Integrating Network Pharmacology and Transcriptomic Strategies to Explore the Pharmacological Mechanism of Hydroxysafflor Yellow A in Delaying Liver Aging
Abstract
:1. Introduction
2. Results
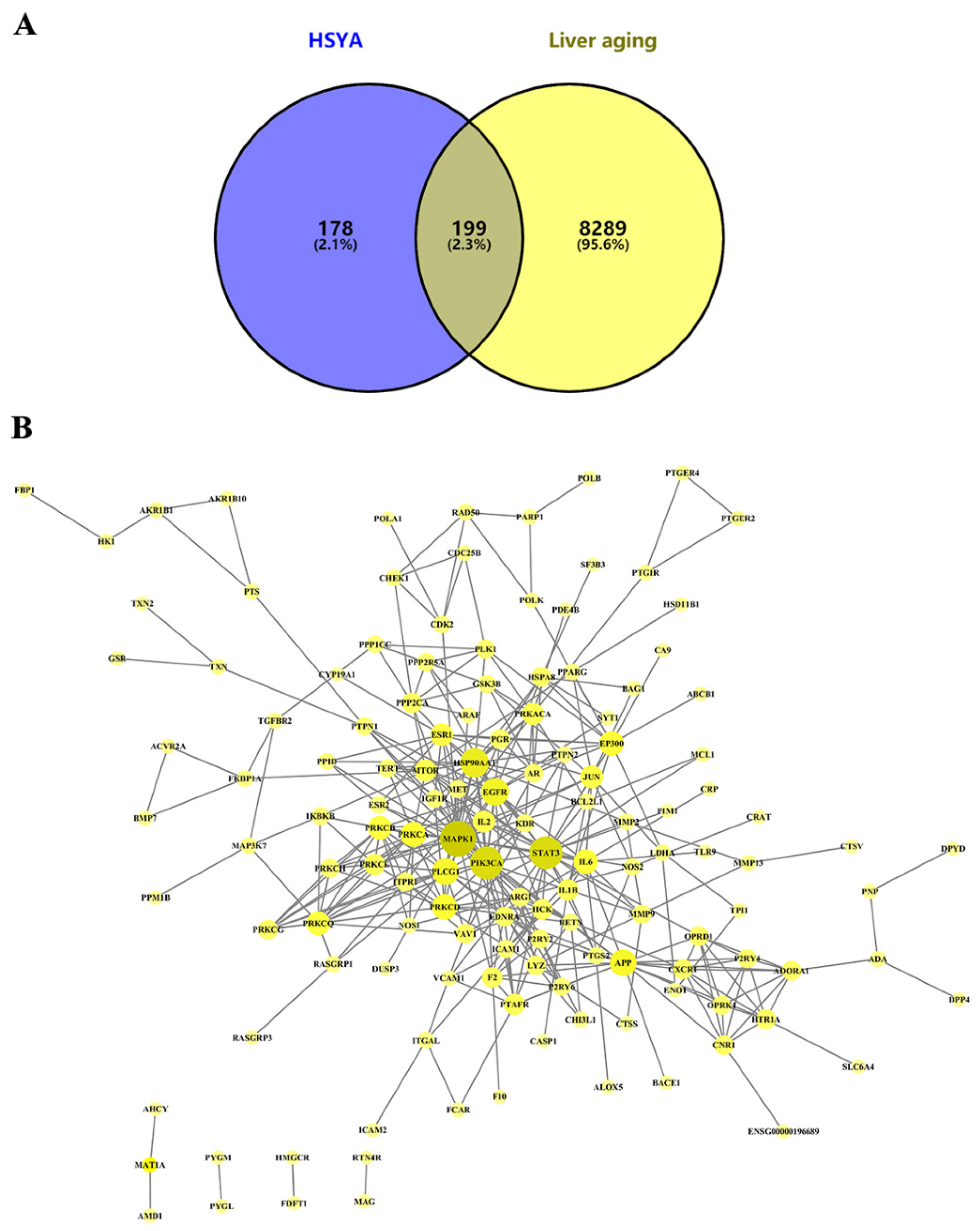
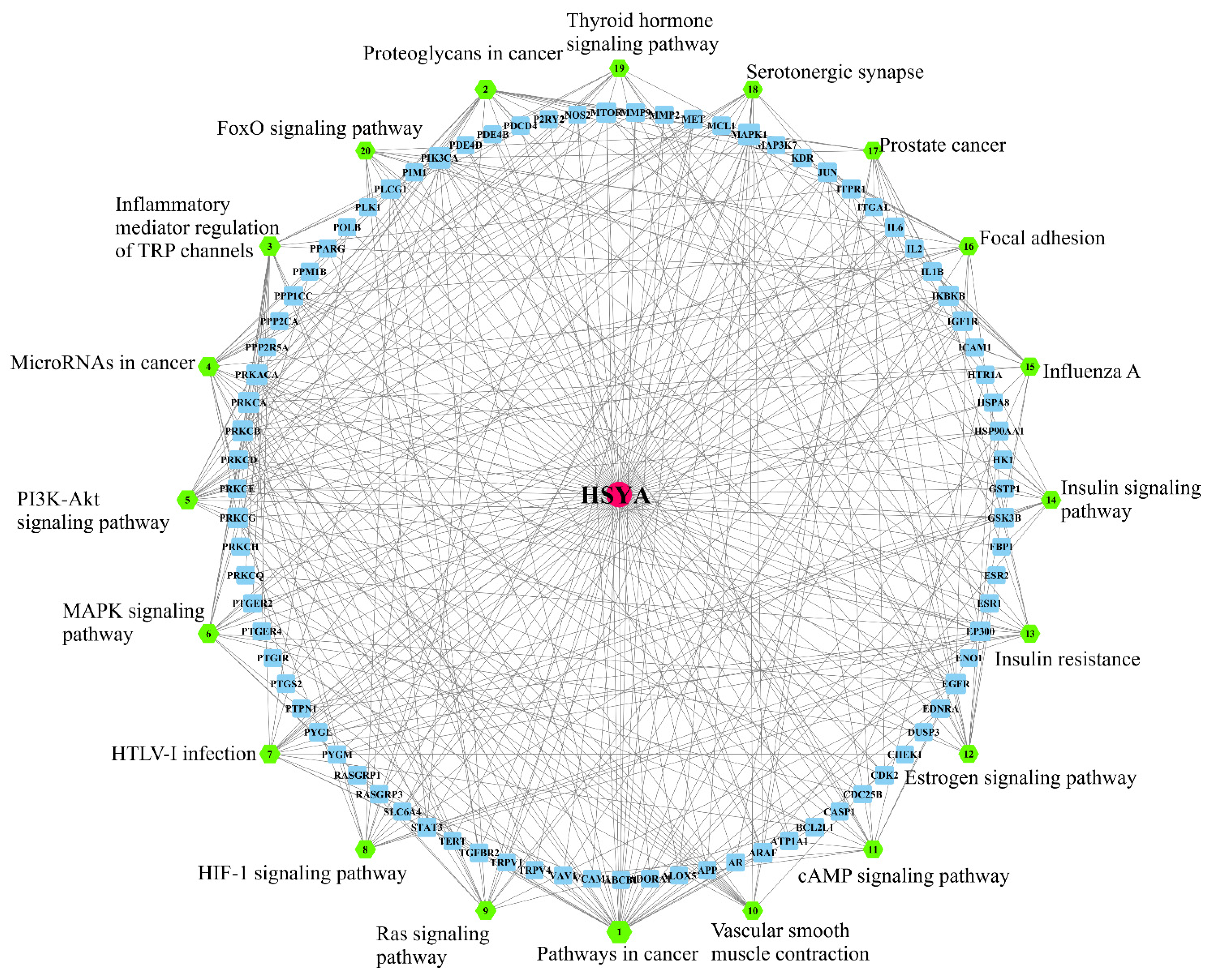
2.1. Network Pharmacological Analysis
2.1.1. Target Prediction and PPI Network Analysis of HSYA Delay of LA
2.1.2. Functional Enrichment Analysis of DEGs
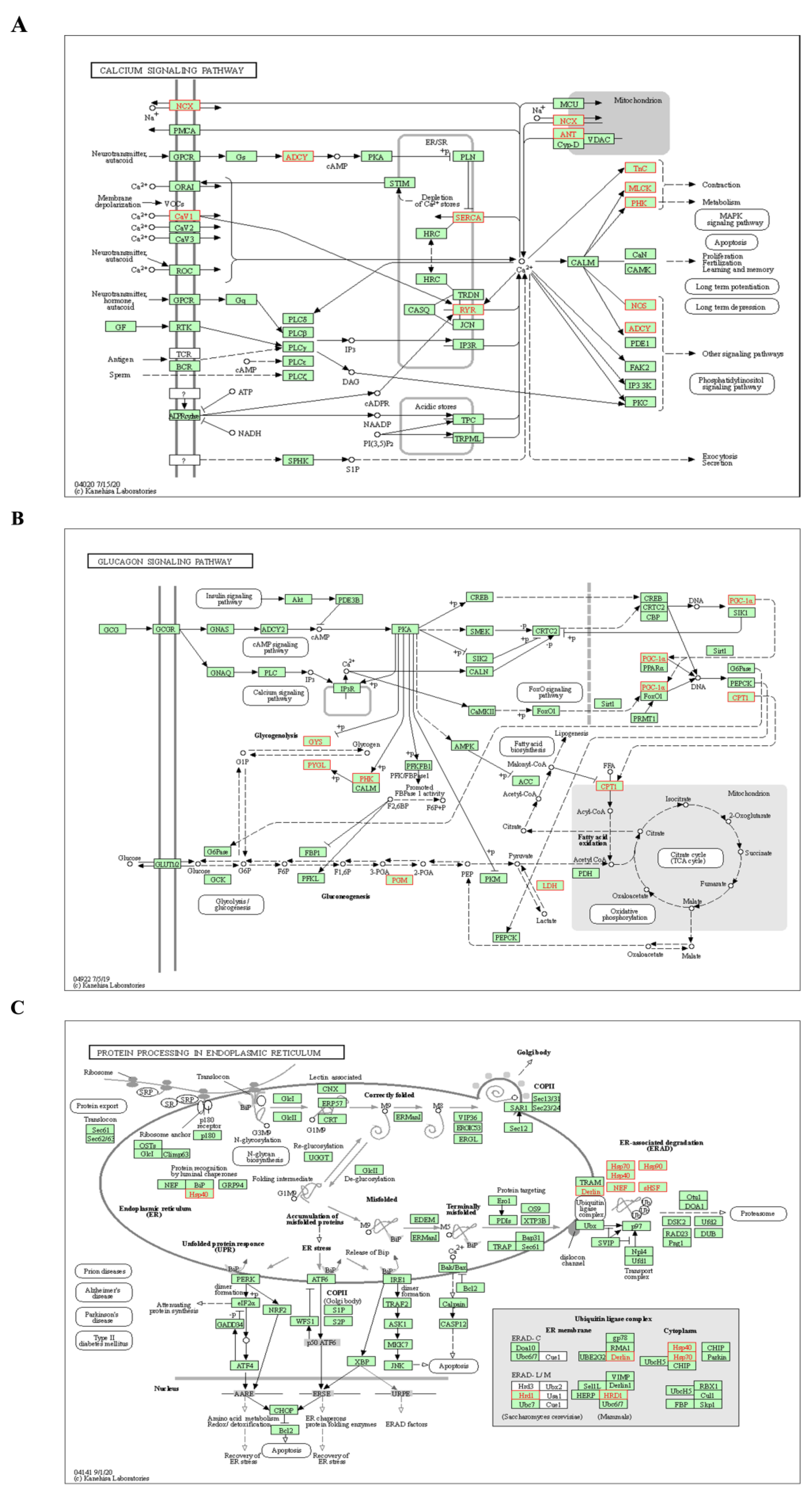
2.1.3. HSYA Target Pathway Analysis
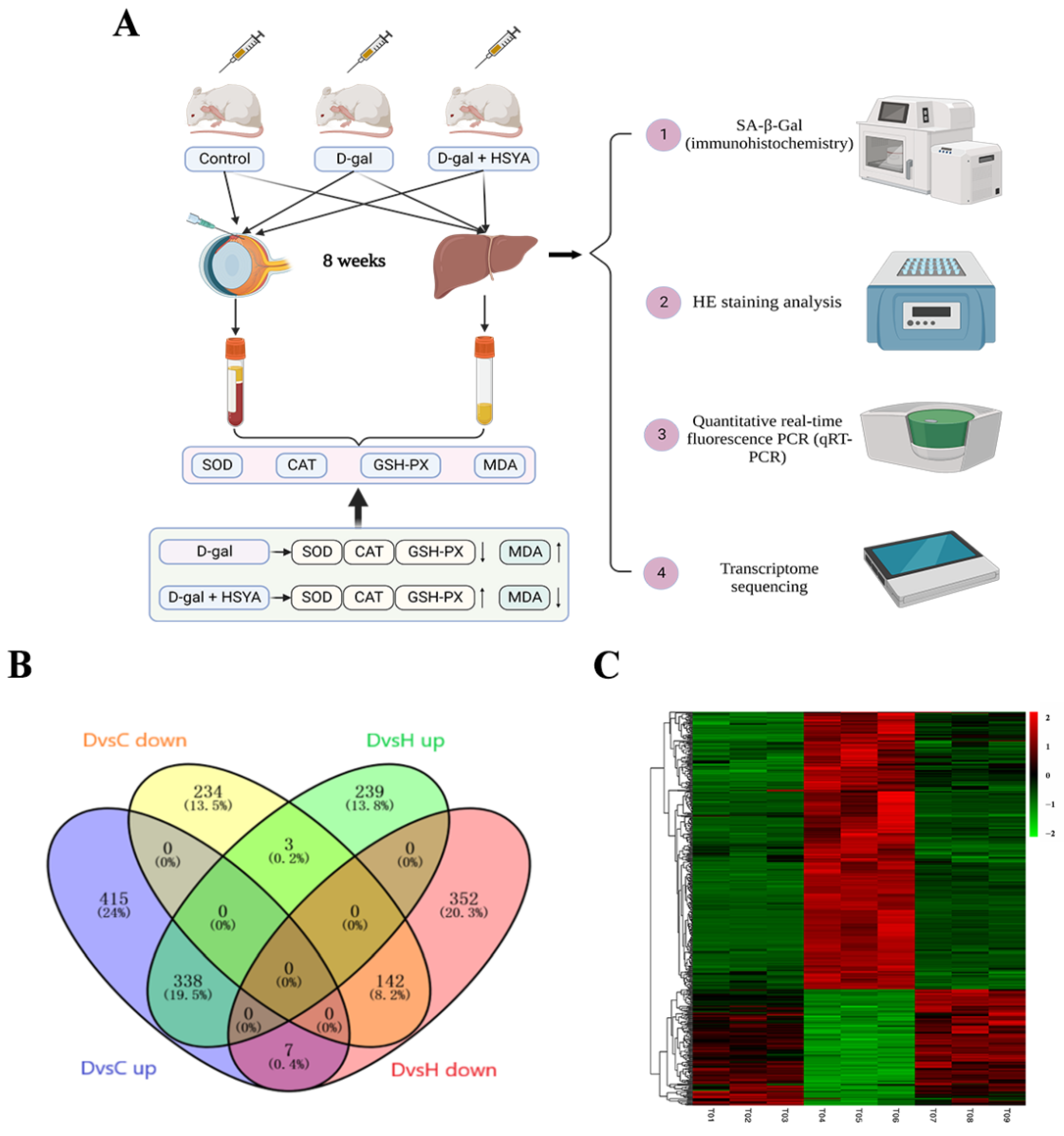
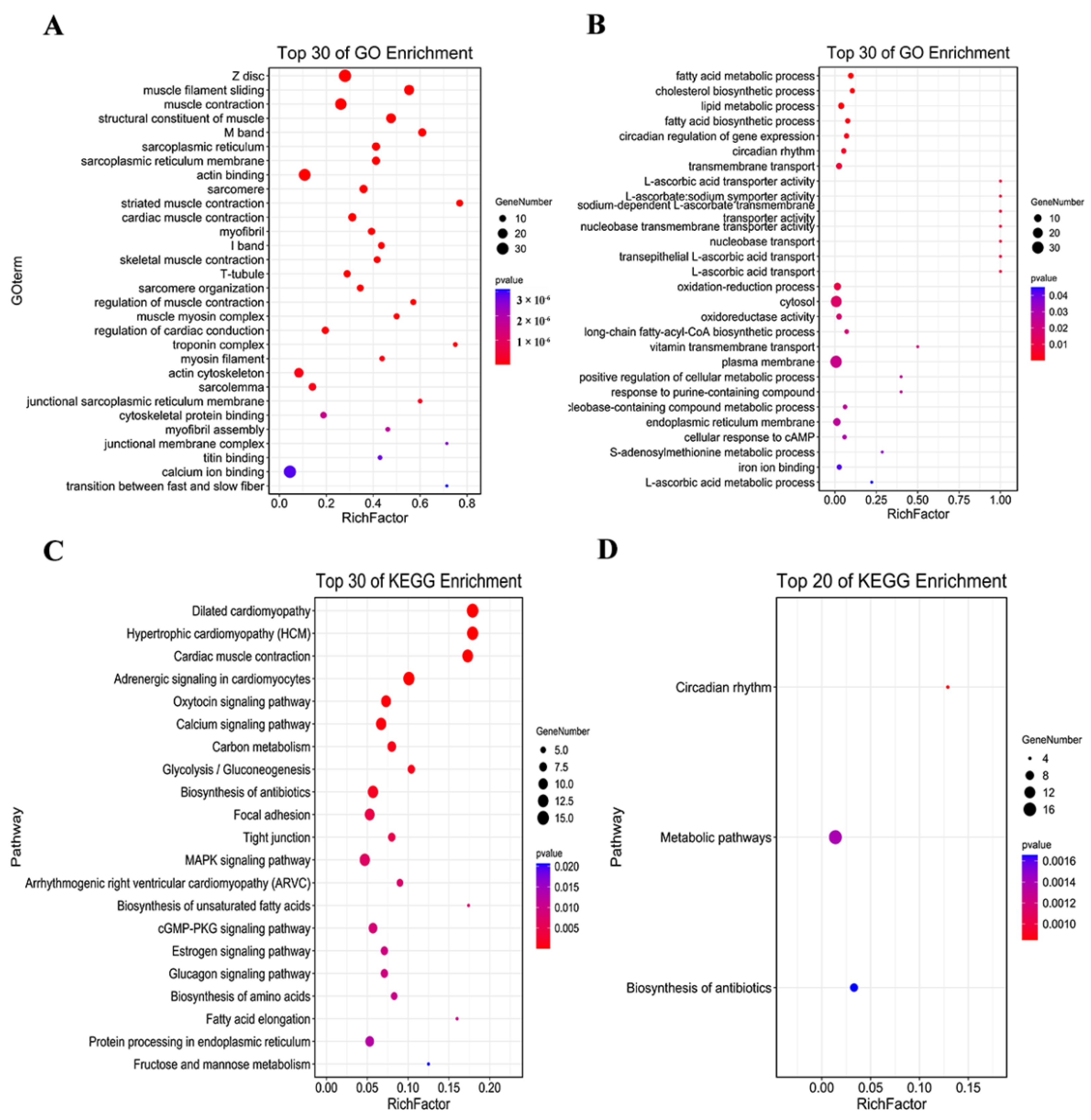
2.2. Transcriptomic Analysis
2.2.1. Identification of DEGs
2.2.2. GO and KEGG Pathway Enrichment Analyses
2.2.3. Analysis of KEGG Pathways Related to HSYA Delay of LA
2.3. Preliminary Verification of HSYA Mechanism of Delaying Liver Aging
2.3.1. Determination of Target Genes and Pathways
2.3.2. Mechanism of HSYA Delay of Liver Aging
2.3.3. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Reagents and Materials
4.2. Collection of Targets for HSYA Delay of Liver Aging
4.3. Network Construction and Analysis
4.4. Animal Experiments and Sample Collection
4.5. Transcriptomics Sequencing and Data Analysis
4.6. RNA Extraction and qRT-PCR Analysis
4.7. Western Blotting
4.8. Enzyme-Linked Immunosorbent Assay (ELISA)
4.9. Molecular Docking
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HSYA | hydroxysafflor yellow A |
| LA | liver aging |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DEGs | differentially expressed genes |
| qRT-PCR | quantitative real-time PCR |
| WHO | World Health Organization |
| D-gal | D-galactose |
| PPI | protein–protein interaction |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| NIMNT | NIM molecular network tool |
| FDR | false discovery rate |
| BCA | bicinchoninic acid |
| PVDF | polyvinylidene fluoride |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| HRP | horse radish peroxidase |
References
- Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, S.A.; Glorioso, J.M.; Nyberg, S.L. Liver regeneration. Transl. Res. 2014, 163, 352–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, N.J.; Kang, S.; Lockwood, G.P.; Le Couteur, D.G.; Cogger, V.C. Hallmarks of Aging in the Liver. Comput. Struct. Biotechnol. J. 2019, 17, 1151–1161. [Google Scholar] [CrossRef]
- Gokarn, R.; Solon-Biet, S.; Youngson, N.A.; Wahl, D.; Cogger, V.C.; McMahon, A.C.; Cooney, G.J.; Ballard, J.; Raubenheimer, D.; Morris, M.J.; et al. The Relationship Between Dietary Macronutrients and Hepatic Telomere Length in Aging Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 446–449. [Google Scholar] [CrossRef]
- Gambino, V.; De Michele, G.; Venezia, O.; Migliaccio, P.; Dall’Olio, V.; Bernard, L.; Minardi, S.P.; Della Fazia, M.A.; Bartoli, D.; Servillo, G.; et al. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell 2013, 12, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Maslov, A.Y.; Ganapathi, S.; Westerhof, M.; Quispe-Tintaya, W.; White, R.R.; Van Houten, B.; Reiling, E.; Dollé, M.E.; van Steeg, H.; Hasty, P.; et al. DNA damage in normally and prematurely aged mice. Aging Cell 2013, 12, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.H.; Kisseleva, T.; Brenner, D.A. Aging and liver disease. Curr. Opin. Gastroenterol. 2015, 31, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Yan, L.; Liu, Y.; Qu, C.; Ni, J.; Li, H. The Burden and Trends of Primary Liver Cancer Caused by Specific Etiologies from 1990 to 2017 at the Global, Regional, National, Age, and Sex Level Results from the Global Burden of Disease Study 2017. Liver Cancer 2020, 9, 563–582. [Google Scholar] [CrossRef]
- Eurostat. Ageing Europe. Looking at the Lives of Older People in the EU; Publications Office of the European Union: Luxembourg, 2019; Available online: https://ec.europa.eu/eurostat/cache/infographs/elderly/index.html (accessed on 1 June 2021).
- Zhang, L.L.; Tian, K.; Tang, Z.H.; Chen, X.J.; Bian, Z.X.; Wang, Y.T.; Lu, J.J. Phytochemistry and Pharmacology of Carthamus tinctorius L. Am. J. Chin Med. 2016, 44, 197–226. [Google Scholar] [CrossRef]
- Delshad, E.; Yousefi, M.; Sasannezhad, P.; Rakhshandeh, H.; Ayati, Z. Medical uses of Carthamus tinctorius L. (Safflower): A comprehensive review from Traditional Medicine to Modern Medicine. Electron. Phys. 2018, 10, 6672–6681. [Google Scholar] [CrossRef]
- Cui, Q.; Ma, Y.H.; Yu, H.Y.; Zhang, Y.L.; Qin, X.D.; Ge, S.Q.; Zhang, G.W. Systematic analysis of the mechanism of hydroxysafflor yellow A for treating ischemic stroke based on network pharmacology technology. Eur. J. Pharmacol. 2021, 908, 174360. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, Z.; He, W.; Chen, H.; Lai, Z.; Duan, Y.; Cao, X.; Tao, J.; Xu, C.; Zhang, Q.; et al. Hydroxysafflor Yellow A Confers Neuroprotection from Focal Cerebral Ischemia by Modulating the Crosstalk Between JAK2/STAT3 and SOCS3 Signaling Pathways. Cell. Mol. Neurobiol. 2020, 40, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Yu, L.J.; Hui, X.C.; Wu, Z.Z.; Yin, K.L.; Yang, H.; Xu, Y. Hydroxy-safflor yellow A attenuates Aβ1-42-induced inflammation by modulating the JAK2/STAT3/NF-κB pathway. Brain Res. 2014, 1563, 72–80. [Google Scholar] [CrossRef]
- He, Y.; Liu, Q.; Li, Y.; Yang, X.; Wang, W.; Li, T.; Zhang, W.; Cui, Y.; Wang, C.; Lin, R. Protective effects of hydroxysafflor yellow A (HSYA) on alcohol-induced liver injury in rats. J. Physiol. Biochem. 2015, 71, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, J.; Zhu, J.; Wang, D.; Chen, S.; Bai, X. Hydroxysafflor yellow A inhibits angiogenesis of hepatocellular carcinoma via blocking ERK/MAPK and NF-κB signaling pathway in H22 tumor-bearing mice. Eur. J. Pharmacol. 2015, 754, 105–114. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; He, P.; Ni, Z.; Hu, Y.; Xu, H.; Dai, H. Hydroxysafflor yellow A protects methylglyoxal-induced injury in the cultured human brain microvascular endothelial cells. Neurosci. Lett. 2013, 549, 146–150. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Sun, Y.; Liu, L.; Bai, X.; Wang, D.; Li, H. Restorative effects of hydroxysafflor yellow A on hepatic function in an experimental regression model of hepatic fibrosis induced by carbon tetrachloride. Mol. Med. Rep. 2017, 15, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Kong, S.Z.; Shi, X.G.; Feng, X.X.; Li, W.J.; Liu, W.H.; Chen, Z.W.; Xie, J.H.; Lai, X.P.; Zhang, S.X.; Zhang, X.J.; et al. Inhibitory effect of hydroxysafflor yellow a on mouse skin photoaging induced by ultraviolet irradiation. Rejuv. Res. 2013, 16, 404–413. [Google Scholar] [CrossRef] [Green Version]
- Min, F.; Sun, H.; Wang, B.; Ahmad, N.; Guo, H.; Gao, H.; Gao, Y.; Liu, X.; Li, H. Hepatoprotective effects of hydroxysafflor yellow A in D-galactose-treated aging mice. Eur. J. Pharmacol. 2020, 881, 173214. [Google Scholar] [CrossRef]
- Lee, S. Systems Biology-A Pivotal Research Methodology for Understanding the Mechanisms of Traditional Medicine. J. Pharmacopunct. 2015, 18, 11–18. [Google Scholar] [CrossRef]
- Cai, F.F.; Bian, Y.Q.; Wu, R.; Sun, Y.; Chen, X.L.; Yang, M.D.; Zhang, Q.R.; Hu, Y.; Sun, M.Y.; Su, S.B. Yinchenhao decoction suppresses rat liver fibrosis involved in an apoptosis regulation mechanism based on network pharmacology and transcriptomic analysis. Biomed. Pharmacother. 2019, 114, 108863–108873. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Yuan, H.; Ma, Q.; Cui, H.; Liu, G.; Zhao, X.; Li, W.; Piao, G. How Can Synergism of Traditional Medicines Benefit from Network Pharmacology? Molecules 2017, 22, 1135. [Google Scholar] [CrossRef] [Green Version]
- Lan, S.; Duan, J.; Zeng, N.; Yu, B.; Yang, X.; Ning, H.; Huang, Y.; Rao, Y. Network pharmacology-based screening of the active ingredients and mechanisms of Huangqi against aging. Medicine 2021, 100, e25660. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, H.; Song, S.; Xu, H.; Chen, Y.; Tian, S.; Zhang, Y.; Zhang, Q. Systematic Understanding of Anti-Aging Effect of Coenzyme Q10 on Oocyte Through a Network Pharmacology Approach. Front. Endocrinol. 2022, 13, 813772. [Google Scholar] [CrossRef]
- Wu, B.; Xiao, X.; Li, S.; Zuo, G. Transcriptomics and metabonomics of the anti-aging properties of total flavones of Epimedium in relation to lipid metabolism. J. Ethnopharmacol. 2019, 229, 73–80. [Google Scholar] [CrossRef]
- Izquierdo, M.C.; Sanz, A.B.; Sánchez-Niño, M.D.; Pérez-Gómez, M.V.; Ruiz-Ortega, M.; Poveda, J.; Ruiz-Andrés, O.; Ramos, A.M.; Moreno, J.A.; Egido, J.; et al. Acute kidney injury transcriptomics unveils a relationship between inflammation and ageing. Nefrologia 2012, 32, 715–723. [Google Scholar] [CrossRef]
- Fonseca Costa, S.S.; Robinson-Rechavi, M.; Ripperger, J.A. Single-cell transcriptomics allows novel insights into aging and circadian processes. Brief. Funct. Genom. 2020, 19, 343–349. [Google Scholar] [CrossRef]
- Malone, J.H.; Oliver, B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011, 9, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Hsin, K.Y.; Ghosh, S.; Kitano, H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS ONE 2013, 8, e83922. [Google Scholar] [CrossRef]
- Xie, Y.; Guo, Y.; Cao, S.; Xue, M.; Fan, Z.; Gao, C.; Jin, B. Hydroxysafflor Yellow A Attenuates Hydrogen Peroxide-Induced Oxidative Damage on Human Umbilical Vein Endothelial Cells. Evid-Based. Compl. Alt. 2020, 2020, 8214128. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Dong, H.; Zhao, X.; Li, Z.; Li, X. Hydroxysafflor yellow A protects against chronic carbon tetrachloride-induced liver fibrosis. Eur. J. Pharmacol. 2011, 660, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhang, Z.; Ma, Y.; Jin, R.; Yi, B.; Yang, D.; Ma, L. Mechanism of hydroxysafflor yellow A on acute liver injury based on transcriptomics. Front. Pharmacol. 2022, 13, 966759. [Google Scholar] [CrossRef]
- Selli, C.; Erac, Y.; Kosova, B.; Erdal, E.S.; Tosun, M. Silencing of TRPC1 regulates store-operated calcium entry and proliferation in Huh7 hepatocellular carcinoma cells. Biomed. Pharmacother. 2015, 71, 194–200. [Google Scholar] [CrossRef]
- Yang, J.; Wang, R.; Cheng, X.; Qu, H.; Qi, J.; Li, D.; Xing, Y.; Bai, Y.; Zheng, X. The vascular dilatation induced by Hydroxysafflor yellow A (HSYA) on rat mesenteric artery through TRPV4-dependent calcium influx in endothelial cells. J. Ethnopharmacol. 2020, 256, 112790. [Google Scholar] [CrossRef]
- Wang, D.; Wang, E.; Li, Y.; Teng, Y.; Li, H.; Jiao, L.; Wu, W. Anti-Aging Effect of Momordica charantia L. on d-Galactose-Induced Subacute Aging in Mice by Activating PI3K/AKT Signaling Pathway. Molecules 2022, 27, 4502. [Google Scholar] [CrossRef]
- Gong, P.; Wang, D.; Cui, D.; Yang, Q.; Wang, P.; Yang, W.; Chen, F. Anti-aging function and molecular mechanism of Radix Astragali and Radix Astragali preparata via network pharmacology and PI3K/Akt signaling pathway. Phytomedicine 2021, 84, 153509. [Google Scholar] [CrossRef]
- Song, H.; Wang, D.; Li, J.; Yang, Y.; Mu, X.; Bai, X. Hydroxysaffl or yellow A inhibits proliferation and migration of human hepa-tocellular carcinoma cells and promotes apoptosis via PI3K pathway. Tumor 2018, 38, 830–839. [Google Scholar]
- Jiang, G.; Zhang, B.B. Glucagon and regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E671–E678. [Google Scholar] [CrossRef] [Green Version]
- Kishnani, P.S.; Goldstein, J.; Austin, S.L.; Arn, P.; Bachrach, B.; Bali, D.S.; Chung, W.K.; El-Gharbawy, A.; Brown, L.M.; Kahler, S.; et al. Diagnosis and management of glycogen storage diseases type VI and IX: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2019, 21, 772–789. [Google Scholar] [CrossRef] [PubMed]
- Kakhlon, O.; Ferreira, I.; Solmesky, L.J.; Khazanov, N.; Lossos, A.; Alvarez, R.; Yetil, D.; Pampou, S.; Weil, M.; Senderowitz, H.; et al. Guaiacol as a drug candidate for treating adult polyglucosan body disease. JCI Insight 2018, 3, e99694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyzaei, Z.; Shamsaeefar, A.; Kazemi, K.; Nikeghbalian, S.; Bahador, A.; Dehghani, M.; Malekhosseini, S.A.; Geramizadeh, B. Liver transplantation in glycogen storage disease: A single-center experience. Orphanet J. Rare Dis. 2022, 17, 127. [Google Scholar] [CrossRef]
- Rao, R.V.; Ellerby, H.M.; Bredesen, D.E. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004, 11, 372–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Xu, F.; Liang, H.; Cao, H.; Cai, M.; Xu, W.; Weng, J. SIRT1/HSF1/HSP pathway is essential for exenatide-alleviated, lipid-induced hepatic endoplasmic reticulum stress. Hepatology 2017, 66, 809–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.R.; Xie, X.F.; Cao, X.Y.; Li, G.M.; Yin, Y.P.; Peng, C. Research progress on mechanism of Carthamus tinctorius in ischemic stroke therapy. China J. Chin. Mater. Med. 2022, 47, 4574–4582. [Google Scholar] [CrossRef]
- Kao, E.; Shinohara, M.; Feng, M.; Lau, M.Y.; Ji, C. Human immunodeficiency virus protease inhibitors modulate Ca2+ homeostasis and potentiate alcoholic stress and injury in mice and primary mouse and human hepatocytes. Hepatology 2012, 56, 594–604. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhang, Y.; Li, Y.; Guo, C.; Fan, Z.; Li, Y.; Yang, M.; Zhou, X.; Sun, Z.; Wang, J. Integrative proteomics and metabolomics approach to elucidate metabolic dysfunction induced by silica nanoparticles in hepatocytes. J. Hazard. Mater. 2022, 434, 128820. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.; Duan, C.; Zhang, J.; Li, X. Hepatoxicity mechanism of cantharidin-induced liver LO2 cells by LC-MS metabolomics combined traditional approaches. Toxicol. Lett. 2020, 333, 49–61. [Google Scholar] [CrossRef]
- Razandi, M.; Pedram, A.; Park, S.T.; Levin, E.R. Proximal events in signaling by plasma membrane estrogen receptors. J. Biol. Chem. 2003, 278, 2701–2712. [Google Scholar] [CrossRef] [Green Version]
- Li, T. Molecular Mechanisms Underlying the Estrogen Regulated Lipid Homeostasis in the Liver. Ph.D. Thesis, Duke University, Durham, NC, USA, 2007. [Google Scholar]
- Wu, L.; Liu, Y.; Zhao, Y.; Li, M.; Guo, L. Targeting DUSP7 signaling alleviates hepatic steatosis, inflammation and oxidative stress in high fat diet (HFD)-fed mice via suppression of TAK1. Free Radic. Biol. Med. 2020, 153, 140–158. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, T.X.; Huang, X.; Li, Y.; Sun, T.; Zang, S.; Guan, K.L.; Xiong, Y.; Liu, J.; Yuan, H.X. Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Int. 2020, 40, 1378–1394. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Tan, Y.; Hu, R.; Xia, Y.; Xia, G. Mechanism of recipe for treatment of ovulatory infertility: A network pharmacology-based study and clinical observations. J. South. Med. Univ. 2021, 41, 319–328. [Google Scholar] [CrossRef]
- Cai, C.; Song, X.; Yu, C. Identification of genes in hepatocellular carcinoma induced by non-alcoholic fatty liver disease. Cancer Biomark. 2020, 29, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, D.A.; Vafiadaki, E.; Sanoudou, D.; Kranias, E.G. Histidine-rich calcium binding protein: The new regulator of sarcoplasmic reticulum calcium cycling. J. Mol. Cell. Cardiol. 2011, 50, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Zvaritch, E.; Tupling, A.R.; Rice, W.J.; de Leon, S.; Rudnicki, M.; McKerlie, C.; Banwell, B.L.; MacLennan, D.H. Targeted disruption of the ATP2A1 gene encoding the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1 (SERCA1) impairs diaphragm function and is lethal in neonatal mice. J. Biol. Chem. 2003, 278, 13367–13375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berridge, M.J. The Inositol Trisphosphate/Calcium Signaling Pathway in Health and Disease. Physiol. Rev. 2016, 96, 1261–1296. [Google Scholar] [CrossRef] [Green Version]
- Negash, A.A.; Olson, R.M.; Griffin, S.; Gale, M., Jr. Modulation of calcium signaling pathway by hepatitis C virus core protein stimulates NLRP3 inflammasome activation. PLoS Pathog. 2019, 15, e1007593. [Google Scholar] [CrossRef]
- Francis, S.H.; Busch, J.L.; Corbin, J.D.; Sibley, D. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev. 2010, 62, 525–563. [Google Scholar] [CrossRef]
- Kelly, S.P.; Dawson-Scully, K. Natural polymorphism in protein kinase G modulates functional senescence in Drosophilamelanogaster. J. Exp. Biol. 2019, 222, jeb199364. [Google Scholar] [CrossRef] [Green Version]
- García-Villafranca, J.; Guillén, A.; Castro, J. Involvement of nitric oxide/cyclic GMP signaling pathway in the regulation of fatty acid metabolism in rat hepatocytes. Biochem. Pharmacol. 2003, 65, 807–812. [Google Scholar] [CrossRef]
- França, M.; Peixoto, C.A. cGMP signaling pathway in hepatic encephalopathy neuroinflammation and cognition. Int. Immunopharmacol. 2020, 79, 106082. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.K.; Wu, B.N.; Chiu, E.Y.; Tseng, C.J.; Yeh, J.L.; Liu, C.P.; Chai, C.Y.; Chen, I.J. NO donor KMUP-1 improves hepatic ischemia-reperfusion and hypoxic cell injury by inhibiting oxidative stress and pro-inflammatory signaling. Int. J. Immunopath. Phar. 2013, 26, 93–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Staay, F.J.; Rutten, K.; Bärfacker, L.; Devry, J.; Erb, C.; Heckroth, H.; Karthaus, D.; Tersteegen, A.; van Kampen, M.; Blokland, A.; et al. The novel selective PDE9 inhibitor BAY 73-6691 improves learning and memory in rodents. Neuropharmacology 2008, 55, 908–918. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Lv, Y.N.; Tan, Y.S.; Shen, K.; Zhai, K.F.; Chen, H.L.; Kou, J.P.; Yu, B.Y. An integrated pathway interaction network for the combination of four effective compounds from ShengMai preparations in the treatment of cardio-cerebral ischemic diseases. Acta Pharmacol. Sin. 2015, 36, 1337–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 9, D1138–D1143. [Google Scholar] [CrossRef]
- Zhao, F.; Guochun, L.; Yang, Y.; Shi, L.; Xu, L.; Yin, L. A network pharmacology approach to determine active ingredients and rationality of herb combinations of Modified-Simiaowan for treatment of gout. J. Ethnopharmacol. 2015, 168, 1–16. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Li, Q.; Du, G.; Meng, G.; Sun, J.; Chen, S. An Integration of Network Pharmacology and Experimental Verification to Investigate the Mechanism of Guizhi to Treat Nephrotic Syndrome. Front. Pharmacol. 2021, 12, 755421. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Han, L.; Zhang, X.; Wu, R.; Cheng, X.; Zhou, W.; Zhang, Y. Knowledge-Based Neuroendocrine Immunomodulation (NIM) Molecular Network Construction and Its Application. Molecules 2018, 23, 1312. [Google Scholar] [CrossRef] [Green Version]
- Altermann, E.; Klaenhammer, T.R. PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genom. 2005, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Velasco, A.; Sánchez, A.; Martínez, I.; Santaclara, F.J.; Pérez-Martín, R.I.; Sotelo, C.G. Development of a real-time PCR method for the identification of Atlantic mackerel (Scomber scombrus). Food Chem. 2013, 141, 2006–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]



| Target | PDB ID | Compound | Binding Affinity (kcal/mol) |
|---|---|---|---|
| NOS1 | 5uo2 | HSYA | −8.8 |
| HSP90AA1 | 6gr5 | HSYA | −7.5 |
| ATP2A1 | none | HSYA | −7.0 |
| Gene | Primer Sequences | |
|---|---|---|
| HSP90AA1 | Forward primer | ACCTTTGCCTTTCAGGCAGAA |
| Reverse primer | CCGATGAATTGGAGATGAGCTC | |
| ATP2A1 | Forward primer | ACCTTTGCCTTTCAGGCAGAA |
| Reverse primer | CCGATGAATTGGAGATGAGCTC | |
| CRAT | Forward primer | AGAAGCTAAGCCCTGATGCCTT |
| Reverse primer | GCGCAGAGAGGCACTTTCATAC | |
| NOS1 | Forward primer | TCCTAAATCCAGCCGATCGAC |
| Reverse primer | CATTCACGAGGTCCTCATGGTT | |
| GAPDH | Forward primer | AGGTCGGTGTGAACGGATTTG |
| Reverse primer | GGGGTCGTTGATGGCAACA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, J.; Sun, S.; Min, F.; Hu, X.; Zhang, Y.; Cheng, Y.; Li, H.; Wang, X.; Liu, X. Integrating Network Pharmacology and Transcriptomic Strategies to Explore the Pharmacological Mechanism of Hydroxysafflor Yellow A in Delaying Liver Aging. Int. J. Mol. Sci. 2022, 23, 14281. https://doi.org/10.3390/ijms232214281
Kong J, Sun S, Min F, Hu X, Zhang Y, Cheng Y, Li H, Wang X, Liu X. Integrating Network Pharmacology and Transcriptomic Strategies to Explore the Pharmacological Mechanism of Hydroxysafflor Yellow A in Delaying Liver Aging. International Journal of Molecular Sciences. 2022; 23(22):14281. https://doi.org/10.3390/ijms232214281
Chicago/Turabian StyleKong, Jie, Siming Sun, Fei Min, Xingli Hu, Yuan Zhang, Yan Cheng, Haiyan Li, Xiaojie Wang, and Xin Liu. 2022. "Integrating Network Pharmacology and Transcriptomic Strategies to Explore the Pharmacological Mechanism of Hydroxysafflor Yellow A in Delaying Liver Aging" International Journal of Molecular Sciences 23, no. 22: 14281. https://doi.org/10.3390/ijms232214281






