Current Knowledge on Bee Innate Immunity Based on Genomics and Transcriptomics
Abstract
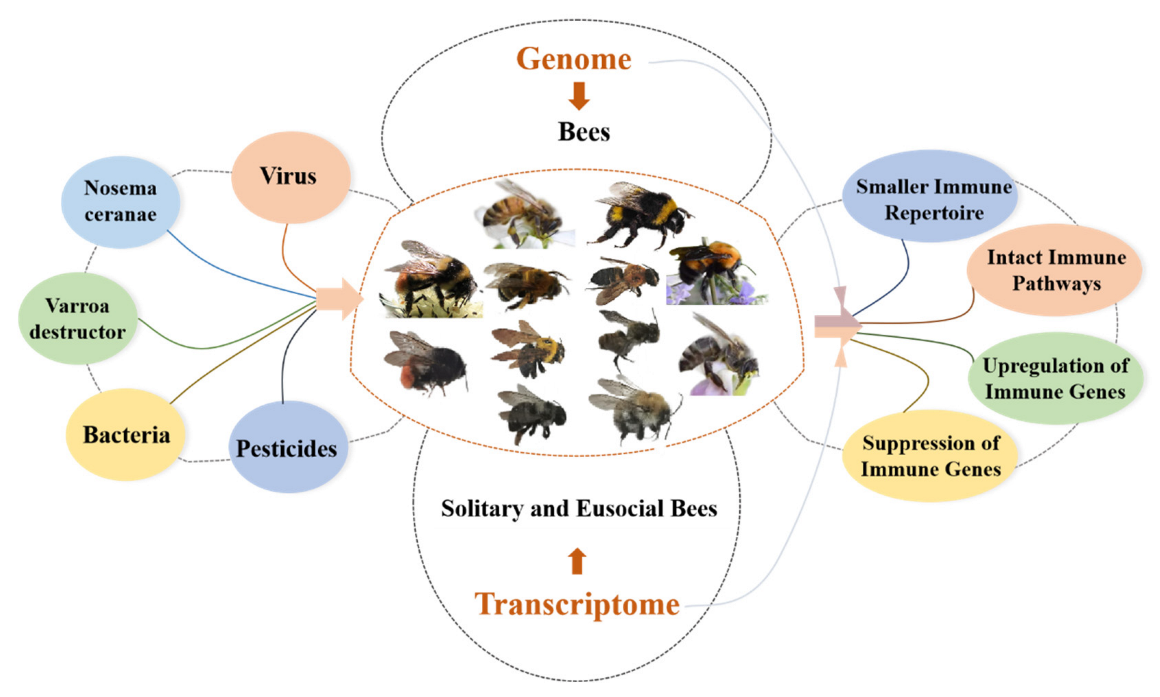
:1. Introduction
2. Genomic Perspective of Innate Bee Immunity
3. Transcriptomic Perspective of Innate Bee Immune Response
3.1. Immune Responses to Viruses
3.2. Immune Response to Parasites
3.3. Immune Response to Bacteria
3.4. Immune Suppression Due to Pesticides
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors Associated with Honey Bee Colony Losses: A Mini-Review. Vet. Sci. 2020, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Bayo, F.; Goulson, D.; Pennacchio, F.; Nazzi, F.; Goka, K.; Desneux, N. Are bee diseases linked to pesticides?—A brief review. Environ. Int. 2016, 89–90, 7–11. [Google Scholar] [CrossRef]
- Cameron, S.A.; Sadd, B.M. Global Trends in Bumble Bee Health. Annu. Rev. Entomol. 2020, 65, 209–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kluser, S.; Peduzzi, P. Global Pollinator Decline: A Literature Review; UNEP/GRIDEurope: Geneva, Switzerland, 2007; Volume 8, 10p. [Google Scholar]
- LeBuhn, G.; Vargas Luna, J. Pollinator decline: What do we know about the drivers of solitary bee declines? Curr. Opin. Insect Sci. 2021, 46, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef]
- Branstetter, M.G.; Childers, A.K.; Cox-Foster, D.; Hopper, K.R.; Kapheim, K.M.; Toth, A.L.; Worley, K.C. Genomes of the Hymenoptera. Curr. Opin. Insect Sci. 2018, 25, 65–75. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Robinson, G.E. The power and promise of applying genomics to honey bee health. Curr. Opin. Insect Sci. 2015, 10, 124–132. [Google Scholar] [CrossRef] [Green Version]
- Consortium, H.G.S. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAfee, A.; Harpur, B.A.; Michaud, S.; Beavis, R.C.; Kent, C.F.; Zayed, A.; Foster, L.J. Toward an Upgraded Honey Bee (Apis mellifera L.) Genome Annotation Using Proteogenomics. J. Proteome Res. 2016, 15, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.; Jung, J.W.; Choi, B.-S.; Jayakodi, M.; Lee, J.; Lim, J.; Yu, Y.; Choi, Y.-S.; Lee, M.-L.; Park, Y. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genom. 2015, 16, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, Q.; Sun, L.; Zheng, H.; Zeng, Z.; Wang, S.; Xu, S.; Zheng, H.; Chen, Y.; Shi, Y.; Wang, Y.; et al. Genomic and transcriptomic analysis of the Asian honeybee Apis cerana provides novel insights into honeybee biology. Sci. Rep. 2018, 8, 822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, L.; Shi, P.; Song, H.; Tang, X.; Zhou, J.; Yang, J.; Yang, M.; Xu, J. De Novo Genome Assembly of Chinese Plateau Honeybee Unravels Intraspecies Genetic Diversity in the Eastern Honeybee, Apis cerana. Insects 2021, 12, 891. [Google Scholar] [CrossRef]
- Sun, C.; Huang, J.; Wang, Y.; Zhao, X.; Su, L.; Thomas, G.W.; Zhao, M.; Zhang, X.; Jungreis, I.; Kellis, M. Genus-wide characterization of bumblebee genomes provides insights into their evolution and variation in ecological and behavioral traits. Mol. Biol. Evol. 2021, 38, 486–501. [Google Scholar] [CrossRef]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; De Graaf, D.C.; Dearden, P.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.; Hasselmann, M.; Lozier, J.D. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef] [Green Version]
- Oppenheim, S.; Cao, X.; Rueppel, O.; Krongdang, S.; Phokasem, P.; DeSalle, R.; Goodwin, S.; Xing, J.; Chantawannakul, P.; Rosenfeld, J.A. Whole genome sequencing and assembly of the Asian honey bee Apis dorsata. Genome Biol. Evol. 2020, 12, 3677–3683. [Google Scholar] [CrossRef] [Green Version]
- Cremer, S.; Armitage, S.A.O.; Schmid-Hempel, P. Social Immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef]
- Xu, J.; James, R. Genes related to immunity, as expressed in the alfalfa leafcutting bee, Megachile rotundata, during pathogen challenge. Insect Mol. Biol. 2009, 18, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Kapheim, K.M.; Pan, H.; Li, C.; Salzberg, S.L.; Puiu, D.; Magoc, T.; Robertson, H.M.; Hudson, M.E.; Venkat, A.; Fischman, B.J. Genomic signatures of evolutionary transitions from solitary to group living. Science 2015, 348, 1139–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werren, J.H.; Richards, S.; Desjardins, C.A.; Niehuis, O.; Gadau, J.; Colbourne, J.K.; Group, N.G.W.; Beukeboom, L.W.; Desplan, C.; Elsik, C.G. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 2010, 327, 343–348. [Google Scholar] [CrossRef]
- Xiao, J.-H.; Yue, Z.; Jia, L.-Y.; Yang, X.-H.; Niu, L.-H.; Wang, Z.; Zhang, P.; Sun, B.-F.; He, S.-M.; Li, Z.; et al. Obligate mutualism within a host drives the extreme specialization of a fig wasp genome. Genome Biol. 2013, 14, R141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barribeau, S.M.; Sadd, B.M.; du Plessis, L.; Brown, M.J.F.; Buechel, S.D.; Cappelle, K.; Carolan, J.C.; Christiaens, O.; Colgan, T.J.; Erler, S.; et al. A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 2015, 16, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.-S.; Luo, A.; Zhang, F.; Niu, Z.-Q.; Wu, Q.-T.; Xiong, M.; Orr, M.C.; Zhu, C.-D. The First Draft Genome of the Plasterer Bee Colletes gigas (Hymenoptera: Colletidae: Colletes). Genome Biol. Evol. 2020, 12, 860–866. [Google Scholar] [CrossRef]
- Wey, B.; Heavner, M.E.; Wittmeyer, K.T.; Briese, T.; Hopper, K.R.; Govind, S. Immune suppressive extracellular vesicle proteins of Leptopilina heterotoma are encoded in the wasp genome. G3 Genes Genomes Genet. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Heraghty, S.D.; Sutton, J.M.; Pimsler, M.L.; Fierst, J.L.; Strange, J.P.; Lozier, J.D. De Novo Genome Assemblies for Three North American Bumble Bee Species: Bombus bifarius, Bombus vancouverensis, and Bombus vosnesenskii. G3 Genes Genomes Genet. 2020, 10, 2585–2592. [Google Scholar] [CrossRef]
- Tvedte, E.S.; Walden, K.K.; McElroy, K.E.; Werren, J.H.; Forbes, A.A.; Hood, G.R.; Logsdon, J.M., Jr.; Feder, J.L.; Robertson, H.M. Genome of the parasitoid wasp Diachasma alloeum, an emerging model for ecological speciation and transitions to asexual reproduction. Genome Biol. Evol. 2019, 11, 2767–2773. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Pan, H.; Li, C.; Blatti, C., III; Harpur, B.A.; Ioannidis, P.; Jones, B.M.; Kent, C.F.; Ruzzante, L.; Sloofman, L. Draft genome assembly and population genetics of an agricultural pollinator, the solitary alkali bee (Halictidae: Nomia melanderi). G3 Genes Genomes Genet. 2019, 9, 625–634. [Google Scholar] [CrossRef]
- Yin, C.; Li, M.; Hu, J.; Lang, K.; Chen, Q.; Liu, J.; Guo, D.; He, K.; Dong, Y.; Luo, J. The genomic features of parasitism, polyembryony and immune evasion in the endoparasitic wasp Macrocentrus cingulum. BMC Genom. 2018, 19, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haddad, N.J.; Adjlane, N.; Saini, D.; Menon, A.; Krishnamurthy, V.; Jonklaas, D.; Tomkins, J.P.; Loucif-Ayad, W.; Horth, L. Whole-genome sequencing of north African honey bee Apis mellifera intermissa to assess its beneficial traits. Entomol. Res. 2018, 48, 174–186. [Google Scholar] [CrossRef]
- Geib, S.M.; Liang, G.H.; Murphy, T.D.; Sim, S.B. Whole genome sequencing of the braconid parasitoid wasp Fopius arisanus, an important biocontrol agent of pest tepritid fruit flies. G3 Genes Genomes Genet. 2017, 7, 2407–2411. [Google Scholar] [CrossRef] [Green Version]
- Brand, P.; Saleh, N.; Pan, H.; Li, C.; Kapheim, K.M.; Ramírez, S.R. The nuclear and mitochondrial genomes of the facultatively eusocial orchid bee Euglossa dilemma. G3 Genes Genomes Genet. 2017, 7, 2891–2898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Standage, D.S.; Berens, A.J.; Glastad, K.M.; Severin, A.J.; Brendel, V.P.; Toth, A.L. Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol. Ecol. 2016, 25, 1769–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehan, S.M.; Glastad, K.M.; Lawson, S.P.; Hunt, B.G. The genome and methylome of a subsocial small carpenter bee, Ceratina calcarata. Genome Biol. Evol. 2016, 8, 1401–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haddad, N.J.; Loucif-Ayad, W.; Adjlane, N.; Saini, D.; Manchiganti, R.; Krishnamurthy, V.; AlShagoor, B.; Batainh, A.M.; Mugasimangalam, R. Draft genome sequence of the Algerian bee Apis mellifera intermissa. Genom. Data 2015, 4, 24–25. [Google Scholar] [CrossRef] [Green Version]
- Kocher, S.D.; Li, C.; Yang, W.; Tan, H.; Yi, S.V.; Yang, X.; Hoekstra, H.E.; Zhang, G.; Pierce, N.E.; Yu, D.W. The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes. Genome Biol. 2013, 14, R142. [Google Scholar] [CrossRef] [Green Version]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [Green Version]
- DeGrandi-Hoffman, G.; Chen, Y. Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci. 2015, 10, 170–176. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee Viruses: Ecology, Pathogenicity, and Impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Daughenbaugh, K.F.; Parekh, F.; Pizzorno, M.C.; Flenniken, M.L. Honey Bee and Bumble Bee Antiviral Defense. Viruses 2018, 10, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhu, L.; Rao, L.; Zhao, L.; Wang, Y.; Wu, X.; Zheng, H.; Liao, X. Nano- and micro-polystyrene plastics disturb gut microbiota and intestinal immune system in honeybee. Sci. Total Environ. 2022, 842, 156819. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Gauthier, L.; Ribière, M.; Chen, Y.P. Honey bee viruses and their effect on bee and colony health. In Honey Bee Colony Health; CRC Press: Boca Raton, FL, USA, 2011; pp. 71–102. [Google Scholar] [CrossRef]
- Yañez, O.; Piot, N.; Dalmon, A.; de Miranda, J.R.; Chantawannakul, P.; Panziera, D.; Amiri, E.; Smagghe, G.; Schroeder, D.; Chejanovsky, N. Bee Viruses: Routes of Infection in Hymenoptera. Front. Microbiol. 2020, 11, 943. [Google Scholar] [CrossRef]
- Galbraith, D.A.; Yang, X.; Niño, E.L.; Yi, S.; Grozinger, C. Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees (Apis mellifera). PLoS Pathog. 2015, 11, e1004713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li-Byarlay, H.; Boncristiani, H.; Howell, G.; Herman, J.; Clark, L.; Strand, M.K.; Tarpy, D.; Rueppell, O. Transcriptomic and Epigenomic Dynamics of Honey Bees in Response to Lethal Viral Infection. Front. Genet. 2020, 11, 566320. [Google Scholar] [CrossRef]
- Doublet, V.; Paxton, R.J.; McDonnell, C.M.; Dubois, E.; Nidelet, S.; Moritz, R.F.A.; Alaux, C.; Le Conte, Y. Brain transcriptomes of honey bees (Apis mellifera) experimentally infected by two pathogens: Black queen cell virus and Nosema ceranae. Genom. Data 2016, 10, 79–82. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Fannon, J.M.; Moore, J.D.; Wood, G.R.; Evans, D.J. The Iflaviruses Sacbrood virus and Deformed wing virus evoke different transcriptional responses in the honeybee which may facilitate their horizontal or vertical transmission. PeerJ 2016, 4, e1591. [Google Scholar] [CrossRef]
- Kim, W.J.; Lee, S.-H.; Kim, J.H.; Fang, Y.; Ha, K.B.; Park, D.H.; Choi, J.Y.; Je, Y.H. Differential gene expressions of innate immune related genes of the Asian honeybee, Apis cerana, latently infected with sacbrood virus. J. Asia-Pac. Entomol. 2017, 20, 17–21. [Google Scholar] [CrossRef]
- Deng, Y.; Zhao, H.; Shen, S.; Yang, S.; Yang, D.; Deng, S.; Hou, C. Identification of Immune Response to Sacbrood Virus Infection in Apis cerana Under Natural Condition. Front. Genet. 2020, 11, 587509. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, Z.; Zhuang, M.; Wang, L.; Li, K.; Yao, J.; Yang, H.; Huang, J.; Hao, Y.; Ying, F. Transcriptome profiling reveals a novel mechanism of antiviral immunity upon sacbrood virus infection in honey bee larvae (Apis cerana). Front. Microbiol. 2021, 12, 615893. [Google Scholar] [CrossRef] [PubMed]
- Pizzorno, M.C.; Field, K.; Kobokovich, A.L.; Martin, P.L.; Gupta, R.A.; Mammone, R.; Rovnyak, D.; Capaldi, E.A. Transcriptomic responses of the honey bee brain to infection with deformed wing virus. Viruses 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- Amiri, E.; Herman, J.J.; Strand, M.K.; Tarpy, D.R.; Rueppell, O. Egg transcriptome profile responds to maternal virus infection in honey bees, Apis mellifera. Infect. Genet. Evol. 2020, 85, 104558. [Google Scholar] [CrossRef]
- Traynor, K.S.; Mondet, F.; de Miranda, J.R.; Techer, M.; Kowallik, V.; Oddie, M.A.Y.; Chantawannakul, P.; McAfee, A. Varroa destructor: A Complex Parasite, Crippling Honey Bees Worldwide. Trends Parasitol. 2020, 36, 592–606. [Google Scholar] [CrossRef]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic Parasite-Pathogen Interactions Mediated by Host Immunity Can Drive the Collapse of Honeybee Colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef] [Green Version]
- Annoscia, D.; Brown, S.P.; Di Prisco, G.; De Paoli, E.; Del Fabbro, S.; Frizzera, D.; Zanni, V.; Galbraith, D.A.; Caprio, E.; Grozinger, C.M.; et al. Haemolymph removal by Varroa mite destabilizes the dynamical interaction between immune effectors and virus in bees, as predicted by Volterra’s model. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190331. [Google Scholar] [CrossRef] [Green Version]
- Doublet, V.; Poeschl, Y.; Gogol-Döring, A.; Alaux, C.; Annoscia, D.; Aurori, C.; Barribeau, S.M.; Bedoya-Reina, O.C.; Brown, M.J.F.; Bull, J.C.; et al. Unity in defence: Honeybee workers exhibit conserved molecular responses to diverse pathogens. BMC Genom. 2017, 18, 207. [Google Scholar] [CrossRef] [Green Version]
- Noël, A.; Le Conte, Y.; Mondet, F. Varroa destructor: How does it harm Apis mellifera honey bees and what can be done about it? Emerg. Top. Life Sci. 2020, 4, 45–57. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef] [Green Version]
- Zanni, V.; Galbraith, D.A.; Annoscia, D.; Grozinger, C.M.; Nazzi, F. Transcriptional signatures of parasitization and markers of colony decline in Varroa-infested honey bees (Apis mellifera). Insect Biochem. Mol. Biol. 2017, 87, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navajas, M.; Migeon, A.; Alaux, C.; Martin-Magniette, M.L.; Robinson, G.E.; Evans, J.D.; Cros-Arteil, S.; Crauser, D.; Le Conte, Y. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genom. 2008, 9, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Conte, Y.; Alaux, C.; Martin, J.F.; Harbo, J.R.; Harris, J.W.; Dantec, C.; Séverac, D.; Cros-Arteil, S.; Navajas, M. Social immunity in honeybees (Apis mellifera): Transcriptome analysis of varroa-hygienic behaviour. Insect Mol. Biol. 2011, 20, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Alaux, C.; Dantec, C.; Parrinello, H.; Le Conte, Y. Nutrigenomics in honey bees: Digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genom. 2011, 12, 496. [Google Scholar] [CrossRef] [Green Version]
- Aufauvre, J.; Misme-Aucouturier, B.; Viguès, B.; Texier, C.; Delbac, F.; Blot, N. Transcriptome Analyses of the Honeybee Response to Nosema ceranae and Insecticides. PLoS ONE 2014, 9, e91686. [Google Scholar] [CrossRef] [PubMed]
- Badaoui, B.; Fougeroux, A.; Petit, F.; Anselmo, A.; Gorni, C.; Cucurachi, M.; Cersini, A.; Granato, A.; Cardeti, G.; Formato, G.; et al. RNA-sequence analysis of gene expression from honeybees (Apis mellifera) infected with Nosema ceranae. PLoS ONE 2017, 12, e0173438. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Chen, H.; Du, Y.; Zhou, D.; Geng, S.; Wang, H.; Wan, J.; Xiong, C.; Zheng, Y.; Guo, R. Genome-Wide Identification of Long Non-Coding RNAs and Their Regulatory Networks Involved in Apis mellifera ligustica Response to Nosema ceranae Infection. Insects 2019, 10, 245. [Google Scholar] [CrossRef] [Green Version]
- Xing, W.; Zhou, D.; Long, Q.; Sun, M.; Guo, R.; Wang, L. Immune response of eastern honeybee worker to Nosema ceranae infection revealed by transcriptomic investigation. Insects 2021, 12, 728. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, J.; Fan, X.; Long, Q.; Chen, H.; Ye, Y.; Zhang, K.; Ren, Z.; Zhang, Y.; Niu, Q.; et al. CircRNA-regulated immune response of Asian honey bee workers to microsporidian infection. bioRxiv 2022. [Google Scholar] [CrossRef]
- Riddell, C.E.; Lobaton Garces, J.D.; Adams, S.; Barribeau, S.M.; Twell, D.; Mallon, E.B. Differential gene expression and alternative splicing in insect immune specificity. BMC Genom. 2014, 15, 1031. [Google Scholar] [CrossRef] [Green Version]
- Colgan, T.J.; Carolan, J.C.; Sumner, S.; Blaxter, M.L.; Brown, M.J.F. Infection by the castrating parasitic nematode Sphaerularia bombi changes gene expression in Bombus terrestris bumblebee queens. Insect Mol. Biol. 2020, 29, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, N.; MacPhail, V.J.; Colla, S.R.; Zayed, A. Conservation genomics reveals pesticide and pathogen exposure in the declining bumble bee Bombus terricola. Mol. Ecol. 2021, 30, 4220–4230. [Google Scholar] [CrossRef] [PubMed]
- Richard, F.-J.; Holt, H.L.; Grozinger, C.M. Effects of immunostimulation on social behavior, chemical communication and genome-wide gene expression in honey bee workers (Apis mellifera). BMC Genom. 2012, 13, 558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornman, R.S.; Lopez, D.; Evans, J.D. Transcriptional response of honey bee larvae infected with the bacterial pathogen Paenibacillus larvae. PLoS ONE 2013, 8, e65424. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Wang, Y.; Zhu, S.; Zhou, H.; Gou, C.; Dong, W.; Wang, Y.; Gao, Y.; Ma, H. Transcriptional profiling reveals the molecular bases of immune regulation in Apis mellifera in response to Ascosphaera apis infection. Entomol. Res. 2019, 49, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Guo, R.; Xu, X.; Xiong, C.; Liang, Q.; Zheng, Y.; Luo, Q.; Zhang, Z.; Huang, Z.; Kumar, D.; et al. Uncovering the immune responses of Apis mellifera ligustica larval gut to Ascosphaera apis infection utilizing transcriptome sequencing. Gene 2017, 621, 40–50. [Google Scholar] [CrossRef]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef]
- Lang, H.; Duan, H.; Wang, J.; Zhang, W.; Guo, J.; Zhang, X.; Hu, X.; Zheng, H. Specific Strains of Honeybee Gut Lactobacillus Stimulate Host Immune System to Protect against Pathogenic Hafnia alvei. Microbiol. Spectr. 2022, 10, e01896-21. [Google Scholar] [CrossRef]
- Wang, K.; Li, J.; Zhao, L.; Mu, X.; Wang, C.; Wang, M.; Xue, X.; Qi, S.; Wu, L. Gut microbiota protects honey bees (Apis mellifera L.) against polystyrene microplastics exposure risks. J. Hazard. Mater. 2021, 402, 123828. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, X.; Zhao, H.; Yang, S.; Gao, J.; Wu, Y.; Diao, Q.; Hou, C. Microplastic Polystyrene Ingestion Promotes the Susceptibility of Honeybee to Viral Infection. Environ. Sci. Technol. 2021, 55, 11680–11692. [Google Scholar] [CrossRef] [PubMed]
- Whiting, A.R. The Biology of the Parasitic Wasp Mormoniella vitripennis [=Nasonia brevicornis] (Walker). Q. Rev. Biol. 1967, 42, 333–406. [Google Scholar] [CrossRef]
- Sackton, T.B.; Werren, J.H.; Clark, A.G. Characterizing the Infection-Induced Transcriptome of Nasonia vitripennis Reveals a Preponderance of Taxonomically-Restricted Immune Genes. PLoS ONE 2013, 8, e83984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.-C.; Chang, Y.-W.; Lu, K.-H.; Yang, E.-C. Gene expression changes in honey bees induced by sublethal imidacloprid exposure during the larval stage. Insect Biochem. Mol. Biol. 2017, 88, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; He, J.; Zhao, X.; Chaimanee, V.; Huang, W.F.; Nie, H.; Zhao, Y.; Su, S. Differential physiological effects of neonicotinoid insecticides on honey bees: A comparison between Apis mellifera and Apis cerana. Pestic. Biochem. Physiol. 2017, 140, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bebane, P.S.A.; Hunt, B.J.; Pegoraro, M.; Jones, A.R.C.; Marshall, H.; Rosato, E.; Mallon, E.B. The effects of the neonicotinoid imidacloprid on gene expression and DNA methylation in the buff-tailed bumblebee Bombus terrestris. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, Z.; Huang, Q.; Zeng, Z.J. The effects of sublethal doses of imidacloprid and deltamethrin on honeybee foraging time and the brain transcriptome. J. Appl. Entomol. 2022, 146, 1169–1177. [Google Scholar] [CrossRef]
- Christen, V.; Schirrmann, M.; Frey, J.E.; Fent, K. Global Transcriptomic Effects of Environmentally Relevant Concentrations of the Neonicotinoids Clothianidin, Imidacloprid, and Thiamethoxam in the Brain of Honey Bees (Apis mellifera). Environ. Sci. Technol. 2018, 52, 7534–7544. [Google Scholar] [CrossRef]
- Mobley, M.W.; Gegear, R.J. One size does not fit all: Caste and sex differences in the response of bumblebees (Bombus impatiens) to chronic oral neonicotinoid exposure. PLoS ONE 2018, 13, e0200041. [Google Scholar] [CrossRef]
- Morfin, N.; Goodwin, P.H.; Guzman-Novoa, E. Interaction of Varroa destructor and Sublethal Clothianidin Doses during the Larval Stage on Subsequent Adult Honey Bee (Apis mellifera L.) Health, Cellular Immunity, Deformed Wing Virus Levels and Differential Gene Expression. Microorganisms 2020, 8, 858. [Google Scholar] [CrossRef]
- Shi, T.F.; Wang, Y.F.; Liu, F.; Qi, L.; Yu, L.S. Sublethal Effects of the Neonicotinoid Insecticide Thiamethoxam on the Transcriptome of the Honey Bees (Hymenoptera: Apidae). J. Econ. Entomol. 2017, 110, 2283–2289. [Google Scholar] [CrossRef] [PubMed]
- Fent, K.; Schmid, M.; Hettich, T.; Schmid, S. The neonicotinoid thiacloprid causes transcriptional alteration of genes associated with mitochondria at environmental concentrations in honey bees. Environ. Pollut. 2020, 266, 115297. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Dong, J.; Guo, H.; Xiao, M.; Wang, D. Identification of long noncoding RNAs reveals the effects of dinotefuran on the brain in Apis mellifera (Hymenopptera: Apidae). BMC Genom. 2021, 22, 502. [Google Scholar] [CrossRef] [PubMed]
- Christen, V.; Fent, K. Exposure of honey bees (Apis mellifera) to different classes of insecticides exhibit distinct molecular effect patterns at concentrations that mimic environmental contamination. Environ. Pollut. 2017, 226, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ. Pollut. 2020, 258, 113671. [Google Scholar] [CrossRef] [PubMed]
- Kablau, A.; Eckert, J.H.; Pistorius, J.; Sharbati, S.; Einspanier, R. Effects of selected insecticidal substances on mRNA transcriptome in larvae of Apis mellifera. Pestic. Biochem. Physiol. 2020, 170, 104703. [Google Scholar] [CrossRef]
- Korb, J.; Meusemann, K.; Aumer, D.; Bernadou, A.; Elsner, D.; Feldmeyer, B.; Foitzik, S.; Heinze, J.; Libbrecht, R.; Lin, S.; et al. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20190728. [Google Scholar] [CrossRef]
- Dai, J.; Shu, R.; Liu, J.; Xia, J.; Jiang, X.; Zhao, P. Transcriptome analysis of Apis mellifera under benomyl stress to discriminate the gene expression in response to development and immune systems. J. Environ. Sci. Health Part B 2021, 56, 594–605. [Google Scholar] [CrossRef]
- Boncristiani, H.; Underwood, R.; Schwarz, R.; Evans, J.D.; Pettis, J.; vanEngelsdorp, D. Direct effect of acaricides on pathogen loads and gene expression levels in honey bees Apis mellifera. J. Insect Physiol. 2012, 58, 613–620. [Google Scholar] [CrossRef]
- Vannette, R.L.; Mohamed, A.; Johnson, B.R. Forager bees (Apis mellifera) highly express immune and detoxification genes in tissues associated with nectar processing. Sci. Rep. 2015, 5, 16224. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Liu, Y. Current Knowledge on Bee Innate Immunity Based on Genomics and Transcriptomics. Int. J. Mol. Sci. 2022, 23, 14278. https://doi.org/10.3390/ijms232214278
Zhao X, Liu Y. Current Knowledge on Bee Innate Immunity Based on Genomics and Transcriptomics. International Journal of Molecular Sciences. 2022; 23(22):14278. https://doi.org/10.3390/ijms232214278
Chicago/Turabian StyleZhao, Xiaomeng, and Yanjie Liu. 2022. "Current Knowledge on Bee Innate Immunity Based on Genomics and Transcriptomics" International Journal of Molecular Sciences 23, no. 22: 14278. https://doi.org/10.3390/ijms232214278
APA StyleZhao, X., & Liu, Y. (2022). Current Knowledge on Bee Innate Immunity Based on Genomics and Transcriptomics. International Journal of Molecular Sciences, 23(22), 14278. https://doi.org/10.3390/ijms232214278




