Biofilms and Benign Colonic Diseases
Abstract
1. Introduction
2. Biofilm Formation Based on Studies with Pseudomonas aeruginosa
2.1. Biofilm Structure
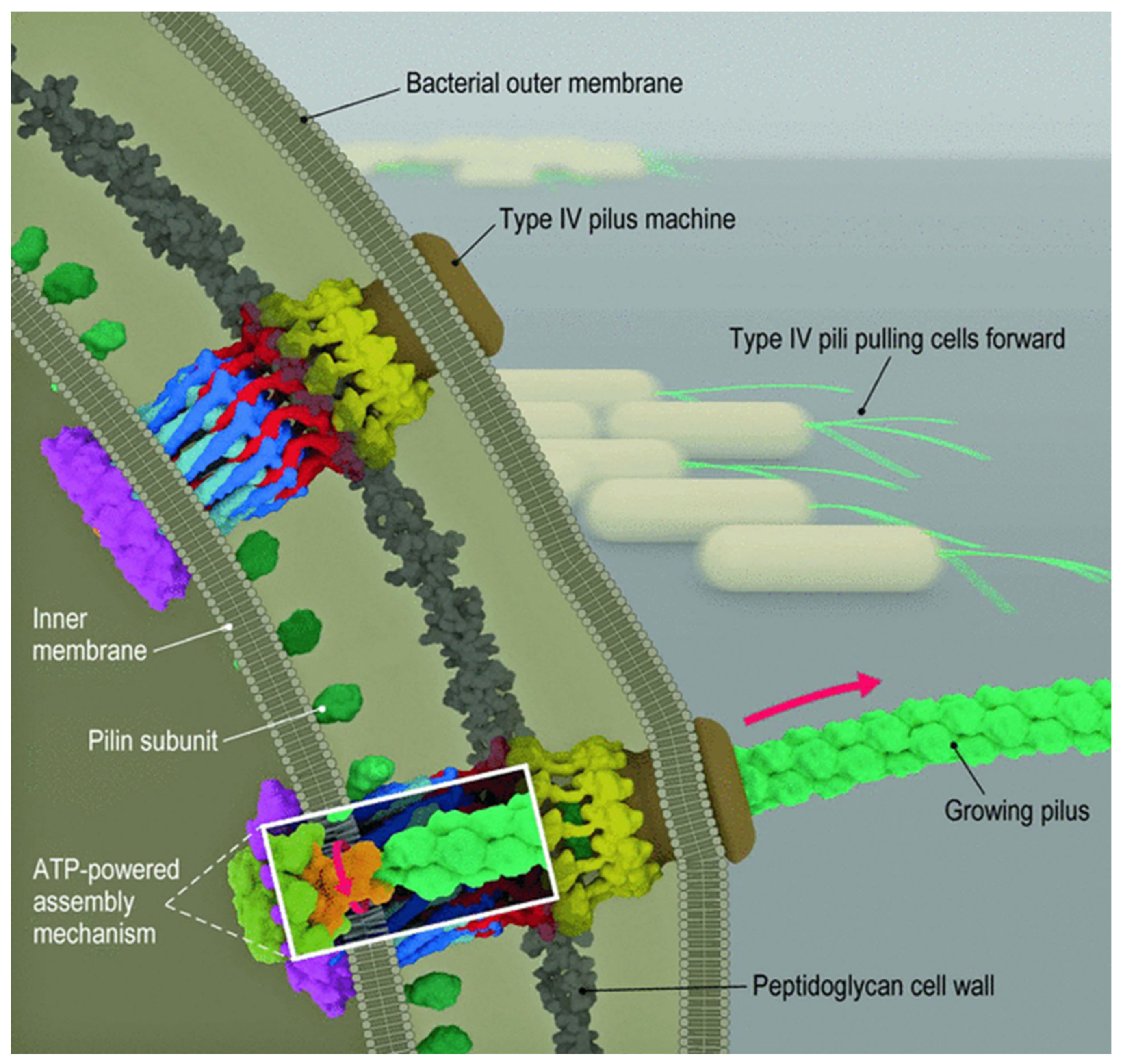
2.2. Bacterial Attachment to Surfaces
3. Quorum Sensing

4. Detection of Biofilms
4.1. In Vitro Techniques for Biofilm Detection
4.1.1. Direct Observation
Light Microscope
Fluorescence In Situ Hybridization (FISH)
Electron and Confocal
4.1.2. Indirect Observation
Roll Plate Method
Congo Red Agar Test
Tube Biofilm Formation Test
Microtiter Plate Assay
Polymerase Chain Reaction
4.2. In Vivo Techniques for Detection of Biofilms
Low-Coherence Interferometry and Optical Coherence Tomography
5. Microbiome and Biofilms in the Colon
5.1. Metabolic Activity in Biofilms
5.2. Host Defenses at the Colonic Surface
5.3. Bacterial Protection in Biofilms [110]
5.4. Other Factors Relevant to Biofilm Formation and Stability in the Colon
5.5. Colonic Biofilms and Disease
6. Inflammatory Bowel Disease (IBD)
6.1. Alteration of Gut Microbiome in IBD
6.2. Microbiome–Host Immune System Interactions and the Pathogenesis in IBD
6.3. Biofilms and IBD
7. Irritable Bowel Syndrome (IBS)
7.1. Alterations in the Gut Microbiota
7.2. Microbiome–Host Immune System Interactions and Pathogenesis in IBS
7.3. Bacterial Biofilm and IBS
8. Clostridium Difficile Colitis
9. Management of Colonic Biofilms
9.1. Antibiotics
9.2. Probiotics
9.3. Other Therapies
10. Critique
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
- Microbiota: a collection of microorganisms, including fungi, viruses, and bacteria, present in a defined environment, such as the gut.
- Microbiome: the entire habitat that includes the microorganisms, their genomes and genes, and the surrounding environment, both cellular and acellular.
- Planktonic bacteria: free-floating bacteria in solutions.
- Biofilms: aggregations of microbial communities in a polymeric matrix that adhere to either biological or nonbiological surfaces.
- Genetic dysbiosis: change in intestinal microbiota (in terms of species and their concentration) or misrecognition by the body.
- Classification hierarchy: domain, kingdom, phylum, class, order, family, genus, species.
- 16 S ribosomal RNA DNA gene: The 16S rRNA gene sequence is about 1550 base pairs long and has both variable and conserved regions. It has nine highly variable regions used for classification and quantitation of microbes and bacteria in complex mixtures.
- Pili: short, filamentous projections on the surface of many (particularly Gram-negative) bacteria used to adhere to other bacteria or to animal cells or to surfaces
- AIEC: adherent, invasive E. coli species that can invade intestinal epithelial cells and can survive and grow in macrophages.
- Methogens: microorganisms in the domain Archaea which use CO2 to produce methane.
- Short-chain fatty acids: fatty acids with fewer than six carbon atoms derived from intestinal microbial fermentation of indigestible food particles.
- Butyric acid: CH3CH2CH2CO2H, molar mass: 88.11 g/mol.
References
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Gadkari, J.; Bhattacharya, S.; Shrivastav, A. Importance and application of biofilm in microbe assisted bioremediation. In Development in Waste Water Treatment Research and Processes; Shah, M.P., Rodriguez-Couto, S., Riti Thapar, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 154–173. [Google Scholar]
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019, 27, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- Monroe, D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007, 5, e307. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Hutchison, J.B.; Melaugh, G.; Rodesney, C.; Roberts, A.E.; Irie, Y.; Jensen, P.; Diggle, S.P.; Allen, R.J.; Gordon, V.; et al. Role of Multicellular Aggregates in Biofilm Formation. mBio 2016, 7, e00237. [Google Scholar] [CrossRef]
- Paula, A.J.; Hwang, G.; Koo, H. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat. Commun. 2020, 11, 1354. [Google Scholar] [CrossRef]
- Reichhardt, C.; Parsek, M.R. Confocal Laser Scanning Microscopy for Analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 2019, 10, 677. [Google Scholar] [CrossRef]
- Stewart, P.S. Diffusion in biofilms. J. Bacteriol. 2003, 185, 1485–1491. [Google Scholar] [CrossRef]
- Rooney, L.M.; Amos, W.B.; Hoskisson, P.A.; McConnell, G. Intra-colony channels in E. coli function as a nutrient uptake system. ISME J. 2020, 14, 2461–2473. [Google Scholar] [CrossRef]
- Chang, C.Y. Surface Sensing for Biofilm Formation in. Front. Microbiol. 2017, 8, 2671. [Google Scholar] [CrossRef]
- Ligthart, K.; Belzer, C.; de Vos, W.M.; Tytgat, H.L.P. Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili. Trends Microbiol. 2020, 28, 340–348. [Google Scholar] [CrossRef]
- O’Neal, L.; Baraquet, C.; Suo, Z.; Dreifus, J.E.; Peng, Y.; Raivio, T.L.; Wozniak, D.J.; Harwood, C.S.; Parsek, M.R. The Wsp system of Pseudomonas aeruginosa links surface sensing and cell envelope stress. Proc. Natl. Acad. Sci. USA 2022, 119, e2117633119. [Google Scholar] [CrossRef]
- Nolan, L.M.; Cavaliere, R.; Turnbull, L.; Whitchurch, C.B. Extracellular ATP inhibits twitching motility-mediated biofilm expansion by Pseudomonas aeruginosa. BMC Microbiol. 2015, 15, 55. [Google Scholar] [CrossRef]
- Webster, S.S.; Wong, G.C.L.; O’Toole, G.A. The Power of Touch: Type 4 Pili, the von Willebrand A Domain, and Surface Sensing by Pseudomonas aeruginosa. J. Bacteriol. 2022, 204, e0008422. [Google Scholar] [CrossRef]
- Armbruster, C.R.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Lee, J.Y.; Tsolis, R.M.; Bäumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, eabp9960. [Google Scholar] [CrossRef]
- Chew, S.-S.; Tan, L.T.-H.; Law, J.W.-F.; Pusparajah, P.; Goh, B.-H.; Ab Mutalib, N.S.; Lee, L.-H. Targeting Gut Microbial Biofilms—A Key to Hinder Colon Carcinogenesis? Cancers 2020, 12, 2272. [Google Scholar] [CrossRef]
- Chang, Y.W.; Rettberg, L.A.; Treuner-Lange, A.; Iwasa, J.; Søgaard-Andersen, L.; Jensen, G.J. Architecture of the type IVa pilus machine. Science 2016, 351, aad2001. [Google Scholar] [CrossRef]
- Beaussart, A.; Baker, A.E.; Kuchma, S.L.; El-Kirat-Chatel, S.; O’Toole, G.A.; Dufrêne, Y.F. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili. ACS Nano 2014, 8, 10723–10733. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.; Vlamakis, H.; Losick, R.; Kolter, R. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 2007, 10, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L.; Losick, R. Bacterially speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Mukherjee, S.; Jemielita, M.; Stergioula, V.; Tikhonov, M.; Bassler, B.L. Photosensing and quorum sensing are integrated to control Pseudomonas aeruginosa collective behaviors. PLoS Biol. 2019, 17, e3000579. [Google Scholar] [CrossRef]
- De Kievit, T.R.; Iglewski, B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [CrossRef]
- Bronesky, D.; Wu, Z.; Marzi, S.; Walter, P.; Geissmann, T.; Moreau, K.; Vandenesch, F.; Caldelari, I.; Romby, P. Staphylococcus aureus RNAIII and Its Regulon Link Quorum Sensing, Stress Responses, Metabolic Adaptation, and Regulation of Virulence Gene Expression. Annu. Rev. Microbiol. 2016, 70, 299–316. [Google Scholar] [CrossRef]
- Grossman, A.D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 1995, 29, 477–508. [Google Scholar] [CrossRef]
- Solomon, J.M.; Magnuson, R.; Srivastava, A.; Grossman, A.D. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes. Dev. 1995, 9, 547–558. [Google Scholar] [CrossRef]
- Barnard, A.M.; Bowden, S.D.; Burr, T.; Coulthurst, S.J.; Monson, R.E.; Salmond, G.P. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1165–1183. [Google Scholar] [CrossRef]
- Bassler, B.L.; Wright, M.; Silverman, M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994, 13, 273–286. [Google Scholar] [CrossRef]
- Engebrecht, J.; Nealson, K.; Silverman, M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 1983, 32, 773–781. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Quadri, L.E.; Kuipers, O.P.; de Vos, W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997, 24, 895–904. [Google Scholar] [CrossRef]
- Okada, M.; Sato, I.; Cho, S.J.; Iwata, H.; Nishio, T.; Dubnau, D.; Sakagami, Y. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 2005, 1, 23–24. [Google Scholar] [CrossRef]
- Lazar, V.; Holban, A.M.; Curutiu, C.; Chifiriuc, M.C. Modulation of Quorum Sensing and Biofilms in Less Investigated Gram-Negative ESKAPE Pathogens. Front. Microbiol. 2021, 12, 676510. [Google Scholar] [CrossRef]
- Li, Y.H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- Preda, V.G.; Sandulescu, O. Communication is the key: Biofilms, quorum sensing, formation and prevention. Discoveries 2019, 7, e100. [Google Scholar] [CrossRef]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Kaplan, H.B.; Greenberg, E.P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 1985, 163, 1210–1214. [Google Scholar] [CrossRef]
- Novick, R.P.; Projan, S.J.; Kornblum, J.; Ross, H.F.; Ji, G.; Kreiswirth, B.; Vandenesch, F.; Moghazeh, S. The agr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 1995, 248, 446–458. [Google Scholar] [CrossRef]
- Seed, P.C.; Passador, L.; Iglewski, B.H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: An autoinduction regulatory hierarchy. J. Bacteriol. 1995, 177, 654–659. [Google Scholar] [CrossRef]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef]
- Geisinger, E.; George, E.A.; Chen, J.; Muir, T.W.; Novick, R.P. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J. Biol. Chem. 2008, 283, 8930–8938. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Perez, L.J.; Wei, Y.; Kraml, C.; Semmelhack, M.F.; Bassler, B.L. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011, 79, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.; Vikstrom, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant Sci. 2014, 5, 309. [Google Scholar] [CrossRef] [PubMed]
- Landman, C.; Grill, J.P.; Mallet, J.M.; Marteau, P.; Humbert, L.; Le Balc’h, E.; Maubert, M.A.; Perez, K.; Chaara, W.; Brot, L.; et al. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS ONE 2018, 13, e0202587. [Google Scholar] [CrossRef]
- Darkoh, C.; Plants-Paris, K.; Bishoff, D.; DuPont, H.L. Clostridium difficile Modulates the Gut Microbiota by Inducing the Production of Indole, an Interkingdom Signaling and Antimicrobial Molecule. mSystems 2019, 4, e00346-18. [Google Scholar] [CrossRef]
- Lenz, D.H.; Bassler, B.L. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 2007, 63, 859–871. [Google Scholar] [CrossRef]
- Lenz, D.H.; Miller, M.B.; Zhu, J.; Kulkarni, R.V.; Bassler, B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005, 58, 1186–1202. [Google Scholar] [CrossRef]
- Thompson, L.S.; Webb, J.S.; Rice, S.A.; Kjelleberg, S. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2003, 220, 187–195. [Google Scholar] [CrossRef]
- Kolter, R.; Greenberg, E.P. Microbial sciences: The superficial life of microbes. Nature 2006, 441, 300–302. [Google Scholar] [CrossRef]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial Quorum Sensing. Cell 2018, 174, 1328–1328.e1. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef]
- Mooney, J.A.; Pridgen, E.M.; Manasherob, R.; Suh, G.; Blackwell, H.E.; Barron, A.E.; Bollyky, P.L.; Goodman, S.B.; Amanatullah, D.F. Periprosthetic bacterial biofilm and quorum sensing. J. Orthop. Res. 2018, 36, 2331–2339. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Silva, N.B.S.; Marques, L.A.; Roder, D.D.B. Diagnosis of biofilm infections: Current methods used, challenges and perspectives for the future. J. Appl. Microbiol. 2021, 131, 2148–2160. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Wagner, M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 2007, 75, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Continenza, M.A.; Al-Ahmad, A.; Karygianni, L.; Follo, M.; Filippi, A.; Macchiarelli, G. Streptococcus spp. and Fusobacterium nucleatum in tongue dorsum biofilm from halitosis patients: A fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM) study. New Microbiol. 2019, 42, 108–113. [Google Scholar] [PubMed]
- Machado, A.; Castro, J.; Cereija, T.; Almeida, C.; Cerca, N. Diagnosis of bacterial vaginosis by a new multiplex peptide nucleic acid fluorescence in situ hybridization method. PeerJ 2015, 3, e780. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; de Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcon, K.P.; Melo, W.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal Biofilms and Polymicrobial Diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef]
- Schlafer, S.; Meyer, R.L. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods 2017, 138, 50–59. [Google Scholar] [CrossRef]
- Sugimoto, S.; Okuda, K.; Miyakawa, R.; Sato, M.; Arita-Morioka, K.; Chiba, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y.; Sato, C. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 2016, 6, 25889. [Google Scholar] [CrossRef]
- Mohmmed, S.A.; Vianna, M.E.; Penny, M.R.; Hilton, S.T.; Mordan, N.; Knowles, J.C. Confocal laser scanning, scanning electron, and transmission electron microscopy investigation of Enterococcus faecalis biofilm degradation using passive and active sodium hypochlorite irrigation within a simulated root canal model. Microbiologyopen 2017, 6, e00455. [Google Scholar] [CrossRef]
- Benamara, H.; Rihouey, C.; Abbes, I.; Ben Mlouka, M.A.; Hardouin, J.; Jouenne, T.; Alexandre, S. Characterization of membrane lipidome changes in Pseudomonas aeruginosa during biofilm growth on glass wool. PLoS ONE 2014, 9, e108478. [Google Scholar] [CrossRef]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001, 413, 860–864. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 2015, 23, 233–242. [Google Scholar] [CrossRef]
- Maki, D.G.; Jarrett, F.; Sarafin, H.W. A semiquantitative culture method for identification of catheter-related infection in the burn patient. J. Surg. Res. 1977, 22, 513–520. [Google Scholar] [CrossRef]
- Slobbe, L.; El Barzouhi, A.; Boersma, E.; Rijnders, B.J. Comparison of the roll plate method to the sonication method to diagnose catheter colonization and bacteremia in patients with long-term tunnelled catheters: A randomized prospective study. J. Clin. Microbiol. 2009, 47, 885–888. [Google Scholar] [CrossRef]
- Lee, J.S.; Bae, Y.M.; Lee, S.Y.; Lee, S.Y. Biofilm Formation of Staphylococcus aureus on Various Surfaces and Their Resistance to Chlorine Sanitizer. J. Food Sci. 2015, 80, M2279–M2286. [Google Scholar] [CrossRef]
- Abdel Halim, R.M.; Kassem, N.N.; Mahmoud, B.S. Detection of Biofilm Producing Staphylococci among Different Clinical Isolates and Its Relation to Methicillin Susceptibility. Open Access Maced. J. Med. Sci. 2018, 6, 1335–1341. [Google Scholar] [CrossRef]
- Solati, S.M.; Tajbakhsh, E.; Khamesipour, F.; Gugnani, H.C. Prevalence of virulence genes of biofilm producing strains of Staphylococcus epidermidis isolated from clinical samples in Iran. AMB Express 2015, 5, 134. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Neopane, P.; Nepal, H.P.; Shrestha, R.; Uehara, O.; Abiko, Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018, 11, 25–32. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Hola, V.; Di Bonaventura, G.; Djukic, S.; Cirkovic, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Coffey, B.M.; Anderson, G.G. Biofilm formation in the 96-well microtiter plate. Methods Mol. Biol. 2014, 1149, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Shahmoradi, M.; Faridifar, P.; Shapouri, R.; Mousavi, S.F.; Ezzedin, M.; Mirzaei, B. Determining the Biofilm Forming Gene Profile of Staphylococcus aureus Clinical Isolates via Multiplex Colony PCR Method. Rep. Biochem. Mol. Biol. 2019, 7, 181–188. [Google Scholar] [PubMed]
- Klancnik, A.; Toplak, N.; Kovac, M.; Marquis, H.; Jersek, B. Quantification of Listeria monocytogenes cells with digital PCR and their biofilm cells with real-time PCR. J. Microbiol. Methods 2015, 118, 37–41. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Yang, B.; Ma, F.; Lu, P.; Li, L.; Wan, C.; Rayner, S.; Chen, S. Duckweed (Lemna minor) as a model plant system for the study of human microbial pathogenesis. PLoS ONE 2010, 5, e13527. [Google Scholar] [CrossRef]
- Kamareddine, L.; Wong, A.C.N.; Vanhove, A.S.; Hang, S.; Purdy, A.E.; Kierek-Pearson, K.; Asara, J.M.; Ali, A.; Morris, J.G., Jr.; Watnick, P.I. Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol. 2018, 3, 243–252. [Google Scholar] [CrossRef]
- De Bentzmann, S.; Giraud, C.; Bernard, C.S.; Calderon, V.; Ewald, F.; Plesiat, P.; Nguyen, C.; Grunwald, D.; Attree, I.; Jeannot, K.; et al. Unique biofilm signature, drug susceptibility and decreased virulence in Drosophila through the Pseudomonas aeruginosa two-component system PprAB. PLoS Pathog. 2012, 8, e1003052. [Google Scholar] [CrossRef]
- Desai, S.K.; Padmanabhan, A.; Harshe, S.; Zaidel-Bar, R.; Kenney, L.J. Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2019, 116, 12462–12467. [Google Scholar] [CrossRef]
- Rendueles, O.; Ferrieres, L.; Fretaud, M.; Begaud, E.; Herbomel, P.; Levraud, J.P.; Ghigo, J.M. A new zebrafish model of oro-intestinal pathogen colonization reveals a key role for adhesion in protection by probiotic bacteria. PLoS Pathog. 2012, 8, e1002815. [Google Scholar] [CrossRef]
- Zysk, A.M.; Nguyen, F.T.; Oldenburg, A.L.; Marks, D.L.; Boppart, S.A. Optical coherence tomography: A review of clinical development from bench to bedside. J. Biomed. Opt. 2007, 12, 051403. [Google Scholar] [CrossRef]
- Chaney, E.J.; Nguyen, C.T.; Boppart, S.A. Novel method for non-invasive induction of a middle-ear biofilm in the rat. Vaccine 2011, 29, 1628–1633. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Tu, H.; Chaney, E.J.; Stewart, C.N.; Boppart, S.A. Non-invasive optical interferometry for the assessment of biofilm growth in the middle ear. Biomed. Opt. Express 2010, 1, 1104–1116. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Jung, W.; Kim, J.; Chaney, E.J.; Novak, M.; Stewart, C.N.; Boppart, S.A. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc. Natl. Acad. Sci. USA 2012, 109, 9529–9534. [Google Scholar] [CrossRef]
- Dsouza, R.; Spillman, D.R.; Barkalifa, R., Jr.; Monroy, G.L.; Chaney, E.J.; White, K.C.; Boppart, S.A. In vivo detection of endotracheal tube biofilms in intubated critical care patients using catheter-based optical coherence tomography. J. Biophotonics 2019, 12, e201800307. [Google Scholar] [CrossRef]
- Chandra, N.; Srivastava, A.; Kumar, S. Bacterial biofilms in human gastrointestinal tract: An intricate balance between health and inflammatory bowel diseases. World 2019, 3, 26–40. [Google Scholar] [CrossRef]
- Béchon, N.; Ghigo, J.M. Gut biofilms: Bacteroides as model symbionts to study biofilm formation by intestinal anaerobes. FEMS Microbiol. Rev. 2022, 46, fuab054. [Google Scholar] [CrossRef]
- Pumbwe, L.; Skilbeck, C.A.; Nakano, V.; Avila-Campos, M.J.; Piazza, R.M.; Wexler, H.M. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 2007, 43, 78–87. [Google Scholar] [CrossRef]
- Pumbwe, L.; Skilbeck, C.A.; Wexler, H.M. Presence of quorum-sensing systems associated with multidrug resistance and biofilm formation in Bacteroides fragilis. Microb. Ecol. 2008, 56, 412–419. [Google Scholar] [CrossRef]
- Pumbwe, L.; Skilbeck, C.A.; Wexler, H.M. Impact of anatomic site on growth, efflux-pump expression, cell structure, and stress responsiveness of Bacteroides fragilis. Curr. Microbiol. 2007, 55, 362–365. [Google Scholar] [CrossRef]
- Ohland, C.L.; Macnaughton, W.K. Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G807–G819. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Theissig, F.; Engelhardt, H.; Bengmark, S.; Koch, S.; Lochs, H.; Dörffel, Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 2007, 56, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, M.; Lang, M.; Holley, H.; Crepaz, D.; Hausmann, B.; Pjevac, P.; Moser, D.; Haller, F.; Hof, F.; Beer, A.; et al. Mucosal Biofilms Are an Endoscopic Feature of Irritable Bowel Syndrome and Ulcerative Colitis. Gastroenterology 2021, 161, 1245–1256.e20. [Google Scholar] [CrossRef]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017, 3, 34. [Google Scholar] [CrossRef]
- Macfarlane, S.; Macfarlane, G.T. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Environ. Microbiol. 2006, 72, 6204–6211. [Google Scholar] [CrossRef]
- De Vos, W.M. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 2015, 1, 15005. [Google Scholar] [CrossRef]
- Ellermann, M.; Sartor, R.B. Intestinal bacterial biofilms modulate mucosal immune responses. J. Immunol. Sci. 2018, 2, 13–18. [Google Scholar]
- Motta, J.-P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef]
- Anderson, A.C.; Rothballer, M.; Altenburger, M.J.; Woelber, J.P.; Karygianni, L.; Vach, K.; Hellwig, E.; Al-Ahmad, A. Long-Term Fluctuation of Oral Biofilm Microbiota following Different Dietary Phases. Appl. Environ. Microbiol. 2020, 86, e01421-20. [Google Scholar] [CrossRef]
- Akimbekov, N.S.; Digel, I.; Sherelkhan, D.K.; Lutfor, A.B.; Razzaque, M.S. Vitamin D and the Host-Gut Microbiome: A Brief Overview. Acta Histochem. Cytochem. 2020, 53, 33–42. [Google Scholar] [CrossRef]
- Thomas, A.M.; Gleber-Netto, F.O.; Fernandes, G.R.; Amorim, M.; Barbosa, L.F.; Francisco, A.L.; de Andrade, A.G.; Setubal, J.C.; Kowalski, L.P.; Nunes, D.N.; et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 2014, 14, 250. [Google Scholar] [CrossRef]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef]
- Komiya, Y.; Shimomura, Y.; Higurashi, T.; Sugi, Y.; Arimoto, J.; Umezawa, S.; Uchiyama, S.; Matsumoto, M.; Nakajima, A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 2019, 68, 1335–1337. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Tomkovich, S.; Taylor, A.; King, J.; Colovas, J.; Bishop, L.; McBride, K.; Royzenblat, S.; Lesniak, N.A.; Bergin, I.L.; Schloss, P.D. An Osmotic Laxative Renders Mice Susceptible to Prolonged Clostridioides difficile Colonization and Hinders Clearance. mSphere 2021, 6, e0062921. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Moser, C.; Pedersen, H.T.; Lerche, C.J.; Kolpen, M.; Line, L.; Thomsen, K.; Høiby, N.; Jensen, P.Ø. Biofilms and host response—Helpful or harmful. APMIS 2017, 125, 320–338. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef]
- Tomkovich, S.; Dejea, C.M.; Winglee, K.; Drewes, J.L.; Chung, L.; Housseau, F.; Pope, J.L.; Gauthier, J.; Sun, X.; Mühlbauer, M.; et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Investig. 2019, 129, 1699–1712. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Shanahan, F.; Marchesi, J.R. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008, 8, 79. [Google Scholar] [CrossRef]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Naves, P.; Blanco, J.; Aldeguer, X.; Blanco, J.E.; Blanco, M.; Ponte, C.; Soriano, F.; Darfeuille-Michaud, A.; Garcia-Gil, L.J. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 2009, 9, 202. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Aldeguer, X.; Lopez-Siles, M.; González-Huix, F.; López-Oliu, C.; Dahbi, G.; Blanco, J.E.; Blanco, J.; Garcia-Gil, L.J.; Darfeuille-Michaud, A. Molecular diversity of Escherichia coli in the human gut: New ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Sitaraman, S.V.; Babbin, B.A.; Gerner-Smidt, P.; Ribot, E.M.; Garrett, N.; Alpern, J.A.; Akyildiz, A.; Theiss, A.L.; Nusrat, A.; et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab. Investig. 2007, 87, 1042–1054. [Google Scholar] [CrossRef]
- Chassaing, B.; Darfeuille-Michaud, A. The σE pathway is involved in biofilm formation by Crohn’s disease-associated adherent-invasive Escherichia coli. J. Bacteriol. 2013, 195, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Prudent, V.; Demarre, G.; Vazeille, E.; Wery, M.; Quenech’Du, N.; Ravet, A.; Dauverd-Girault, J.; van Dijk, E.; Bringer, M.-A.; Descrimes, M.; et al. The Crohn’s disease-related bacterial strain LF82 assembles biofilm-like communities to protect itself from phagolysosomal attack. Commun. Biol. 2021, 4, 627. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale Laura, P.; Lochs, H. Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef] [PubMed]
- Pittayanon, R.; Lau, J.T.; Yuan, Y.; Leontiadis, G.I.; Tse, F.; Surette, M.; Moayyedi, P. Gut Microbiota in Patients with Irritable Bowel Syndrome—A Systematic Review. Gastroenterology 2019, 157, 97–108. [Google Scholar] [CrossRef]
- Zhuang, X.; Tian, Z.; Li, L.; Zeng, Z.; Chen, M.; Xiong, L. Fecal Microbiota Alterations Associated with Diarrhea-Predominant Irritable Bowel Syndrome. Front. Microbiol. 2018, 9, 1600. [Google Scholar] [CrossRef]
- Carroll, I.M.; Chang, Y.-H.; Park, J.; Sartor, R.B.; Ringel, Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010, 2, 19. [Google Scholar] [CrossRef]
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009, 9, 95. [Google Scholar] [CrossRef]
- Chong, P.P.; Chin, V.K.; Looi, C.Y.; Wong, W.F.; Madhavan, P.; Yong, V.C. The Microbiome and Irritable Bowel Syndrome—A Review on the Pathophysiology, Current Research and Future Therapy. Front. Microbiol. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 521–530.e248. [Google Scholar] [CrossRef]
- Tana, C.; Umesaki, Y.; Imaoka, A.; Handa, T.; Kanazawa, M.; Fukudo, S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol. Motil. 2010, 22, 512-e115. [Google Scholar] [CrossRef]
- Macfarlane, S.; Woodmansey, E.J.; Macfarlane, G.T. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 2005, 71, 7483–7492. [Google Scholar] [CrossRef]
- Wadhwa, A.; Al Nahhas, M.F.; Dierkhising, R.A.; Patel, R.; Kashyap, P.; Pardi, D.S.; Khanna, S.; Grover, M. High risk of post-infectious irritable bowel syndrome in patients with Clostridium difficile infection. Aliment. Pharmacol. Ther. 2016, 44, 576–582. [Google Scholar] [CrossRef]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of Gut Microbiota in Patients with Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef]
- Chassard, C.; Dapoigny, M.; Scott, K.P.; Crouzet, L.; Del’homme, C.; Marquet, P.; Martin, J.C.; Pickering, G.; Ardid, D.; Eschalier, A.; et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012, 35, 828–838. [Google Scholar] [CrossRef]
- Jeffery, I.B.; Das, A.; O’Herlihy, E.; Coughlan, S.; Cisek, K.; Moore, M.; Bradley, F.; Carty, T.; Pradhan, M.; Dwibedi, C.; et al. Differences in Fecal Microbiomes and Metabolomes of People with vs without Irritable Bowel Syndrome and Bile Acid Malabsorption. Gastroenterology 2020, 158, 1016–1028.e8. [Google Scholar] [CrossRef]
- Finn, E.; Andersson, F.L.; Madin-Warburton, M. Burden of Clostridioides difficile infection (CDI)—A systematic review of the epidemiology of primary and recurrent CDI. BMC Infect. Dis. 2021, 21, 456. [Google Scholar] [CrossRef]
- Ðapa, T.; Leuzzi, R.; Ng, Y.K.; Baban, S.T.; Adamo, R.; Kuehne, S.A.; Scarselli, M.; Minton, N.P.; Serruto, D.; Unnikrishnan, M. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 2013, 195, 545–555. [Google Scholar] [CrossRef]
- James, G.A.; Chesnel, L.; Boegli, L.; deLancey Pulcini, E.; Fisher, S.; Stewart, P.S. Analysis of Clostridium difficile biofilms: Imaging and antimicrobial treatment. J. Antimicrob. Chemother. 2018, 73, 102–108. [Google Scholar] [CrossRef]
- Normington, C.; Moura, I.B.; Bryant, J.A.; Ewin, D.J.; Clark, E.V.; Kettle, M.J.; Harris, H.C.; Spittal, W.; Davis, G.; Henn, M.R.; et al. Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. NPJ Biofilms Microbiomes 2021, 7, 16. [Google Scholar] [CrossRef]
- Dubois, T.; Tremblay, Y.D.N.; Hamiot, A.; Martin-Verstraete, I.; Deschamps, J.; Monot, M.; Briandet, R.; Dupuy, B. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ Biofilms Microbiomes 2019, 5, 14. [Google Scholar] [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Auchtung, J.; Endres, B.T.; Ruan, W.; Bassères, E.; Engevik, A.C.; Wu, Q.; Nicholson, M.; Luna, R.A.; et al. Fusobacteriumnucleatum Adheres to Clostridioides difficile via the RadD Adhesin to Enhance Biofilm Formation in Intestinal Mucus. Gastroenterology 2021, 160, 1301–1314.e8. [Google Scholar] [CrossRef]
- Hengzhuang, W.; Wu, H.; Ciofu, O.; Song, Z.; Høiby, N. In Vivo Pharmacokinetics/Pharmacodynamics of Colistin and Imipenem in Pseudomonas aeruginosa Biofilm Infection. Antimicrob. Agents Chemother. 2012, 56, 2683–2690. [Google Scholar] [CrossRef]
- Herrmann, G.; Yang, L.; Wu, H.; Song, Z.; Wang, H.; Høiby, N.; Ulrich, M.; Molin, S.; Riethmüller, J.; Döring, G. Colistin-Tobramycin Combinations Are Superior to Monotherapy Concerning the Killing of Biofilm Pseudomonas aeruginosa. J. Infect. Dis. 2010, 202, 1585–1592. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Kaplan Jeffrey, B.; Izano Era, A.; Gopal, P.; Karwacki Michael, T.; Kim, S.; Bose Jeffrey, L.; Bayles Kenneth, W.; Horswill Alexander, R. Low Levels of β-Lactam Antibiotics Induce Extracellular DNA Release and Biofilm Formation in Staphylococcus aureus. mBio 2012, 3, e00198-12. [Google Scholar] [CrossRef]
- Hengzhuang, W.; Wu, H.; Ciofu, O.; Song, Z.; Høiby, N. Pharmacokinetics/Pharmacodynamics of Colistin and Imipenem on Mucoid and Nonmucoid Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2011, 55, 4469–4474. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Yamaguchi, T.; Ishii, Y.; Chono, K.; Tateda, K. Inhibitory effect of fidaxomicin on biofilm formation in Clostridioides difficile. J. Infect. Chemother. 2020, 26, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.R.; Cheng, J.K.J.; Unnikrishnan, M. Clostridioides difficile biofilms: A mechanism of persistence in the gut? PLoS Pathog. 2021, 17, e1009348. [Google Scholar] [CrossRef] [PubMed]
- Von Rosenvinge, E.C.; O’May, G.A.; Macfarlane, S.; Macfarlane, G.T.; Shirtliff, M.E. Microbial biofilms and gastrointestinal diseases. Pathog. Dis. 2013, 67, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, M.R.M.; Whitchurch, C.B.; Burrows, L.L. Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr. Opin. Microbiol. 2018, 45, 164–169. [Google Scholar] [CrossRef]
- Kafil, H.S.; Mobarez, A.M.; Moghadam, M.F.; Hashemi, Z.s.; Yousefi, M. Gentamicin induces efaA expression and biofilm formation in Enterococcus faecalis. Microb. Pathog. 2016, 92, 30–35. [Google Scholar] [CrossRef]
- Kaplan, J.B. Antibiotic-Induced Biofilm Formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef]
- Ciofu, O.; Mandsberg, L.F.; Wang, H.; Høiby, N. Phenotypes selected during chronic lung infection in cystic fibrosis patients: Implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol. Med. Microbiol. 2012, 65, 215–225. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Goldenberg, J.Z.; Yap, C.; Lytvyn, L.; Lo, C.K.; Beardsley, J.; Mertz, D.; Johnston, B.C. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst. Rev. 2017, 12, CD006095. [Google Scholar] [CrossRef]
- Goodman, C.; Keating, G.; Georgousopoulou, E.; Hespe, C.; Levett, K. Probiotics for the prevention of antibiotic-associated diarrhoea: A systematic review and meta-analysis. BMJ Open 2021, 11, e043054. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Hosseiniyan Khatibi, S.M.; Sharifi, S.; Memar, M.Y.; Zununi Vahed, S. The Battle of Probiotics and Their Derivatives Against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef]
- Ghasemian, A.; Eslami, M.; Shafiei, M.; Najafipour, S.; Rajabi, A. Probiotics and their increasing importance in human health and infection control. Rev. Res. Med. Microbiol. 2018, 29, 153–158. [Google Scholar] [CrossRef]
- Kim, Y.; Oh, S.; Kim, S.H. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Res. Commun. 2009, 379, 324–329. [Google Scholar] [CrossRef]
- Söderling, E.M.; Marttinen, A.M.; Haukioja, A.L. Probiotic Lactobacilli Interfere with Streptococcus mutans Biofilm Formation In Vitro. Curr. Microbiol. 2011, 62, 618–622. [Google Scholar] [CrossRef]
- Matsubara, V.H.; Wang, Y.; Bandara, H.M.H.N.; Mayer, M.P.A.; Samaranayake, L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016, 100, 6415–6426. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Koh, E.; Wong, A.; March, J.C.; Bentley, W.E.; Lee, Y.S.; Chang, M.W. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017, 8, 15028. [Google Scholar] [CrossRef]
- Heumann, A.; Assifaoui, A.; Da Silva Barreira, D.; Thomas, C.; Briandet, R.; Laurent, J.; Beney, L.; Lapaquette, P.; Guzzo, J.; Rieu, A. Intestinal release of biofilm-like microcolonies encased in calcium-pectinate beads increases probiotic properties of Lacticaseibacillus paracasei. NPJ Biofilms Microbiomes 2020, 6, 44. [Google Scholar] [CrossRef]
- Wallis, J.K.; Krömker, V.; Paduch, J.-H. Biofilm Challenge: Lactic Acid Bacteria Isolated from Bovine Udders versus Staphylococci. Foods 2019, 8, 79. [Google Scholar] [CrossRef]
- Tarrah, A.; da Silva Duarte, V.; de Castilhos, J.; Pakroo, S.; Lemos Junior, W.J.F.; Luchese, R.H.; Fioravante Guerra, A.; Rossi, R.C.; Righetto Ziegler, D.; Corich, V.; et al. Probiotic potential and biofilm inhibitory activity of Lactobacillus casei group strains isolated from infant feces. J. Funct. Foods 2019, 54, 489–497. [Google Scholar] [CrossRef]
- Ben Slama, R.; Kouidhi, B.; Zmantar, T.; Chaieb, K.; Bakhrouf, A. Anti-listerial and Anti-biofilm Activities of Potential Probiotic Lactobacillus Strains Isolated from Tunisian Traditional Fermented Food. J. Food Saf. 2013, 33, 8–16. [Google Scholar] [CrossRef]
- Lee, J.-E.; Lee, N.-K.; Paik, H.-D. Antimicrobial and anti-biofilm effects of probiotic Lactobacillus plantarum KU200656 isolated from kimchi. Food Sci. Biotechnol. 2021, 30, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Tazehabadi, M.H.; Algburi, A.; Popov, I.V.; Ermakov, A.M.; Chistyakov, V.A.; Prazdnova, E.V.; Weeks, R.; Chikindas, M.L. Probiotic Bacilli Inhibit Salmonella Biofilm Formation without Killing Planktonic Cells. Front. Microbiol. 2021, 12, 615328. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019, 8, 76. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songtanin, B.; Peterson, C.J.; Molehin, A.J.; Nugent, K. Biofilms and Benign Colonic Diseases. Int. J. Mol. Sci. 2022, 23, 14259. https://doi.org/10.3390/ijms232214259
Songtanin B, Peterson CJ, Molehin AJ, Nugent K. Biofilms and Benign Colonic Diseases. International Journal of Molecular Sciences. 2022; 23(22):14259. https://doi.org/10.3390/ijms232214259
Chicago/Turabian StyleSongtanin, Busara, Christopher J. Peterson, Adebayo J. Molehin, and Kenneth Nugent. 2022. "Biofilms and Benign Colonic Diseases" International Journal of Molecular Sciences 23, no. 22: 14259. https://doi.org/10.3390/ijms232214259
APA StyleSongtanin, B., Peterson, C. J., Molehin, A. J., & Nugent, K. (2022). Biofilms and Benign Colonic Diseases. International Journal of Molecular Sciences, 23(22), 14259. https://doi.org/10.3390/ijms232214259








