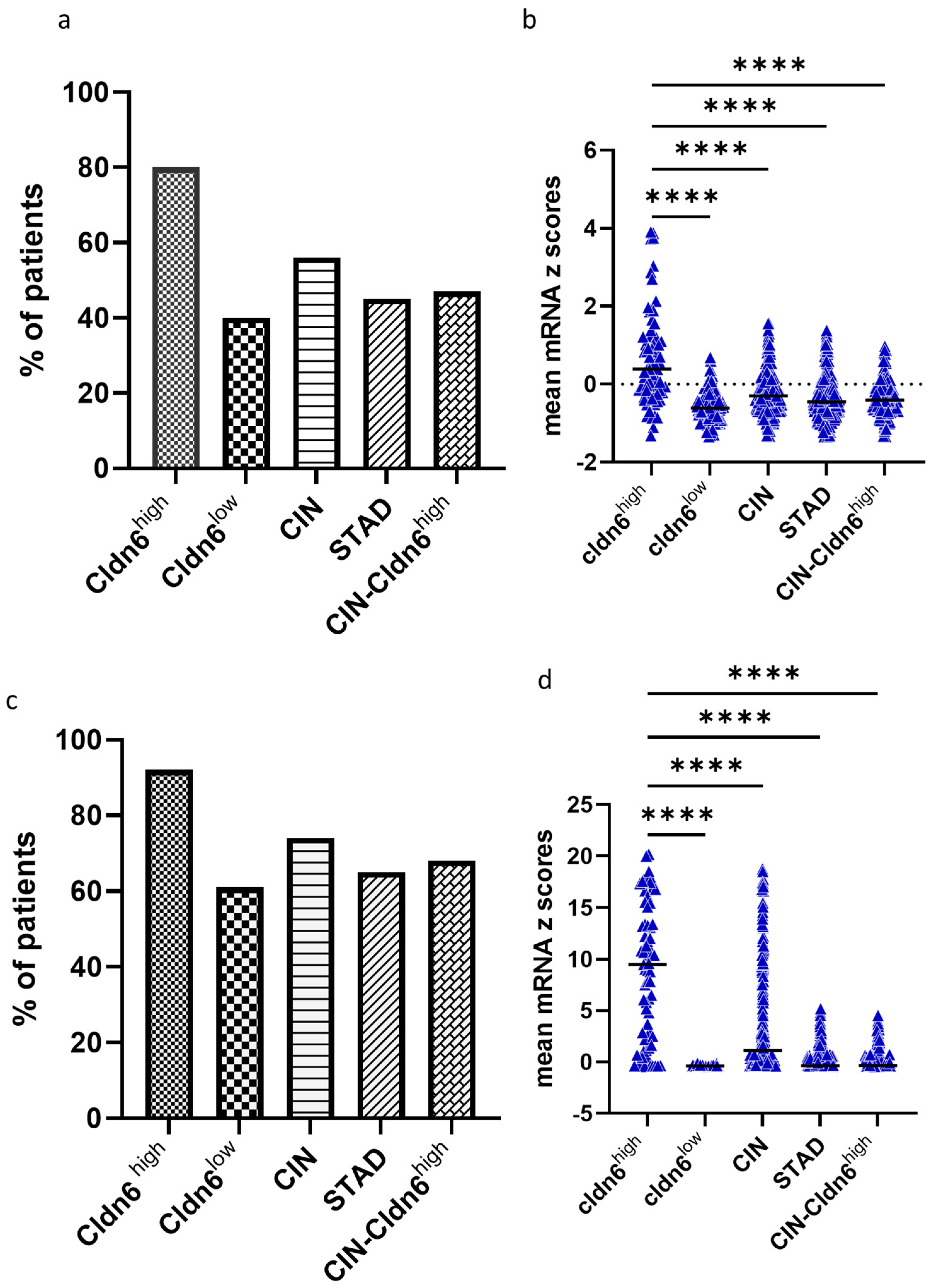
2.1. Cldn6 Expression and Molecular Subtype
2.2. Genomic Alterations and Differential Expression of Genes
2.3. GSEA and Pathway Analysis
2.4. Hub Genes and Prognostic Markers
2.5. HNF-4α/HNF-1α
3. Discussion
4. Materials and Methods
4.1. Database
4.2. Survival Analysis
4.3. Genomic Alterations and Differentially Expressed Genes
4.4. Signal Pathway and Transcription Factor Activity
4.5. Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO) Enrichment
4.6. Gene Interaction Network and Identification of Hub Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.G.; Somer, R.A. Intratumor Heterogeneity: Novel Approaches for Resolving Genomic Architecture and Clonal Evolution. Mol. Cancer Res. 2017, 15, 1127–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-David, U.; Nudel, N.; Benvenisty, N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat. Commun. 2013, 4, 1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadler, C.R.; Bahr-Mahmud, H.; Plum, L.M.; Schmoldt, K.; Kolsch, A.C.; Tureci, O.; Sahin, U. Characterization of the first-in-class T-cell-engaging bispecific single-chain antibody for targeted immunotherapy of solid tumors expressing the oncofetal protein claudin 6. Oncoimmunology 2016, 5, e1091555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendon-Huerta, E.; Teresa, F.; Teresa, G.M.; Xochitl, G.S.; Georgina, A.F.; Veronica, Z.Z.; Montano, L.F. Distribution and expression pattern of claudins 6, 7, and 9 in diffuse- and intestinal-type gastric adenocarcinomas. J. Gastrointest. Cancer 2010, 41, 52–59. [Google Scholar] [CrossRef]
- Ramos, M.; Pereira, M.A.; de Mello, E.S.; Cirqueira, C.D.S.; Zilberstein, B.; Alves, V.A.F.; Ribeiro-Junior, U.; Cecconello, I. Gastric cancer molecular classification based on immunohistochemistry and in situ hybridization: Analysis in western patients after curative-intent surgery. World J. Clin. Oncol. 2021, 12, 688–701. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Zavala-Zendejas, V.E.; Torres-Martinez, A.C.; Salas-Morales, B.; Fortoul, T.I.; Montano, L.F.; Rendon-Huerta, E.P. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Investig. 2011, 29, 1–11. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, Y.; Li, Q.; Zhang, Z.; Zhao, G.; Xu, J. CLDN6 promotes tumor progression through the YAP1-snail1 axis in gastric cancer. Cell Death Dis. 2019, 10, 949. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Li, Y.; Zhang, T.; Han, S.T. Claudin-6 is down-regulated in gastric cancer and its potential pathway. Cancer Biomark. 2020, 28, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Li, M.; Xiang, R.; Zhou, X.; Zhu, L.; Zhai, Y. Expression of CLDN6 in tissues of gastric cancer patients: Association with clinical pathology and prognosis. Oncol. Lett. 2019, 17, 4621–4625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasingam, V.; Diamandis, E.P. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008, 5, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yuan, Q.; Wu, D.; Meng, X.; Wang, B. Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget 2017, 8, 70271–70280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Liu, X.; Zhou, N.; Wu, Q.; Zhou, L.; Li, Q. Gene expression profiling and bioinformatics analysis of gastric carcinoma. Exp. Mol. Pathol. 2014, 96, 361–366. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Y.; Chen, Y.; Jiang, J.; Chen, Y.; Lu, J.; Kong, M.; Mo, F.; Huang, Y.; Zhao, W.; et al. A novel genomic classification system of gastric cancer via integrating multidimensional genomic characteristics. Gastric Cancer 2021, 24, 1227–1241. [Google Scholar] [CrossRef]
- Fenoglio-Preiser, C.M.; Wang, J.; Stemmermann, G.N.; Noffsinger, A. TP53 and gastric carcinoma: A review. Hum. Mutat. 2003, 21, 258–270. [Google Scholar] [CrossRef]
- Lauren, P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Cai, H.; Jing, C.; Chang, X.; Ding, D.; Han, T.; Yang, J.; Lu, Z.; Hu, X.; Liu, Z.; Wang, J.; et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J. Transl. Med. 2019, 17, 189. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef]
- Chen, Z.H.; Yan, S.M.; Chen, X.X.; Zhang, Q.; Liu, S.X.; Liu, Y.; Luo, Y.L.; Zhang, C.; Xu, M.; Zhao, Y.F.; et al. The genomic architecture of EBV and infected gastric tissue from precursor lesions to carcinoma. Genome Med. 2021, 13, 146. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Coto, D.L.; Gil, J.; Hernandez, A.; Herrera-Goepfert, R.; Castro-Romero, I.; Hernandez-Marquez, E.; Arenas-Linares, A.S.; Calderon-Sosa, V.T.; Sanchez-Aleman, M.A.; Mendez-Tenorio, A.; et al. Quantitative proteomics reveals proteins involved in the progression from non-cancerous lesions to gastric cancer. J. Proteom. 2018, 186, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Mun, D.G.; Bhin, J.; Kim, S.; Kim, H.; Jung, J.H.; Jung, Y.; Jang, Y.E.; Park, J.M.; Kim, H.; Jung, Y.; et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell 2019, 35, 111–124.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukita, S.; Furuse, M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann. N. Y. Acad. Sci. 2000, 915, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Rusu, A.D.; Georgiou, M. The multifarious regulation of the apical junctional complex. Open Biol. 2020, 10, 190278. [Google Scholar] [CrossRef]
- Qu, H.; Jin, Q.; Quan, C. CLDN6: From Traditional Barrier Function to Emerging Roles in Cancers. Int. J. Mol. Sci. 2021, 22, 13416. [Google Scholar] [CrossRef]
- Torres-Martinez, A.C.; Gallardo-Vera, J.F.; Lara-Holguin, A.N.; Montano, L.F.; Rendon-Huerta, E.P. Claudin-6 enhances cell invasiveness through claudin-1 in AGS human adenocarcinoma gastric cancer cells. Exp. Cell Res. 2017, 350, 226–235. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell 2007, 12, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Yamada, Y.; Yoshida, T.; Hayashi, K.; Sekiya, T.; Yokota, J.; Hirohashi, S.; Nakatani, K.; Nakano, H.; Sugimura, T.; Terada, M. p53 gene mutations in gastric cancer metastases and in gastric cancer cell lines derived from metastases. Cancer Res. 1991, 51, 5800–5805. [Google Scholar]
- Nemtsova, M.V.; Kalinkin, A.I.; Kuznetsova, E.B.; Bure, I.V.; Alekseeva, E.A.; Bykov, I.I.; Khorobrykh, T.V.; Mikhaylenko, D.S.; Tanas, A.S.; Kutsev, S.I.; et al. Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci. Rep. 2020, 10, 504. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, A.; Bourgmayer, A.; Kurtz, J.E.; Mellitzer, G.; Gaiddon, C. Isoforms of the p53 Family and Gastric Cancer: A Menage a Trois for an Unfinished Affair. Cancers 2021, 13, 916. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.P.; Gupta, S.; Singh, A.K.; Kumar, S. Emerging Role of Migration and Invasion Enhancer 1 (MIEN1) in Cancer Progression and Metastasis. Front. Oncol. 2019, 9, 868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassilev, B.; Sihto, H.; Li, S.; Holtta-Vuori, M.; Ilola, J.; Lundin, J.; Isola, J.; Kellokumpu-Lehtinen, P.L.; Joensuu, H.; Ikonen, E. Elevated levels of StAR-related lipid transfer protein 3 alter cholesterol balance and adhesiveness of breast cancer cells: Potential mechanisms contributing to progression of HER2-positive breast cancers. Am. J. Pathol. 2015, 185, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Hann, S.S. Biological Roles and Mechanisms of Circular RNA in Human Cancers. OncoTargets Ther. 2020, 13, 2067–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, M.J.; Kim, R.N.; Song, K.; Jeon, S.; Jeong, H.M.; Kim, J.S.; Han, J.; Hong, S.; Oh, E.; Choi, J.S.; et al. Genes co-amplified with ERBB2 or MET as novel potential cancer-promoting genes in gastric cancer. Oncotarget 2017, 8, 92209–92226. [Google Scholar] [CrossRef] [Green Version]
- Lian, Y.; Meng, L.; Ding, P.; Sang, M. Epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification, and non-coding RNAs. Clin. Epigenet. 2018, 10, 115. [Google Scholar] [CrossRef]
- Ogata, K.; Aihara, R.; Mochiki, E.; Ogawa, A.; Yanai, M.; Toyomasu, Y.; Ando, H.; Ohno, T.; Asao, T.; Kuwano, H. Clinical significance of melanoma antigen-encoding gene-1 (MAGE-1) expression and its correlation with poor prognosis in differentiated advanced gastric cancer. Ann. Surg. Oncol. 2011, 18, 1195–1203. [Google Scholar] [CrossRef]
- Zhai, X.; Xu, L.; Zhang, S.; Zhu, H.; Mao, G.; Huang, J. High expression levels of MAGE-A9 are correlated with unfavorable survival in lung adenocarcinoma. Oncotarget 2016, 7, 4871–4881. [Google Scholar] [CrossRef] [Green Version]
- Hatiboglu, G.; Pritsch, M.; Macher-Goeppinger, S.; Zoller, M.; Huber, J.; Haferkamp, A.; Pahernik, S.; Wagener, N.; Hohenfellner, M. Prognostic value of melanoma-associated antigen A9 in renal cell carcinoma. Scand. J. Urol. 2013, 47, 311–322. [Google Scholar] [CrossRef]
- Lin, J.; Lin, L.; Thomas, D.G.; Greenson, J.K.; Giordano, T.J.; Robinson, G.S.; Barve, R.A.; Weishaar, F.A.; Taylor, J.M.; Orringer, M.B.; et al. Melanoma-associated antigens in esophageal adenocarcinoma: Identification of novel MAGE-A10 splice variants. Clin. Cancer Res. 2004, 10, 5708–5716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhai, X.; Wang, G.; Feng, J.; Zhu, H.; Xu, L.; Mao, G.; Huang, J. High expression of MAGE-A9 in tumor and stromal cells of non-small cell lung cancer was correlated with patient poor survival. Int. J. Clin. Exp. Pathol. 2015, 8, 541–550. [Google Scholar] [PubMed]
- Ohta, M.; Tanaka, F.; Sadanaga, N.; Yamaguchi, H.; Inoue, H.; Mori, M. Expression of the TRAG-3 gene in human esophageal cancer: The frequent synchronous expression of MAGE-3 gene. Oncol. Rep. 2006, 15, 1529–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambon, A.; Mandruzzato, S.; Parenti, A.; Macino, B.; Dalerba, P.; Ruol, A.; Merigliano, S.; Zaninotto, G.; Zanovello, P. MAGE, BAGE, and GAGE gene expression in patients with esophageal squamous cell carcinoma and adenocarcinoma of the gastric cardia. Cancer 2001, 91, 1882–1888. [Google Scholar] [CrossRef]
- Glass, M.; Misiak, D.; Bley, N.; Muller, S.; Hagemann, S.; Busch, B.; Rausch, A.; Huttelmaier, S. IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer. Front. Mol. Biosci. 2021, 8, 632219. [Google Scholar] [CrossRef]
- Reinhard, K.; Rengstl, B.; Oehm, P.; Michel, K.; Billmeier, A.; Hayduk, N.; Klein, O.; Kuna, K.; Ouchan, Y.; Woll, S.; et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science 2020, 367, 446–453. [Google Scholar] [CrossRef]
- Du, H.; Yang, X.; Fan, J.; Du, X. Claudin 6: Therapeutic prospects for tumours, and mechanisms of expression and regulation (Review). Mol. Med. Rep. 2021, 24, 677. [Google Scholar] [CrossRef]
- Quan, C.; Lu, S.J. Identification of genes preferentially expressed in mammary epithelial cells of Copenhagen rat using subtractive hybridization and microarrays. Carcinogenesis 2003, 24, 1593–1599. [Google Scholar] [CrossRef] [Green Version]
- Song, P.; Li, Y.; Dong, Y.; Liang, Y.; Qu, H.; Qi, D.; Lu, Y.; Jin, X.; Guo, Y.; Jia, Y.; et al. Estrogen receptor beta inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J. Exp. Clin. Cancer Res. 2019, 38, 354. [Google Scholar] [CrossRef] [Green Version]
- Bolanos-Garcia, V.M.; Miguel, R.N. On the structure and function of apolipoproteins: More than a family of lipid-binding proteins. Prog. Biophys. Mol. Biol. 2003, 83, 47–68. [Google Scholar] [CrossRef]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Zhang, C.J.; Zhu, N.; Du, K.; Yin, Y.F.; Tan, X.; Liao, D.F.; Qin, L. Lipid metabolism and carcinogenesis, cancer development. Am. J. Cancer Res. 2018, 8, 778–791. [Google Scholar] [PubMed]
- Martovetsky, G.; Bush, K.T.; Nigam, S.K. Kidney versus Liver Specification of SLC and ABC Drug Transporters, Tight Junction Molecules, and Biomarkers. Drug Metab. Dispos. 2016, 44, 1050–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, D.D.; Zhou, L.Y.; Tang, H. Hepatocyte nuclear factor 4alpha and cancer-related cell signaling pathways: A promising insight into cancer treatment. Exp. Mol. Med. 2021, 53, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ooi, W.F.; Qamra, A.; Tan, J.; Chua, B.Y.; Ho, S.W.T.; Das, K.; Adam Isa, Z.F.; Li, Z.; Yao, X.; et al. HNF4alpha pathway mapping identifies wild-type IDH1 as a targetable metabolic node in gastric cancer. Gut 2020, 69, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Riscal, R.; Skuli, N.; Simon, M.C. Even Cancer Cells Watch Their Cholesterol! Mol. Cell 2019, 76, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Kahn, B.M.; Lucas, A.; Alur, R.G.; Wengyn, M.D.; Schwartz, G.W.; Li, J.; Sun, K.; Maurer, H.C.; Olive, K.P.; Faryabi, R.B.; et al. The vascular landscape of human cancer. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Ma’ayan, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [Green Version]
- Lehner, B.; Crombie, C.; Tischler, J.; Fortunato, A.; Fraser, A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006, 38, 896–903. [Google Scholar] [CrossRef]










| Type | Cldn6high | Cldn6low |
|---|---|---|
| CIN | 69 | 57 |
| EBV | 0 | 22 |
| GS | 2 | 18 |
| MSI | 0 | 38 |
| POLE | 0 | 3 |
| Gene | Cldn6 high (n = 71) | Cldn6 low (n = 138) | Log Ratio | p-Value | q-Value |
|---|---|---|---|---|---|
| TP53 | 76.06% | 31.88% | 1.25 | 9.51 × 10−10 | 2.062 × 10−5 |
| MIEN1 | 29.58% | 5.80% | 2.35 | 5.917 × 10−6 | 0.0321 |
| STARD3 | 29.58% | 6.52% | 2.18 | 1.454 × 10−5 | 0.0487 |
| PGAP3 | 28.17% | 5.80% | 2.28 | 1.536 × 10−5 | 0.0487 |
| CCNE1 | 26.76% | 5.07% | 2.40 | 1.571 × 10−5 | 0.0487 |
| DOCK3 | 0.00% | 20.29% | <−10 | 3.143 × 10−6 | 0.0266 |
| ARID1A | 9.86% | 39.13% | −1.99 | 3.682 × 10−6 | 0.0266 |
| Gene | Log Ratio | p-Value | q-Value |
|---|---|---|---|
| CLDN6 | 8.49 | 7.14 × 10−41 | 1.35 × 10−36 |
| MAGEA6 | 6.12 | 1.62 × 10−21 | 4.36 × 10−18 |
| IGF2BP1 | 6.12 | 3.63 × 10−24 | 1.71 × 10−20 |
| MAGEA3 | 5.86 | 4.58 × 10−21 | 1.08 × 10−17 |
| MAGEA4 | 5.62 | 2.11 × 10−15 | 6.33 × 10−13 |
| APOA1 | 5.55 | 7.33 × 10−18 | 5.13 × 10−15 |
| MAGEA2 | 5.37 | 1.62 × 10−19 | 2.35 × 10−16 |
| MAGEA9B | 5.04 | 1.11 × 10−16 | 5.00 × 10−14 |
| MAGEA12 | 4.93 | 2.67 × 10−17 | 1.60 × 10−14 |
| CSAG1 | 4.35 | 3.83 × 10−16 | 1.54 × 10−13 |
| COL2A1 | 4.25 | 1.55 × 10−11 | 1.20 × 10−09 |
| APOA4 | 4.2 | 1.29 × 10−12 | 1.44 × 10−10 |
| PRAME | 4.18 | 4.49 × 10−16 | 1.73 × 10−13 |
| MAGEA10 | 4.14 | 2.63 × 10−13 | 3.73 × 10−11 |
| TF | 4.11 | 1.48 × 10−12 | 1.61 × 10−10 |
| FGB | 4.06 | 7.51 × 10−12 | 6.30 × 10−10 |
| TTR | 4.02 | 1.24 × 10−12 | 1.40 × 10−10 |
| APOC3 | 3.96 | 1.33 × 10−15 | 4.24 × 10−13 |
| DSCR8 | 3.91 | 1.82 × 10−13 | 2.77 × 10−11 |
| LGALS7B | 3.89 | 8.11 × 10−16 | 2.94 × 10−13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dwivedi, S.; Hernández-Montes, G.; Montaño, L.F.; Rendón-Huerta, E.P. Chromosomally Unstable Gastric Cancers Overexpressing Claudin-6 Disclose Cross-Talk between HNF1A and HNF4A, and Upregulated Cholesterol Metabolism. Int. J. Mol. Sci. 2022, 23, 13977. https://doi.org/10.3390/ijms232213977
Dwivedi S, Hernández-Montes G, Montaño LF, Rendón-Huerta EP. Chromosomally Unstable Gastric Cancers Overexpressing Claudin-6 Disclose Cross-Talk between HNF1A and HNF4A, and Upregulated Cholesterol Metabolism. International Journal of Molecular Sciences. 2022; 23(22):13977. https://doi.org/10.3390/ijms232213977
Chicago/Turabian StyleDwivedi, Sanyog, Georgina Hernández-Montes, Luis Felipe Montaño, and Erika Patricia Rendón-Huerta. 2022. "Chromosomally Unstable Gastric Cancers Overexpressing Claudin-6 Disclose Cross-Talk between HNF1A and HNF4A, and Upregulated Cholesterol Metabolism" International Journal of Molecular Sciences 23, no. 22: 13977. https://doi.org/10.3390/ijms232213977
APA StyleDwivedi, S., Hernández-Montes, G., Montaño, L. F., & Rendón-Huerta, E. P. (2022). Chromosomally Unstable Gastric Cancers Overexpressing Claudin-6 Disclose Cross-Talk between HNF1A and HNF4A, and Upregulated Cholesterol Metabolism. International Journal of Molecular Sciences, 23(22), 13977. https://doi.org/10.3390/ijms232213977




