Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora
Abstract
:1. Introduction
2. Results
2.1. Identification and Classification of MYBs in C. camphora
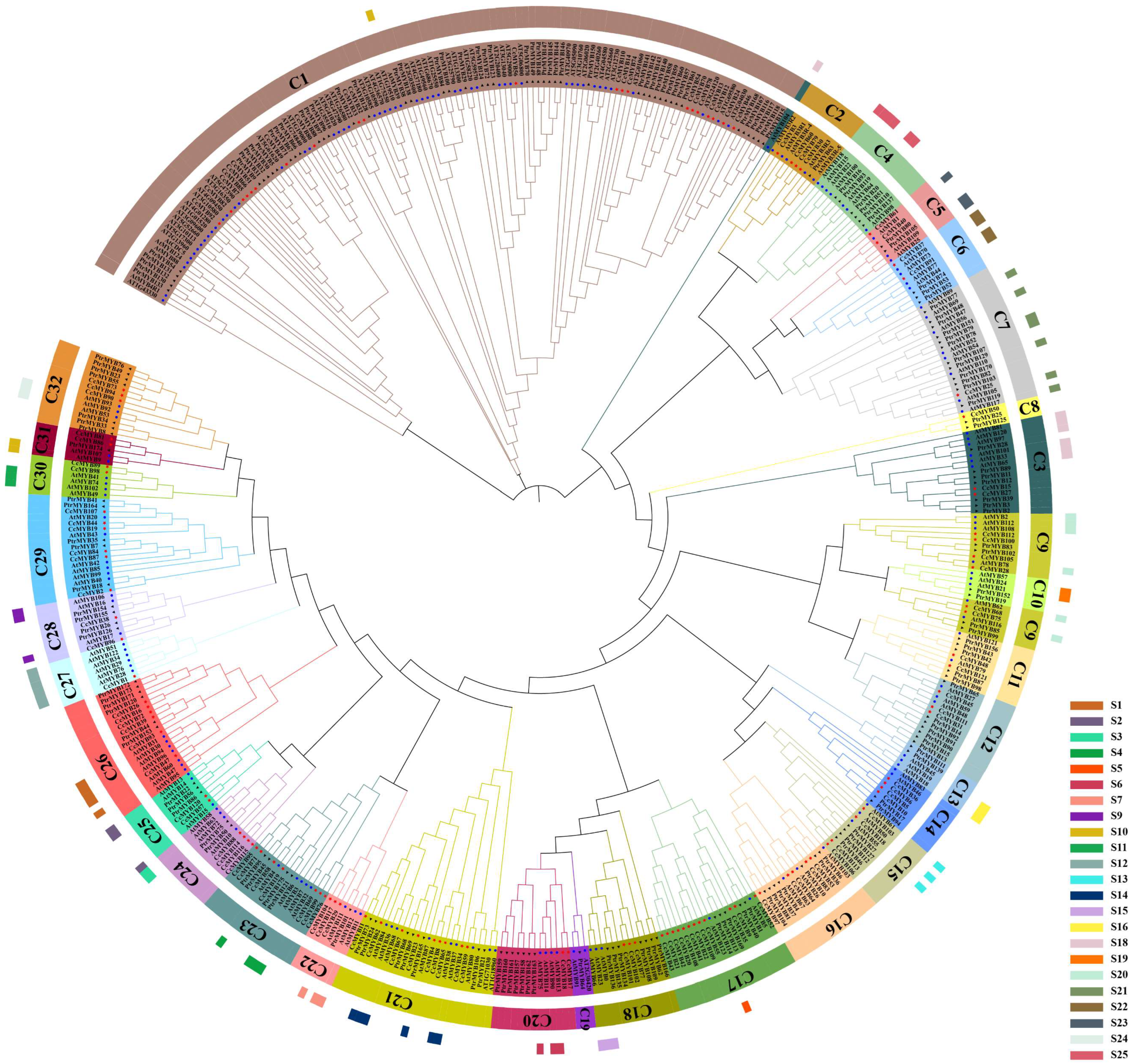
2.2. Conserved Domain and Phylogeny
2.3. Exon–Intron Structure and Cis-Elements of CcMYB Genes
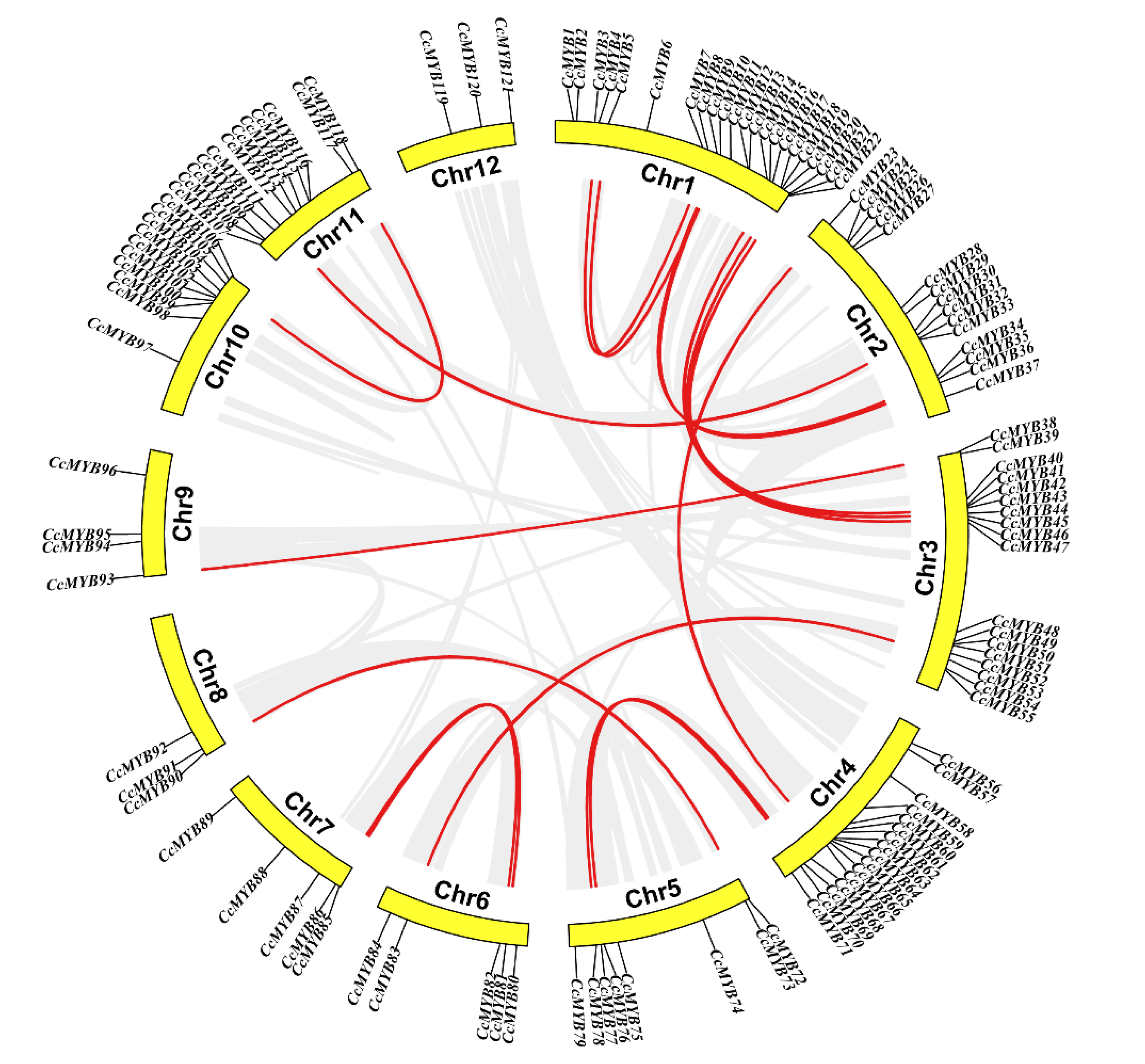
2.4. Chromosome Distribution and Collinearity of CcMYB Genes
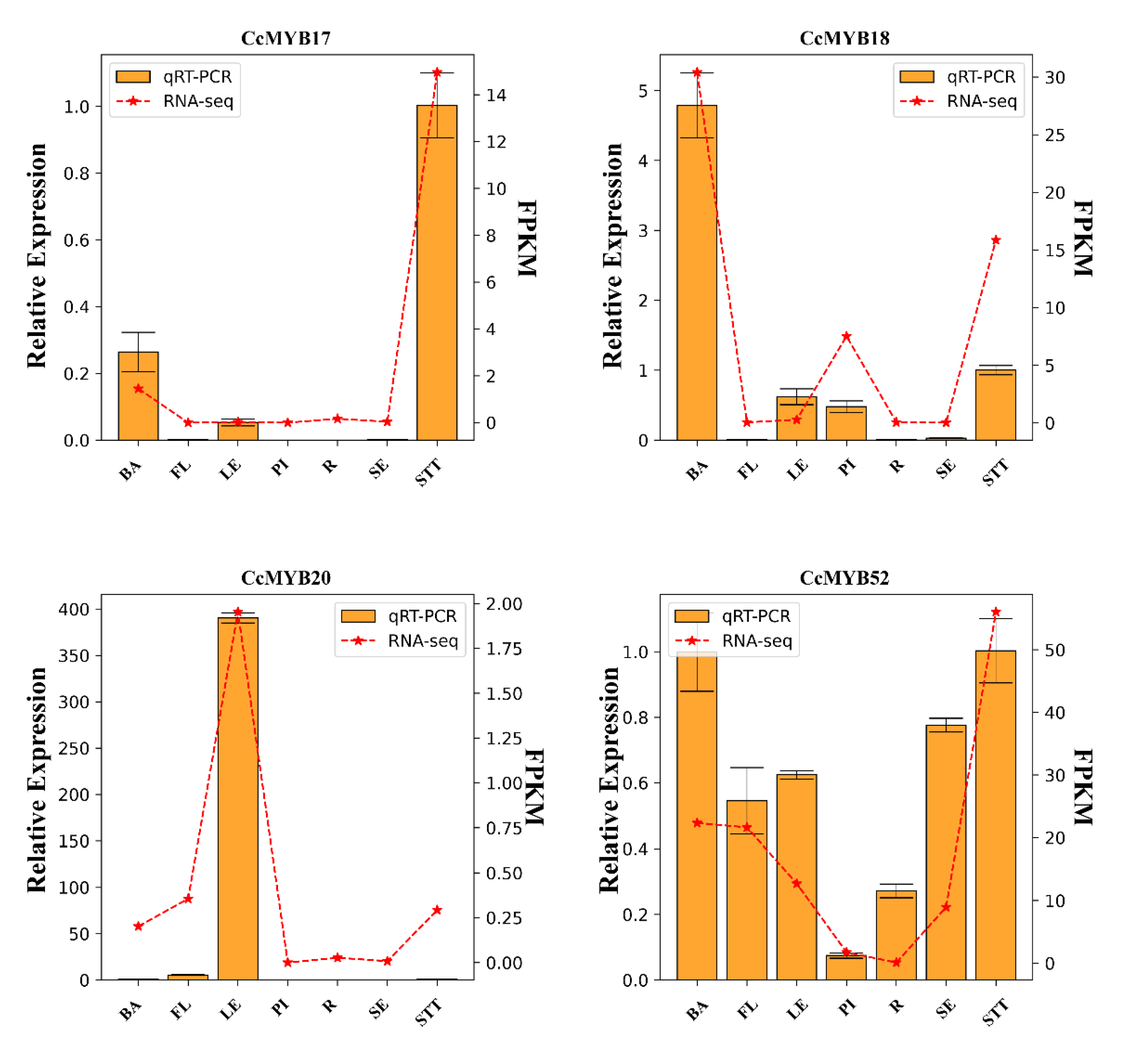
2.5. Tissue-Specific Expression Profiles
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Bioinformatics Analysis of MYB Transcription Factor Genes in C. comphora
4.3. Gene Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Rosinski, J.A.; Atchley, W.R. Molecular Evolution of the Myb Family of Transcription Factors: Evidence for Polyphyletic Origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Kuno, N.; Møller, S.G.; Shinomura, T.; Xu, X.; Chua, N.H.; Furuya, M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 2003, 15, 2476–2488. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene Expression and Yeast Two-Hybrid Studies of 1R-MYB Transcription Factor Mediating Drought Stress Response in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2015, 6, 1117. [Google Scholar] [CrossRef] [Green Version]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Liu, T.; Nan, W.; Diddugodage, C.J.; Niu, Y.; Li, C.; Wang, Y.; Shi, X.; Wang, C.; Wang, J.; et al. Two transcription factors TaPpm1 and TaPpb1 co-regulate anthocyanin biosynthesis in purple pericarps of wheat. J. Exp. Bot. 2018, 69, 2555–2567. [Google Scholar] [CrossRef] [Green Version]
- Marie, A.M.; Komivi, D.; Linhai, W.; Rong, Z.; Yanyan, W.; Ndiaga, C.; Mame, O.S.; Zhang, X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes 2017, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Peter, S.M.; Konrad, W.; Birthe, B.K.; Meike, B. Specificity of MYB interactions relies on motifs in ordered and disordered contexts. Nucleic Acids Res. 2019, 47, 9592–9608. [Google Scholar] [CrossRef] [Green Version]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis : Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Thiedig, K.; Weisshaar, B.; Stracke, R. Functional and evolutionary analysis of the Arabidopsis 4R-MYB protein SNAPc4 as part of the SNAP complex. Plant Physiol. 2021, 185, 1002–1020. [Google Scholar] [CrossRef]
- Wong, M.W.; Henry, R.W.; Ma, B.; Kobayashi, R.; Klages, N.; Matthias, P.; Strubin, M.; Hernandez, N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol. Cell. Biol. 1998, 18, 368–377. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Harding, G.A.; Parise, J.; McNamara-Schroeder, K.J.; Stumph, W.E. Architectural arrangement of cloned proximal sequence element-binding protein subunits on Drosophila U1 and U6 snRNA gene promoters. Mol. Cell. Biol. 2004, 24, 1897–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Ito, M.; Araki, S.; Matsunaga, S.; Itoh, T.; Nishihama, R.; Machida, Y.; Doonan, J.H.; Watanabe, A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 2001, 13, 1891–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raffaele, S.; Rivas, S. Regulate and be regulated: Integration of defense and other signals by the AtMYB30 transcription factor. Front. Plant Sci. 2013, 4, 98. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.-W.; Lee, H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.-Y.; Lee, Y.N.; Kim, S.-G.; Lee, Y.-H.; Park, W.J.; Park, C.-M. The MYB96 Transcription Factor Mediates Abscisic Acid Signaling during Drought Stress Response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Zhang, D.; Liu, C.; Shen, W.; He, J.; Yue, Q.; Niu, C.; Yang, F.; Li, X.; Shen, X.; et al. MdGH3.6 is targeted by MdMYB94 and plays a negative role in apple water-deficit stress tolerance. Plant J. 2022, 109, 1271–1289. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Marsafari, M.; Zha, J.; Koffas, M. Microbial Coculture for Flavonoid Synthesis. Trends Biotechnol. 2020, 38, 686–688. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.E.; Teeri, T.H. Activation of Anthocyanin Biosynthesis in Gerbera hybrida (Asteraceae) Suggests Conserved Protein-Protein and Protein-Promoter Interactions between the Anciently Diverged Monocots and Eudicots. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laitinen, R.A.E.; Ainasoja, M.; Broholm, S.K.; Teeri, T.H.; Elomaa, P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 2008, 59, 3691–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakatsuka, T.; Haruta, K.S.; Pitaksutheepong, C.; Abe, Y.; Kakizaki, Y.; Yamamoto, K.; Shimada, N.; Yamamura, S.; Nishihara, M. Identification and Characterization of R2R3-MYB and bHLH Transcription Factors Regulating Anthocyanin Biosynthesis in Gentian Flowers. Plant Cell Physiol. 2008, 49, 1818–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, C.-Y.; Yeh, K.-W. Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 2008, 66, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Higginson, T.; Li, S.F.; Parish, R.W. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003, 35, 177–192. [Google Scholar] [CrossRef]
- Tominaga, R.; Iwata, M.; Okada, K.; Wada, T. Functional Analysis of the Epidermal-Specific MYB Genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 2007, 19, 2264–2277. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xu, H.; Jiang, S.; Zhang, Z.; Lu, N.; Qiu, H.; Qu, C.; Wang, Y.; Wu, S.; Chen, X. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The Grapevine R2R3-MYB Transcription Factor VvMYBF1 Regulates Flavonol Synthesis in Developing Grape Berries. Plant Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gao, L.; Shan, Y.; Liu, Y.; Tian, Y.; Xia, T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 141, 7–16. [Google Scholar] [CrossRef]
- Yang, X.; Guo, T.; Li, J.; Chen, Z.; Guo, B.; An, X. Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int. J. Biol. Macromol. 2021, 191, 359–376. [Google Scholar] [CrossRef]
- MacMillan, C.P.; Birke, H.; Chuah, A.; Brill, E.; Tsuji, Y.; Ralph, J.; Dennis, E.S.; Llewellyn, D.; Pettolino, F.A. Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls. BMC Genom. 2017, 18, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, T.F.; Qi, H.R.; Luan, X.Y.; Xu, W.L.; Yu, F.X.; Zhong, Y.D.; Xu, M. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 2022, 20, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, C.; Zhang, R.; Ye, S.; Zhao, Z.; Huang, Y.; Xu, X.; Lan, W.; Yang, D. Metabolomics analysis to evaluate the antibacterial activity of the essential oil from the leaves of Cinnamomum camphora (Linn.). Presl. J. Ethnopharmacol. 2020, 253, 112652. [Google Scholar] [CrossRef]
- Singh, R.; Jawaid, T. Cinnamomum camphora (Kapur): Review. Pharmacogn. J. 2012, 4, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yan, W. Conservation and applications of camphor tree (Cinnamomum camphora) in China: Ethnobotany and genetic resources. Genet. Resour. Crop Evol. 2016, 63, 1049–1061. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, J.; Zhang, B.; Jin, X.; Zhang, H.; Jin, Z. Transcriptional Analysis of Metabolic Pathways and Regulatory Mechanisms of Essential Oil Biosynthesis in the Leaves of Cinnamomum camphora (L.). Presl. Front. Genet. 2020, 11, 598714. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.-A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Xie, C.; Huang, Y.; An, W.; Liu, S.; Huang, S.; Zheng, X. Metabolism and transcriptome profiling provides insight into the genes and transcription factors involved in monoterpene biosynthesis of borneol chemotype of Cinnamomum camphora induced by mechanical damage. PeerJ 2021, 9, e11465. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xie, C.; Zhan, T.; Li, L.; Liu, S.; Huang, Y.; An, W.; Zheng, X.; Huang, S. Genome-Wide Identification and Functional Characterization of the Trans-Isopentenyl Diphosphate Synthases Gene Family in Cinnamomum camphora. Front. Plant Sci. 2021, 12, 708697. [Google Scholar] [CrossRef] [PubMed]
- Ralf, S.; Martin, W.; Bernd, W. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and Diversification of the Populus R2R3-MYB Family of Transcription Factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Chen, Y.; Wu, H.; Yin, T. Genome-Wide Comparative Analysis of R2R3 MYB Gene Family in Populus and Salix and Identification of Male Flower Bud Development-Related Genes. Front. Plant Sci. 2021, 12, 721558. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Jones, D.C.; Li, W.; Xie, F.; Ma, J.; Sun, R.; Wang, Q.; Zhu, S.; Zhang, B. Genome-Wide Identification of R2R3-MYB Genes and Expression Analyses During Abiotic Stress in Gossypium raimondii. Sci. Rep. 2016, 6, 22980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C.; et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.K.; Rao, G.Y. Insights into the Diversification and Evolution of R2R3-MYB Transcription Factors in Plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Achaz, G.; Coissac, E.; Viari, A.; Netter, P. Analysis of Intrachromosomal Duplications in Yeast Saccharomyces cerevisiae: A Possible Model for Their Origin. Mol. Biol. Evol. 2000, 17, 1268–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.D.; Xu, C.Y.; Zheng, Y.J.; Wu, Y.F.; Zhang, Y.T.; Zhang, T.; Xiong, Z.Y.; Yang, H.K.; Li, J.; Fu, C.; et al. Chromosome-level genome assembly and resequencing of camphor tree (Cinnamomum camphora) provides insight into phylogeny and diversification of terpenoid and triglyceride biosynthesis of Cinnamomum. Hortic. Res. 2022, uhac216. [Google Scholar] [CrossRef]
- Li, Y.; Lin, W.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Song, X.H.; Yang, Q.S.; Liu, Y.; Li, J.J.; Chang, X.C.; Xian, L.H.; Zhang, J. Genome-wide identification of Pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis. Int. J. Biol. Macromol. 2021, 192, 16–27. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the Poplar R2R3-MYB Gene Family and Over-Expression of PsnMYB108 Confers Salt Tolerance in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef] [PubMed]
- Klempnauer, K.H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.G.; Lam-Son, P.T.; Lu, K.; Huang, Y.B.; Jia-Na, L. The Evolutionary History of R2R3-MYB Proteins Across 50 Eukaryotes: New Insights Into Subfamily Classification and Expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Segarra, G.; Van der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 Type MYB Transcription Factor Is Involved in the Cold Regulation of CBF Genes and in Acquired Freezing Tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mekapogu, M.; Vasamsetti, B.M.; Kwon, O.K.; Ahn, M.S.; Lim, S.H.; Jung, J.A. Anthocyanins in Floral Colors: Biosynthesis and Regulation in Chrysanthemum Flowers. Int. J. Mol. Sci. 2020, 21, 6537. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 2020, 145, 341–354. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, Z.; Li, Y.; Kang, Y.; Song, X.; Wang, J.; Zhou, Y. Genome-Wide Identification and Expression Analysis of MYB Transcription Factor Superfamily in Dendrobium catenatum. Front Genet. 2021, 12, 714696. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, C.; Gong, X.; Luan, X.; Wu, Z.; Li, H.; Liu, Q.; Xu, M.; Yu, F. Transcriptome and metabolome analyses reveal a key role of the anthocyanin biosynthetic pathway cascade in the pigmentation of a Cinnamomum camphora red bark mutant (‘Gantong 1’). Ind. Crops Prod. 2022, 175, 114236. [Google Scholar] [CrossRef]
- Chen, L.; Sun, P.; Wang, T.; Chen, K.; Jia, Q.; Wang, H.; Li, Y. Diverse Mechanisms of Antidiabetic Effects of the Different Procyanidin Oligomer Types of Two Different Cinnamon Species on db/db Mice. J. Agric. Food Chem. 2012, 60, 9144–9150. [Google Scholar] [CrossRef] [PubMed]
- Brownfield, L.; Hafidh, S.; Borg, M.; Sidorova, A.; Mori, T.; Twell, D. A Plant Germline-Specific Integrator of Sperm Specification and Cell Cycle Progression. PLoS Genet. 2009, 5, e1000430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx., C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [PubMed]






| Gene ID | MYB Name | No. of Domains | AT Hit | Gene ID | MYB Name | No. of Domains | AT Hit |
|---|---|---|---|---|---|---|---|
| Ccam01T000258.1 | CcMYB1 | 2 | AtMYB29 | Ccam04T001684.1 | CcMYB62 | 2 | AtMYB84 |
| Ccam01T000296.1 | CcMYB2 | 2 | AtMYB40 | Ccam04T001846.1 | CcMYB63 | 2 | AtMYB26 |
| Ccam01T000688.1 | CcMYB3 | 3 | AtMYB3R1 | Ccam04T001846.2 | CcMYB64 | 2 | AtMYB26 |
| Ccam01T000754.1 | CcMYB4 | 2 | AtMYB38 | Ccam04T001876.1 | CcMYB65 | 1 | AT2G36890 |
| Ccam01T000934.1 | CcMYB5 | 2 | AtMYB46 | Ccam04T001898.1 | CcMYB66 | 1 | AT5G47390 |
| Ccam01T001469.1 | CcMYB6 | 2 | AtMYB46 | Ccam04T001984.1 | CcMYB67 | 2 | AtMYB36 |
| Ccam01T001922.1 | CcMYB7 | 1 | AT5G47390 | Ccam04T002420.1 | CcMYB68 | 2 | AtMYB116 |
| Ccam01T001979.1 | CcMYB8 | 2 | AtMYB38 | Ccam04T002616.1 | CcMYB69 | 2 | AtMYB4 |
| Ccam01T002141.1 | CcMYB9 | 2 | AtMYB5 | Ccam04T002681.1 | CcMYB70 | part | AT3G24120 |
| Ccam01T002197.1 | CcMYB10 | 2 | AtMYB46 | Ccam04T002944.1 | CcMYB71 | 2 | AtMYB4 |
| Ccam01T002391.1 | CcMYB11 | 2 | AtMYB84 | Ccam05T000046.1 | CcMYB72 | 2 | AtMYB94 |
| Ccam01T002796.1 | CcMYB12 | part | AT3G24120 | Ccam05T000101.1 | CcMYB73 | 2 | AtMYB93 |
| Ccam01T002796.2 | CcMYB13 | part | AT3G24120 | Ccam05T000964.2 | CcMYB74 | part | AT2G01060 |
| Ccam01T003262.1 | CcMYB14 | part | AT2G01060 | Ccam05T001874.1 | CcMYB75 | 2 | AtMYB116 |
| Ccam01T003298.1 | CcMYB15 | 2 | AtMYB33 | Ccam05T002107.1 | CcMYB76 | 2 | AtMYB4 |
| Ccam01T003334.1 | CcMYB16 | 2 | AtMYB60 | Ccam05T002146.1 | CcMYB77 | 2 | AtMYB66 |
| Ccam01T003511.1 | CcMYB17 | 2 | AtMYB113 | Ccam05T002218.1 | CcMYB78 | part | AT3G24120 |
| Ccam01T003512.1 | CcMYB18 | 2 | AtMYB113 | Ccam05T002668.1 | CcMYB79 | 3 | AtMYB3R5 |
| Ccam01T003530.1 | CcMYB19 | 2 | AtMYB43 | Ccam06T000233.1 | CcMYB80 | 1 | AT5G61620 |
| Ccam01T003681.1 | CcMYB20 | 2 | AtMYB123 | Ccam06T000471.1 | CcMYB81 | 2 | AtMYB9 |
| Ccam01T003682.1 | CcMYB21 | 2 | AtMYB123 | Ccam06T000579.1 | CcMYB82 | 1 | AT5G47390 |
| Ccam01T003684.1 | CcMYB22 | 2 | AtMYB123 | Ccam06T001498.1 | CcMYB83 | 2 | AtMYB103 |
| Ccam02T000146.1 | CcMYB23 | 2 | AtMYB36 | Ccam06T001794.1 | CcMYB84 | 2 | AtMYB42 |
| Ccam02T000623.1 | CcMYB24 | 0 | AT3G04030 | Ccam07T000059.1 | CcMYB85 | 1 | AT5G47390 |
| Ccam02T000634.1 | CcMYB25 | 2 | AtMYB105 | Ccam07T000142.1 | CcMYB86 | 2 | AtMYB9 |
| Ccam02T000715.1 | CcMYB26 | 2 | AtMYB60 | Ccam07T000603.1 | CcMYB87 | 2 | AtMYB42 |
| Ccam02T000749.1 | CcMYB27 | 2 | AtMYB65 | Ccam07T001311.1 | CcMYB88 | 2 | AtMYB58 |
| Ccam02T001376.1 | CcMYB28 | 2 | AtMYB78 | Ccam07T002058.1 | CcMYB89 | 2 | AtMYB102 |
| Ccam02T001462.2 | CcMYB29 | 2 | AtMYB111 | Ccam08T000090.1 | CcMYB90 | 5 | AtMYB93 |
| Ccam02T001819.1 | CcMYB30 | 3 | AtMYB3R5 | Ccam08T000196.1 | CcMYB91 | 2 | AtMYB73 |
| Ccam02T001906.1 | CcMYB31 | 2 | AtMYB59 | Ccam08T000630.1 | CcMYB92 | 2 | AT3G10580 |
| Ccam02T001944.1 | CcMYB32 | 1 | AT1G15720 | Ccam09T000028.1 | CcMYB93 | 2 | AtMYB30 |
| Ccam02T001944.2 | CcMYB33 | 1 | AT1G15720 | Ccam09T000765.1 | CcMYB94 | 2 | AtMYB93 |
| Ccam02T002770.1 | CcMYB34 | 2 | AtMYB35 | Ccam09T000873.1 | CcMYB95 | 2 | AtMYB4 |
| Ccam02T002809.1 | CcMYB35 | 2 | AtMYB5 | Ccam09T001430.1 | CcMYB96 | 2 | AtMYB17 |
| Ccam02T002880.1 | CcMYB36 | 2 | AtMYB46 | Ccam10T000428.1 | CcMYB97 | 2 | AtMYB67 |
| Ccam02T003202.1 | CcMYB37 | 2 | AtMYB44 | Ccam10T000876.1 | CcMYB98 | 2 | AtMYB102 |
| Ccam03T000003.1 | CcMYB38 | 2 | AtMYB106 | Ccam10T001218.1 | CcMYB99 | 2 | AtMYB4 |
| Ccam03T000033.1 | CcMYB39 | 2 | AtMYB94 | Ccam10T001249.1 | CcMYB100 | 2 | AtMYB21 |
| Ccam03T001088.1 | CcMYB40 | 2 | AtMYB109 | Ccam10T001299.1 | CcMYB101 | 2 | AtMYB23 |
| Ccam03T001159.1 | CcMYB41 | 2 | AtMYB5 | Ccam10T001299.2 | CcMYB102 | 2 | AtMYB23 |
| Ccam03T001160.1 | CcMYB42 | 2 | AtMYB123 | Ccam10T001421.1 | CcMYB103 | 2 | AtMYB55 |
| Ccam03T001161.1 | CcMYB43 | 2 | AtMYB123 | Ccam10T001628.1 | CcMYB104 | 2 | AtMYB111 |
| Ccam03T001249.1 | CcMYB44 | 2 | AtMYB20 | Ccam10T001729.1 | CcMYB105 | 2 | AtMYB78 |
| Ccam03T001287.1 | CcMYB45 | 2 | AtMYB59 | Ccam10T001828.1 | CcMYB106 | 2 | AtMYB61 |
| Ccam03T001320.1 | CcMYB46 | part | AT3G04030 | Ccam11T000037.1 | CcMYB107 | 2 | AtMYB43 |
| Ccam03T001354.1 | CcMYB47 | 2 | AtMYB60 | Ccam11T000138.1 | CcMYB108 | 2 | AtMYB123 |
| Ccam03T002316.1 | CcMYB48 | 2 | AtMYB71 | Ccam11T000141.1 | CcMYB109 | 2 | AtMYB5 |
| Ccam03T002477.1 | CcMYB49 | 1 | AT5G47390 | Ccam11T000460.1 | CcMYB110 | part | AT2G01060 |
| Ccam03T002495.1 | CcMYB50 | 2 | AtMYB33 | Ccam11T000608.1 | CcMYB111 | 2 | AtMYB59 |
| Ccam03T002497.1 | CcMYB51 | 1 | AT5G47390 | Ccam11T000931.1 | CcMYB112 | 2 | AtMYB78 |
| Ccam03T002631.1 | CcMYB52 | 2 | AtMYB5 | Ccam11T001000.1 | CcMYB113 | 2 | AtMYB61 |
| Ccam03T002754.1 | CcMYB53 | 2 | AtMYB103 | Ccam11T001081.1 | CcMYB114 | 2 | AtMYB15 |
| Ccam03T002940.1 | CcMYB54 | part | AT3G24120 | Ccam11T001150.1 | CcMYB115 | 2 | AtMYB15 |
| Ccam03T002949.1 | CcMYB55 | 2 | AtMYB123 | Ccam11T001151.1 | CcMYB116 | 2 | AtMYB15 |
| Ccam04T000335.1 | CcMYB56 | part | AT5G29000 | Ccam11T001744.1 | CcMYB117 | 2 | AtMYB12 |
| Ccam04T000453.1 | CcMYB57 | 2 | AtMYB15 | Ccam11T001870.1 | CcMYB118 | 2 | AtMYB61 |
| Ccam04T000811.1 | CcMYB58 | 2 | AtMYB4 | Ccam12T000355.1 | CcMYB119 | 2 | AtMYB4 |
| Ccam04T001106.1 | CcMYB59 | 2 | AtMYB80 | Ccam12T000791.1 | CcMYB120 | 2 | AtMYB15 |
| Ccam04T001450.1 | CcMYB60 | 3 | AtMYB3R4 | Ccam12T001449.1 | CcMYB121 | 2 | AtMYB71 |
| Ccam04T001597.1 | CcMYB61 | 2 | AtMYB109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, X.; Xu, W.; Zhang, J.; Shen, T.; Chen, C.; Xi, M.; Zhong, Y.; Xu, M. Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora. Int. J. Mol. Sci. 2022, 23, 14279. https://doi.org/10.3390/ijms232214279
Luan X, Xu W, Zhang J, Shen T, Chen C, Xi M, Zhong Y, Xu M. Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora. International Journal of Molecular Sciences. 2022; 23(22):14279. https://doi.org/10.3390/ijms232214279
Chicago/Turabian StyleLuan, Xiaoyue, Wenlin Xu, Jiaqi Zhang, Tengfei Shen, Caihui Chen, Mengli Xi, Yongda Zhong, and Meng Xu. 2022. "Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora" International Journal of Molecular Sciences 23, no. 22: 14279. https://doi.org/10.3390/ijms232214279





