Adaptation of Helicoverpa armigera to Soybean Peptidase Inhibitors Is Associated with the Transgenerational Upregulation of Serine Peptidases
Abstract
:1. Introduction
2. Results
2.1. H. armigera Larvae Are Resistant to SPIs
2.2. SPI Ingestion Results in Marked Changes in Gene Expression in the H. armigera Midgut
2.3. Differential Expression of Digestive Peptidases May Be Associated with the Adaptation of H. armigera to SPIs
2.4. Transgenerational Persistence of the Transcriptional Profile Induced by SPI Exposure
2.5. DNA Methylation May Be Involved in the Molecular Response of H. armigera to SPI Ingestion
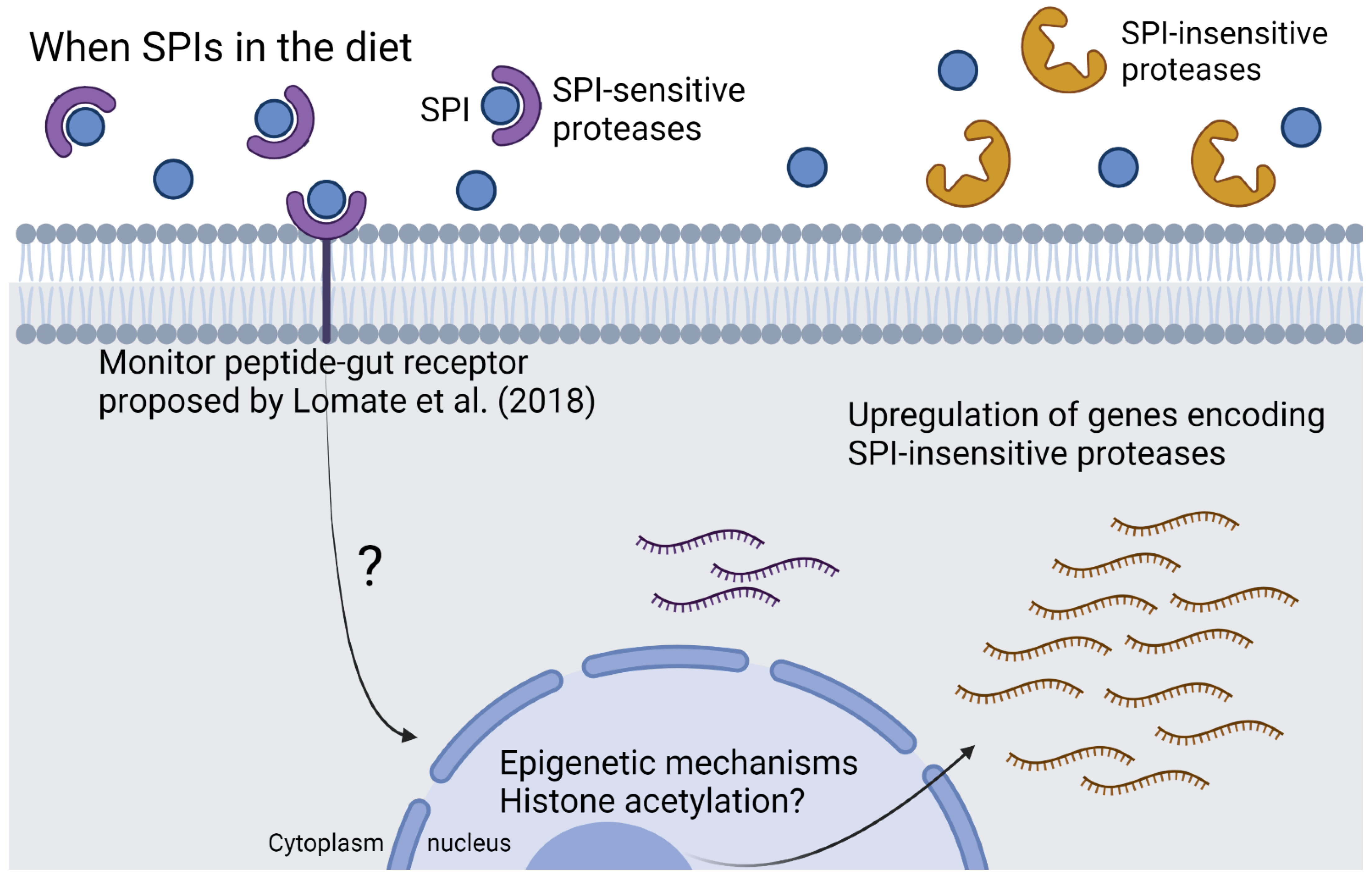
3. Discussion
4. Materials and Methods
4.1. Extraction of Soybean Proteinase Inhibitors (SPIs)
4.2. Inhibitory Activity of SPIs
4.3. Insect Rearing and Feeding Assays
4.4. RNA-Seq Analysis
4.5. Identification of Serine Peptidase Gene Sequences
4.6. CpG Island Mapping
4.7. Functional Analysis of DNA Methylation in CpG Islands of Serine Peptidase Genes
4.8. Genome-Wide Characterization of DNA Methylation
4.9. RT-qPCR Experiments
4.10. Functional Annotation and Gene Ontology Enrichment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kriticos, D.J.; Ota, N.; Hutchison, W.D.; Beddow, J.; Walsh, T.; Tay, W.T.; Borchert, D.M.; Paula-Moreas, S.V.; Czepak, C.; Zalucki, M.P. The potential distribution of invading Helicoverpa armigera in North America: Is it just a matter of time? PLoS ONE 2015, 10, e0119618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reigada, C.; Guimarães, K.F.; Parra, J.R.P. Relative Fitness of Helicoverpa armigera (Lepidoptera: Noctuidae) on Seven Host Plants: A Perspective for IPM in Brazil. J. Insect Sci. 2016, 16, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malka, O.; Feldmesser, E.; Brunschot, S.; Santos-Garcia, D.; Han, W.-H.; Seal, S.; Colvin, J.; Morin, S. The Molecular Mechanisms That Determine Different Degrees of Polyphagy in the Bemisia Tabaci Species Complex. Evol. Appl. 2021, 14, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.R. Why so Many Polyphagous Fruit Flies (Diptera: Tephritidae)? A Further Contribution to the ‘Generalism’ Debate. Biol. J. Linn. Soc. 2016, 120, 245–257. [Google Scholar] [CrossRef]
- Souza, T.P.; Dias, R.O.; Castelhano, E.C.; Brandão, M.M.; Moura, D.S.; Silva-filho, M.C. Comparative Biochemistry and Physiology, Part B Comparative Analysis of Expression pro Fi Ling of the Trypsin and Chymotrypsin Genes from Lepidoptera Species with Different Levels of Sensitivity to Soybean Peptidase Inhibitors. Comp. Biochem. Physiol. Part B 2016, 196–197, 67–73. [Google Scholar] [CrossRef]
- de la Paz Celorio-Mancera, M.; Wheat, C.W.; Vogel, H.; Söderlind, L.; Janz, N.; Nylin, S. Mechanisms of Macroevolution: Polyphagous Plasticity in Butterfly Larvae Revealed by RNA-Seq. Mol. Ecol. 2013, 22, 4884–4895. [Google Scholar] [CrossRef]
- Paulillo, L.C.M.S.; Lopes, A.R.; Cristofoletti, P.T.; Parra, J.R.P.; Terra, W.R.; Silva-Filho, M.C. Changes in Midgut Endopeptidase Activity of Spodoptera frugiperda (Lepidoptera: Noctuidae) Are Responsible for Adaptation to Soybean Proteinase Inhibitors. J. Econ. Entomol. 2000, 93, 892–896. [Google Scholar] [CrossRef]
- Brioschi, D.; Nadalini, L.D.; Bengtson, M.H.; Sogayar, M.C.; Moura, D.S.; Silva-Filho, M.C. General up Regulation of Spodoptera frugiperda Trypsins and Chymotrypsins Allows Its Adaptation to Soybean Proteinase Inhibitor. Insect Biochem. Mol. Biol. 2007, 37, 1283–1290. [Google Scholar] [CrossRef]
- Dias, R.O.; Tramontano, A.; Via, A.; Brand, M.M.; Silva-filho, M.C.; Brandão, M.M.; Tramontano, A.; Silva-filho, M.C. Digestive Peptidase Evolution in Holometabolous Insects Led to a Divergent Group of Enzymes in Lepidoptera. Insect Biochem. Mol. Biol. 2015, 58, 1–11. [Google Scholar] [CrossRef]
- Kuwar, S.S.; Pauchet, Y.; Vogel, H.; Heckel, D.G. Adaptive Regulation of Digestive Serine Proteases in the Larval Midgut of Helicoverpa armigera in Response to a Plant Protease Inhibitor. Insect Biochem. Mol. Biol. 2015, 59, 18–29. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Zeng, R. Insect Response to Plant Defensive Protease Inhibitors. Annu. Rev. Entomol. 2015, 60, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Chikate, Y.R.; Tamhane, V.A.; Joshi, R.S.; Gupta, V.S.; Giri, A.P. Differential Protease Activity Augments Polyphagy in Helicoverpa armigera. Insect Mol. Biol. 2013, 22, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Meriño-Cabrera, Y.; Oliveira Mendes, T.A.; Castro, J.G.S.; Barbosa, S.L.; Macedo, M.L.R.; Almeida Oliveira, M.G. Noncompetitive Tight-binding Inhibition of Anticarsia Gemmatalis Trypsins by Adenanthera Pavonina Protease Inhibitor Affects Larvae Survival. Arch. Insect Biochem. Physiol. 2020, 104, e21687. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, S.; Shi, Y.; Yang, Y.; Wu, Y. Global Gene Expression Changes Induced by Knockout of a Protease Gene Cluster in Helicoverpa armigera with CRISPR/Cas9. J. Insect Physiol. 2020, 122, 104023. [Google Scholar] [CrossRef] [PubMed]
- Spit, J.; Holtof, M.; Badisco, L.; Vergauwen, L.; Vogel, E.; Knapen, D.; Vanden Broeck, J. Transcriptional Analysis of The Adaptive Digestive System of The Migratory Locust in Response to Plant Defensive Protease Inhibitors. Sci. Rep. 2016, 6, 32460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomate, P.R.; Dewangan, V.; Mahajan, N.S.; Kumar, Y.; Kulkarni, A.; Saxena, S.; Gupta, V.S.; Giri, A.P. Integrated Transcriptomic and Proteomic Analyses Suggest the Participation of Endogenous Protease Inhibitors in the Regulation of Protease Gene Expression in Helicoverpa armigera. Mol. Cell. Proteom. 2018, 17, 1324–1336. [Google Scholar] [CrossRef] [Green Version]
- Cal, S.; Quesada, V.; Garabaya, C.; López-Otín, C. Polyserase-I, a Human Polyprotease with the Ability to Generate Independent Serine Protease Domains from a Single Translation Product. Proc. Natl. Acad. Sci. USA 2003, 100, 9185–9190. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.; Jiang, H.; Kanost, M.R.; Wang, Y. Serine Proteases and Their Homologs in the Drosophila Melanogaster Genome: An Initial Analysis of Sequence Conservation and Phylogenetic Relationships. Gene 2003, 304, 117–131. [Google Scholar] [CrossRef]
- Perkin, L.C.; Elpidina, E.N.; Oppert, B. RNA Interference and Dietary Inhibitors Induce a Similar Compensation Response in Tribolium Castaneum Larvae. Insect Mol. Biol. 2017, 26, 35–45. [Google Scholar] [CrossRef]
- Zheng, S.; Luo, J.; Zhu, X.; Gao, X.; Hua, H.; Cui, J. Transcriptomic Analysis of Salivary Gland and Proteomic Analysis of Oral Secretion in Helicoverpa armigera under Cotton Plant Leaves, Gossypol, and Tannin Stresses. Genomics 2022, 114, 110267. [Google Scholar] [CrossRef]
- Jin, M.; Liao, C.; Fu, X.; Holdbrook, R.; Wu, K.; Xiao, Y. Adaptive Regulation of Detoxification Enzymes in Helicoverpa armigera to Different Host Plants. Insect Mol. Biol. 2019, 28, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, S.R.; Bezerra, C.d.S.; Lourenço de Oliveira, D.G.; Souza Lima, L.; Maria Neto, S.; Ramalho de Oliveira, C.F.; Valério Verbisck, N.; Rodrigues Macedo, M.L. Novel Peptidase Kunitz Inhibitor from Platypodium Elegans Seeds Is Active against Spodoptera frugiperda Larvae. J. Agric. Food Chem. 2018, 66, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in Lepidopteran Insects. Invertebr. Immun. 2010, 708, 181–204. [Google Scholar]
- Zupanič, N.; Počič, J.; Leonardi, A.; Šribar, J.; Kordiš, D.; Križaj, I. Serine Pseudoproteases in Physiology and Disease. FEBS J. 2022. [Google Scholar] [CrossRef]
- Yang, L.; Xing, B.; Wang, L.; Yuan, L.; Manzoor, M.; Li, F.; Wu, S. Identification of Serine Protease, Serine Protease Homolog and Prophenoloxidase Genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 2021, 24, 1144–1152. [Google Scholar] [CrossRef]
- Piao, S.; Song, Y.-L.; Kim, J.H.; Park, S.Y.; Park, J.W.; Lee, B.L.; Oh, B.-H.; Ha, N.-C. Crystal Structure of a Clip-Domain Serine Protease and Functional Roles of the Clip Domains. EMBO J. 2005, 24, 4404–4414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.S.; Kim, B.Y.; Choo, Y.M.; Jin, B.R. Dual Role of the Serine Protease Homolog BmSPH-1 in the Development and Immunity of the Silkworm Bombyx mori. Dev. Comp. Immunol. 2018, 85, 170–176. [Google Scholar] [CrossRef]
- Wang, R.-X.; Tong, X.-L.; Gai, T.-T.; Li, C.-L.; Qiao, L.; Hu, H.; Han, M.-J.; Xiang, Z.-H.; Lu, C.; Dai, F.-Y. A Serine Protease Homologue Bombyx mori Scarface Induces a Short and Fat Body Shape in Silkworm. Insect Mol. Biol. 2018, 27, 319–332. [Google Scholar] [CrossRef]
- Terra, W.R.; Barroso, I.G.; Dias, R.O.; Ferreira, C. Molecular Physiology of Insect Midgut. In Advances in Insect Physiology; Academic Press Inc.: Cambridge, MA, USA, 2019; Volume 56, pp. 117–163. ISBN 9780081028421. [Google Scholar]
- Mukherjee, K.; Grizanova, E.; Chertkova, E.; Lehmann, R.; Dubovskiy, I.; Vilcinskas, A. Experimental Evolution of Resistance against Bacillus thuringiensis in the Insect Model Host Galleria Mellonella Results in Epigenetic Modifications. Virulence 2017, 8, 1618–1630. [Google Scholar] [CrossRef] [Green Version]
- Walther, M.; Schrahn, S.; Krauss, V.; Lein, S.; Kessler, J.; Jenuwein, T.; Reuter, G. Heterochromatin Formation in Drosophila Requires Genome-Wide Histone Deacetylation in Cleavage Chromatin before Mid-Blastula Transition in Early Embryogenesis. Chromosoma 2020, 129, 83–98. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Cheng, D.; Mon, H.; Tatsuke, T.; Zhu, L.; Xu, J.; Lee, J.M.; Xia, Q.; Kusakabe, T. Genome-Wide Identification of Polycomb Target Genes Reveals a Functional Association of Pho with Scm in Bombyx mori. PLoS ONE 2012, 7, e34330. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tong, X.; Li, C.; Lu, K.; Tan, D.; Hu, H.; Liu, H.; Dai, F. Genome-Wide Identification and Expression Profiling of the C2H2-Type Zinc Finger Protein Genes in the Silkworm Bombyx mori. PeerJ 2019, 7, e7222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafeez, M.; Li, X.; Zhang, J.; Zhang, Z.; Huang, J.; Wang, L.; Khan, M.M.; Shah, S.; Fernández-Grandon, G.M.; Lu, Y. Role of Digestive Protease Enzymes and Related Genes in Host Plant Adaptation of a Polyphagous Pest, Spodoptera frugiperda. Insect Sci. 2021, 28, 611–626. [Google Scholar] [CrossRef]
- Golikhajeh, N.; Naseri, B.; Razmjou, J. Geographic Origin and Host Cultivar Influence on Digestive Physiology of Spodoptera exigua (Lepidoptera: Noctuidae) Larvae. J. Insect Sci. 2017, 17, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppold, A.; Kreß, A.; Vanden Bussche, J.; Diogo, J.B.B.; Kuch, U.; Oehlmann, J.; Vandegehuchte, M.B.B.; Müller, R.; Bussche, J.V.; Diogo, J.B.B.; et al. Epigenetic Alterations and Decreasing Insecticide Sensitivity of the Asian Tiger Mosquito Aedes albopictus. Ecotoxicol. Environ. Saf. 2015, 122, 45–53. [Google Scholar] [CrossRef]
- Chittka, A.; Chittka, L. Epigenetics of Royalty. PLoS Biol. 2010, 8, e1000532. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Twyman, R.M.; Vilcinskas, A. Insects as Models to Study the Epigenetic Basis of Disease. Prog. Biophys. Mol. Biol. 2015, 118, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Faulk, C.; Dolinoy, D.C. Timing Is Everything. Epigenetics 2011, 6, 791–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfrey, K.M.; Lillycrop, K.A.; Burdge, G.C.; Gluckman, P.D.; Hanson, M.A. Epigenetic Mechanisms and the Mismatch Concept of the Developmental Origins of Health and Disease. Pediatr. Res. 2007, 61, 5R–10R. [Google Scholar] [CrossRef]
- Broadway, R.M.; Duffey, S.S.; Pearce, G.; Ryan, C.A. Plant Proteinase Inhibitors: A Defense against Herbivorous Insects? Entomol. Exp. Appl. 1986, 41, 33–38. [Google Scholar] [CrossRef]
- Hines, M.E.; Nielsen, S.S.; Shade, R.E.; Pomeroy, M.A. The Effect of Two Proteinase Inhibitors, E-64 and the Bowman-Birk Inhibitor, on the Developmental Time and Mortality of Acanthoscelides obtectus. Entomol. Exp. Appl. 1990, 57, 201–207. [Google Scholar] [CrossRef]
- Johnston, K.A.; Gatehouse, J.A.; Anstee, J.H. Effects of Soybean Protease Inhibitors on the Growth and Development of Larval Helicoverpa armigera. J. Insect Physiol. 1993, 39, 657–664. [Google Scholar] [CrossRef]
- Kunitz, M. Crystallization of a Trypsin Inhibitor from Soybean. Science 1945, 101, 668–669. [Google Scholar] [CrossRef] [PubMed]
- Kuwar, S.S.; Pauchet, Y.; Heckel, D.G. Effects of Class-specific, Synthetic, and Natural Proteinase Inhibitors on Life-history Traits of the Cotton Bollworm Helicoverpa armigera. Arch. Insect Biochem. Physiol. 2020, 103, e21647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, C.A. Protease Inhibitors in Plants: Genes for Improving Defenses Against Insects and Pathogens. Annu. Rev. Phytopathol. 1990, 28, 425–449. [Google Scholar] [CrossRef]
- Clemente, M.; Corigliano, M.; Pariani, S.; Sánchez-López, E.; Sander, V.; Ramos-Duarte, V. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef] [Green Version]
- Hilder, V.A.; Gatehouse, A.M.R.; Sheerman, S.E.; Barker, R.F.; Boulter, D. A Novel Mechanism of Insect Resistance Engineered into Tobacco. Lett. Nat. 1987, 330, 160–163. [Google Scholar] [CrossRef]
- McManus, M.T.; White, D.W.R.; McGregor, P.G. Accumulation of a Chymotrypsin Inhibitor in Transgenic Tobacco Can Affect the Growth of Insect Pests. Transgenic Res. 1994, 3, 50–58. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.; Kumar, S.; Mittal, P.; Singh, I.K. Protease Inhibitors: Recent Advancement in Its Usage as a Potential Biocontrol Agent for Insect Pest Management. Insect Sci. 2020, 27, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Zhang, J.; Lyu, H.; Song, Q.; Feng, Q.; Xiang, H.; Zheng, S. DNA Methylation Mediates BmDeaf1-Regulated Tissue- and Stage-Specific Expression of BmCHSA-2b in the Silkworm, Bombyx mori. Epigenetics Chromatin 2018, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Jayachandran, B.; Hussain, M.; Asgari, S. An Insect Trypsin-like Serine Protease as a Target of MicroRNA: Utilization of MicroRNA Mimics and Inhibitors by Oral Feeding. Insect Biochem. Mol. Biol. 2013, 43, 398–406. [Google Scholar] [CrossRef]
- Lomate, P.R.; Mahajan, N.S.; Kale, S.M.; Gupta, V.S.; Giri, A.P. Identi Fi Cation and Expression pro Fi Ling of Helicoverpa armigera MicroRNAs and Their Possible Role in the Regulation of Digestive Protease Genes. Insect Biochem. Mol. Biol. 2014, 54, 129–137. [Google Scholar] [CrossRef]
- Chandra, G.; Asokan, R.; Manamohan, M.; Ellango, R.; Sharma, H.C.; Akbar, S.M.D.; Krishna Kumar, N.K. Double-Stranded RNA-Mediated Suppression of Trypsin-Like Serine Protease (t-SP) Triggers Over-Expression of Another t-SP Isoform in Helicoverpa armigera. Appl. Biochem. Biotechnol. 2018, 184, 746–761. [Google Scholar] [CrossRef] [PubMed]
- Hummel, B.C.W. A Modified Spectrophotometric Determination of Chymotrypsin, Trypsin, And Thrombin. Can. J. Biochem. Physiol. 1959, 37, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Mihsfeldt, L.H.; Parra, J.R.P. Biologia de Tuta absoluta (Meyrick, 1917) Em Dieta Artificial. Sci. Agric. 1999, 56, 769–776. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, L.R.; Dias, R.O.; Fuzita, F.J.; Ferreira, C.; Terra, W.R.; Silva-Filho, M.C. The Evolution, Gene Expression Profile, and Secretion of Digestive Peptidases in Lepidoptera Species. Catalysts 2020, 10, 217. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. Genome-Wide High-Resolution Mapping of DNA Methylation Identifies Epigenetic Variation across Embryo and Endosperm in Maize (Zea may). BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI Bioinformatics Web and Programmatic Tools Framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Dahiya, R. MethPrimer: Designing Primers for Methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [Green Version]
- Rohde, C.; Zhang, Y.; Reinhardt, R.; Jeltsch, A. BISMA–Fast and Accurate Bisulfite Sequencing Data Analysis of Individual Clones from Unique and Repetitive Sequences. BMC Bioinform. 2010, 11, 230. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and Validation of Reference Genes for Normalization of Gene Expression Analysis Using Qrt-Pcr in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Van den Hoff, M.J.B.; Moorman, A.F.M. Amplification Efficiency: Linking Baseline and Bias in the Analysis of Quantitative PCR Data. Nucleic Acids Res. 2009, 37, 45. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, 2001–2007. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, E.; Moharramipour, S.; Asgari, S.; Mehrabadi, M. Induction of DNA methyltransferase genes in Helicoverpa armigera following injection of pathogenic bacteria modulates expression of antimicrobial peptides and affects bacterial proliferation. J. Insect Physiol. 2019, 118, 103939. [Google Scholar] [CrossRef] [PubMed]
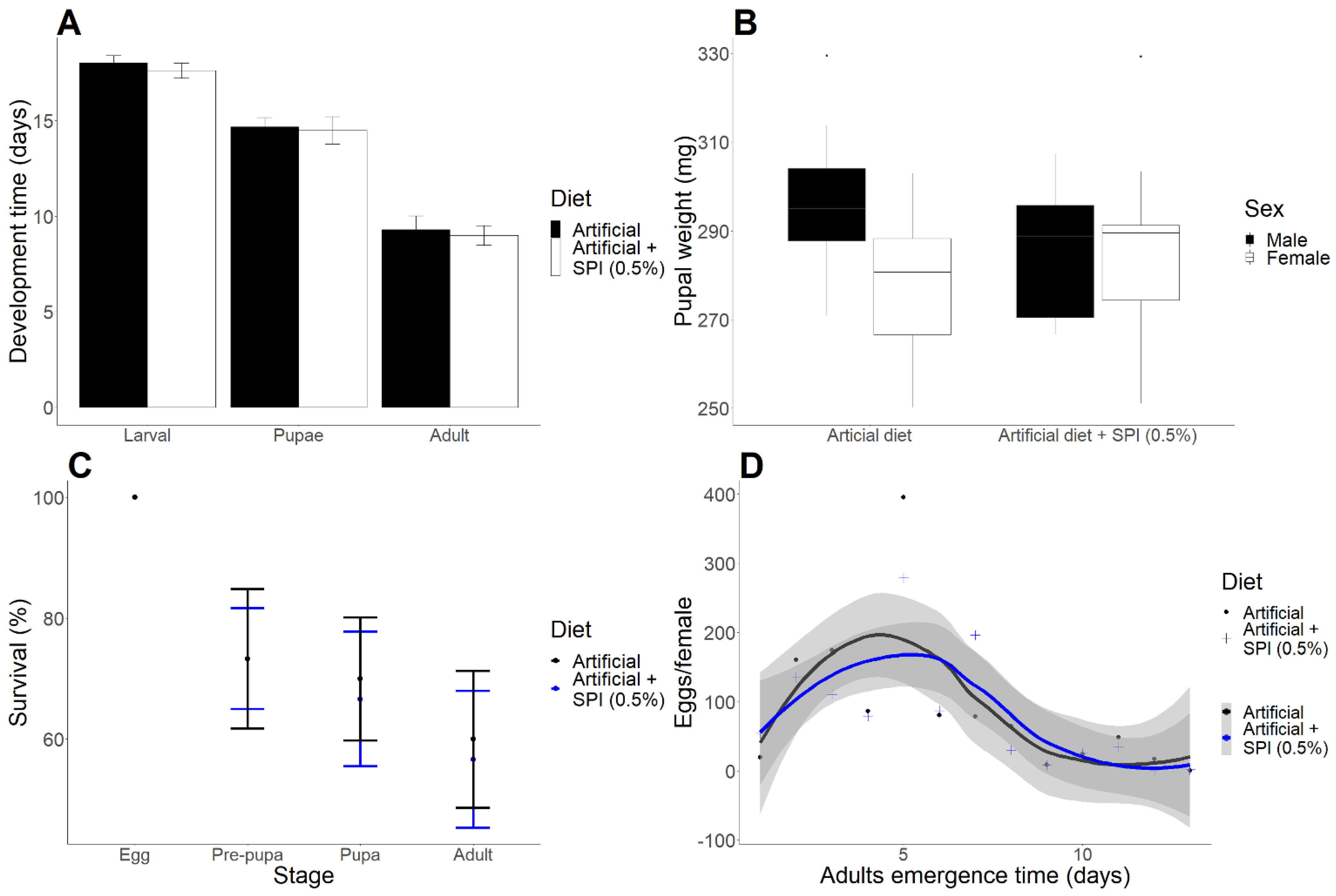
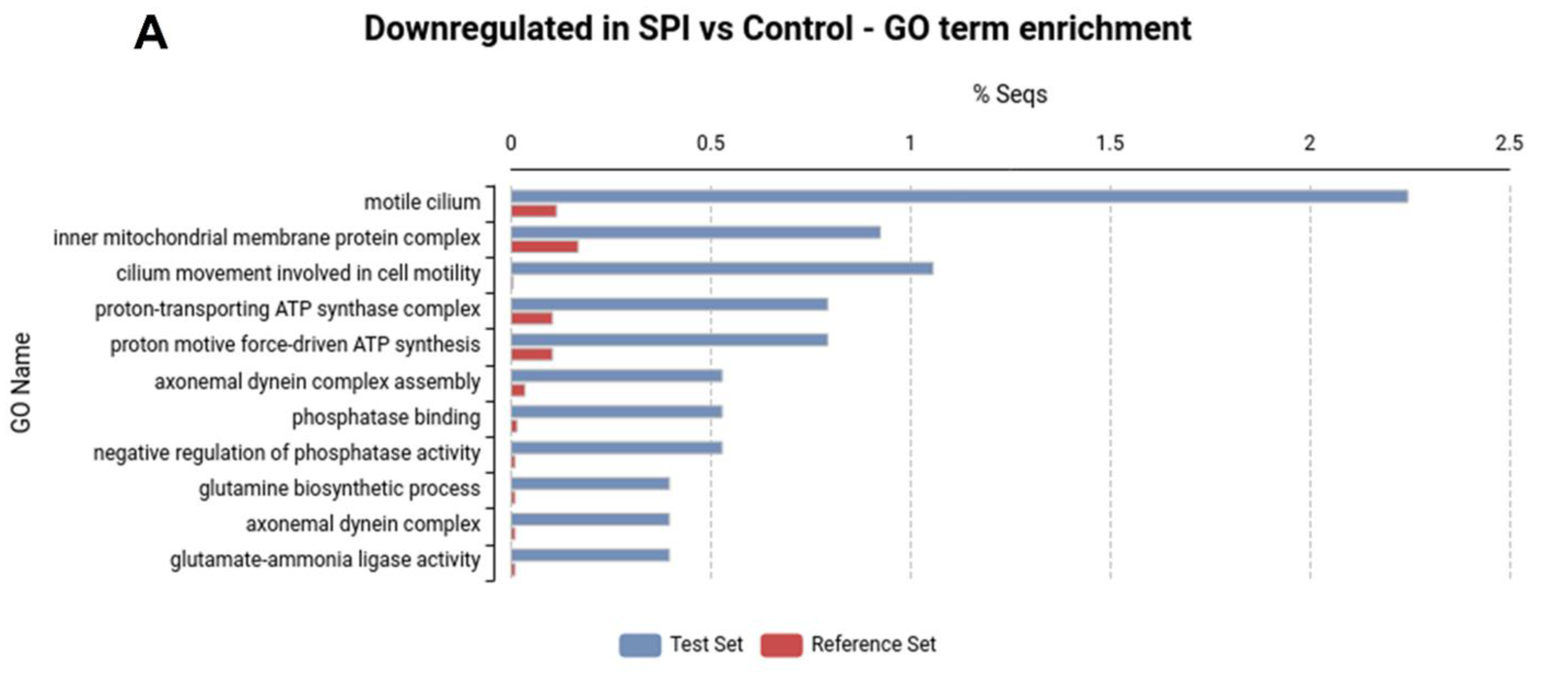
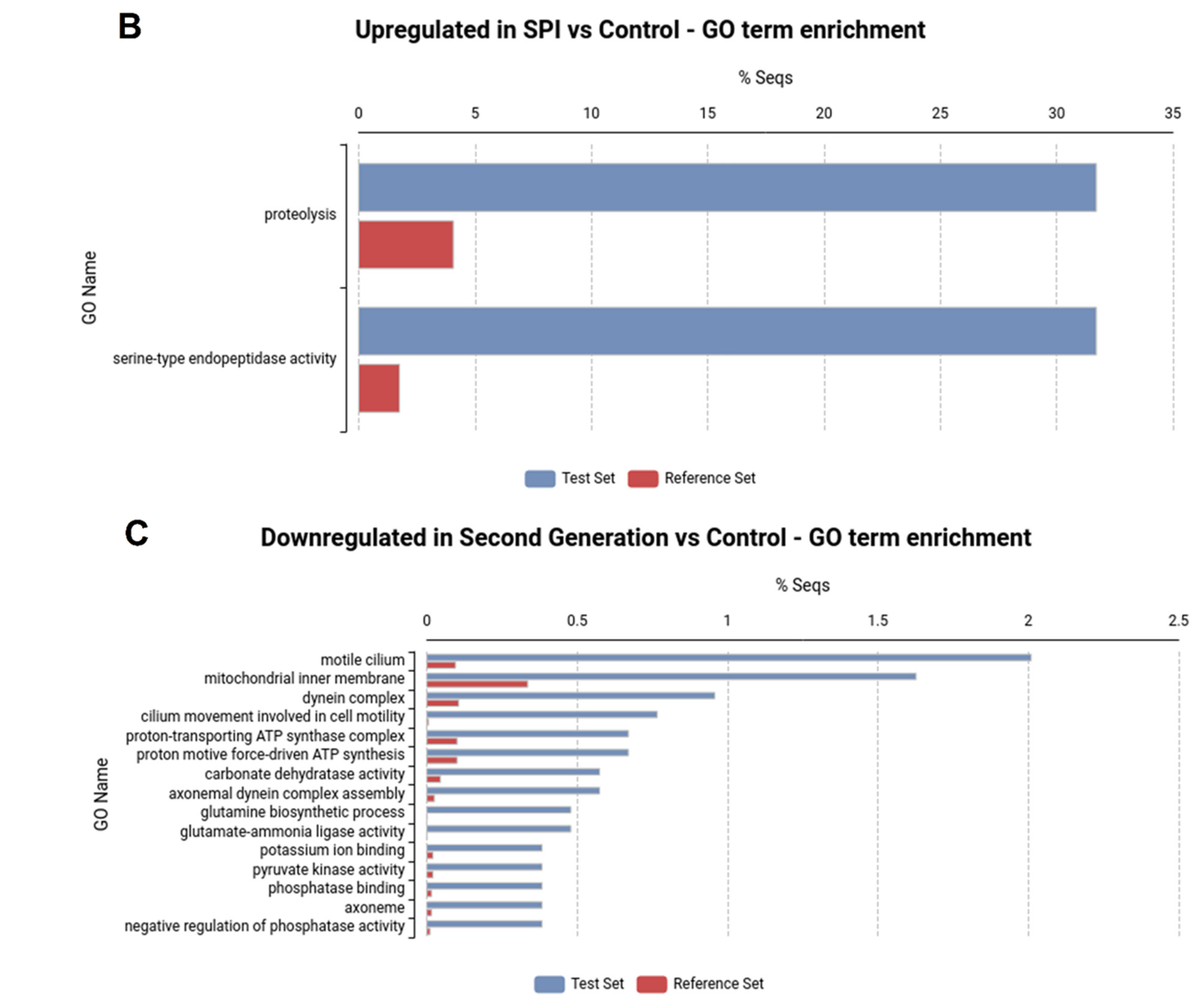

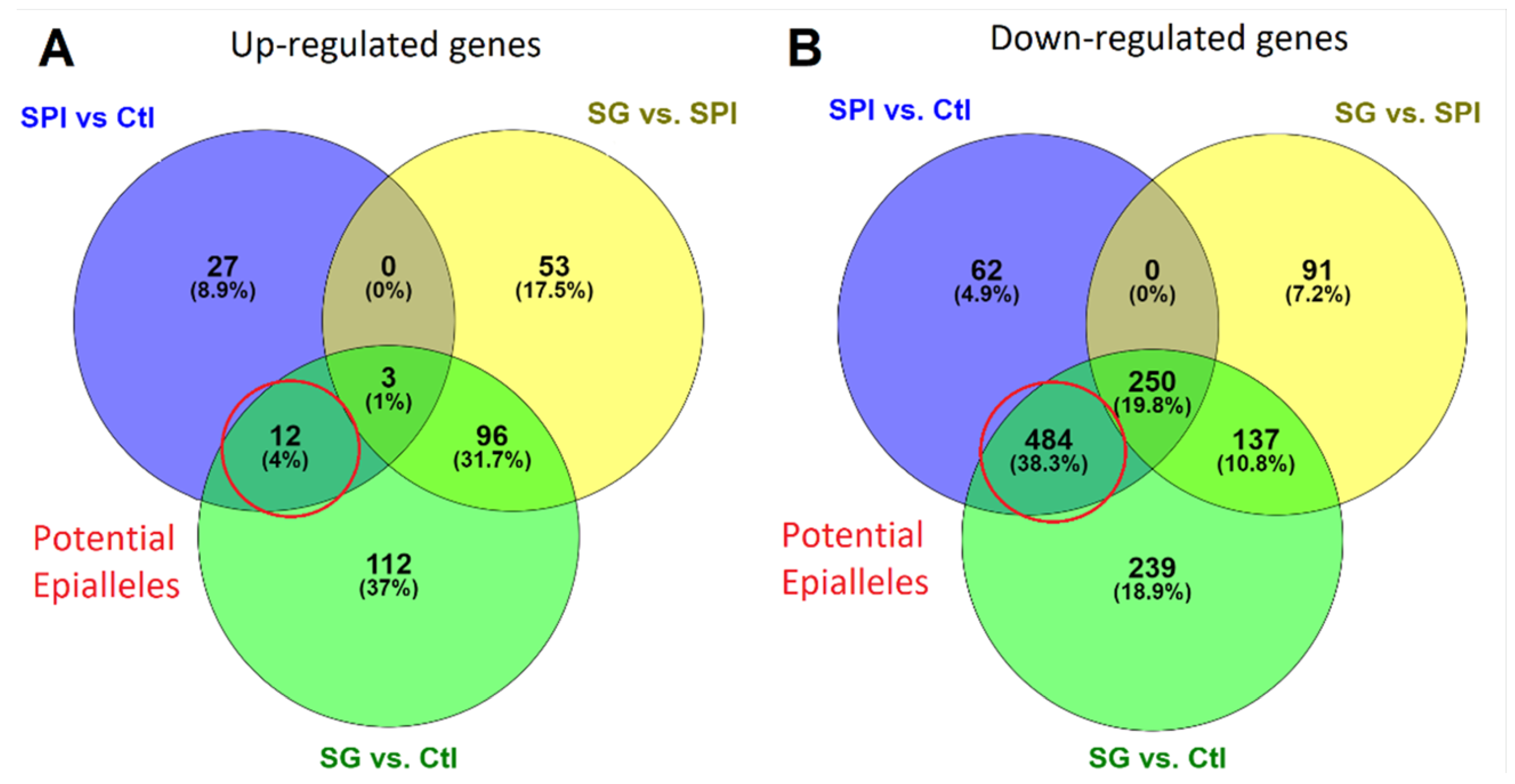
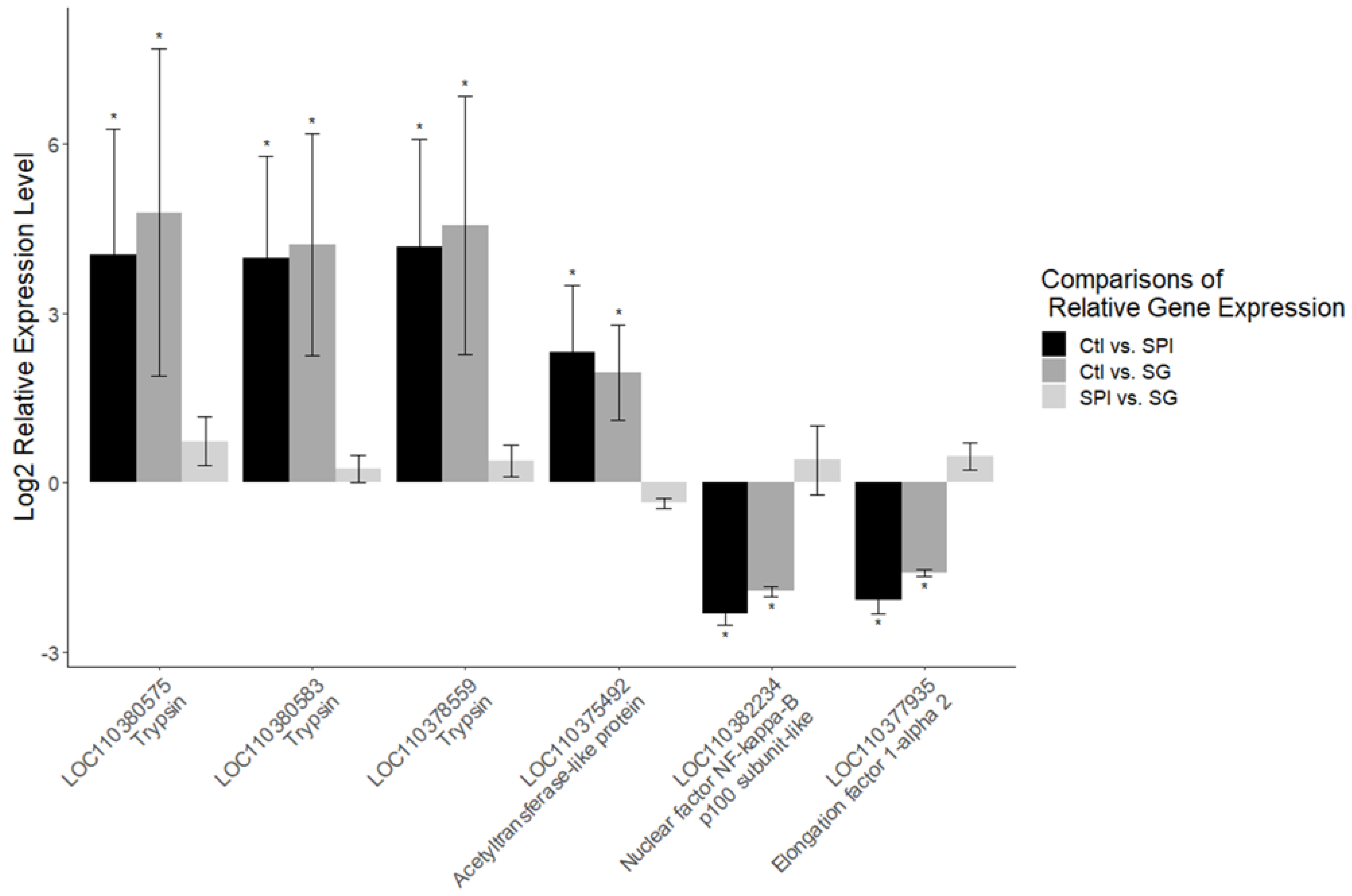
| Locus | SPI vs. Ctl | SG vs. Ctl | Protein Name | ||
|---|---|---|---|---|---|
| Log2 Fold-Change | (p-Adjust < 0.05) | Log2 Fold-Change | (p-Adjust < 0.05) | ||
| LOC110370568 | 1.03 | 0.002 | 1.76 | 0.000 | Diuretic hormone |
| LOC110371086 | 1.03 | 0.000 | 2.48 | 0.000 | Uricase |
| LOC110372133 | 2.03 | 0.000 | 1.50 | 0.012 | Probable aldehyde oxidase 2 |
| LOC110375492 | 1.16 | 0.012 | 1.81 | 0.000 | Uncharacterized protein LOC110375492 |
| LOC110377152 | 1.13 | 0.020 | 1.59 | 0.001 | Uncharacterized protein LOC110377152 |
| LOC110378559 | 4.75 | 0.036 | 6.43 | 0.003 | Trypsin, alkaline C-like |
| LOC110378803 | 1.37 | 0.035 | 1.47 | 0.025 | Gloverin-like |
| LOC110379025 | 1.90 | 0.000 | 1.48 | 0.009 | Trypsin CFT-1-like, partial |
| LOC110380575 | 1.33 | 0.001 | 1.40 | 0.001 | Trypsin, alkaline C-like |
| LOC110380583 | 1.59 | 0.000 | 1.90 | 0.000 | Trypsin CFT-1-like |
| LOC110383800 | 1.87 | 0.000 | 1.69 | 0.001 | Lipoprotein lipase-like |
| LOC110384494 | 1.13 | 0.022 | 1.08 | 0.040 | Serine proteases, trypsin domain (IPR001254) |
| Locus | Ctl vs. SPI | qvalue | Protein | Ctl vs. SG | qvalue |
|---|---|---|---|---|---|
| LOC110384175 | −28.2 | 0.01 | Uncharacterized LOC110384175 | −29.0 | 0.00 |
| LOC110369926 | −30.5 | 0.00 | Pre-rRNA processing protein FTSJ3 | −40.7 | 0.00 |
| LOC110369936 | −29.8 | 0.01 | A-kinase anchor protein 10, mitochondrial | −29.6 | 0.00 |
| LOC110370916 | −34.3 | 0.00 | Cytochrome b-c1 complex subunit 6, mitochondrial-like | −26.5 | 0.01 |
| LOC110371715 | −32.4 | 0.00 | Microprocessor complex subunit DGCR8, transcript variant X1 | −28.6 | 0.00 |
| LOC110372825 | −39.1 | 0.00 | THO complex subunit 4 | −35.0 | 0.00 |
| LOC110372886 | −31.5 | 0.01 | Beta-arrestin−1, transcript variant X1 | −28.3 | 0.00 |
| LOC110374361 | −33.5 | 0.00 | Regulator of nonsense transcripts 1 homolog | −31.4 | 0.00 |
| LOC110374662 | −36.2 | 0.00 | nudC domain-containing protein 1 | −30.2 | 0.00 |
| LOC110374778 | −35.9 | 0.01 | Acetyl-coenzyme A transporter 1 | 32.9 | 0.00 |
| LOC110375095 | 34.5 | 0.00 | Uncharacterized LOC110375095 | 31.5 | 0.00 |
| LOC110375190 | 38.0 | 0.00 | Uncharacterized LOC110375190 | 28.6 | 0.01 |
| LOC110375794 | 37.3 | 0.00 | Voltage-dependent anion-selective channel-like | 32.3 | 0.00 |
| LOC110377217 | −25.2 | 0.01 | Uncharacterized LOC110377217 | −32.9 | 0.00 |
| LOC110380153 | −25.6 | 0.00 | Uncharacterized LOC110380153 | −25.6 | 0.00 |
| LOC110381188 | −29.3 | 0.00 | Uncharacterized LOC110381188 | 25.4 | 0.00 |
| LOC110382008 | 30.0 | 0.01 | Zinc finger protein 2-like | 30.7 | 0.00 |
| LOC110383013 | 34.6 | 0.01 | Thioredoxin-related transmembrane protein 2 homolog, transcript variant X1 | 27.9 | 0.00 |
| LOC110383900 | −27.7 | 0.00 | Thiamine transporter 2-like | −30.8 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velasquez-Vasconez, P.A.; Hunt, B.J.; Dias, R.O.; Souza, T.P.; Bass, C.; Silva-Filho, M.C. Adaptation of Helicoverpa armigera to Soybean Peptidase Inhibitors Is Associated with the Transgenerational Upregulation of Serine Peptidases. Int. J. Mol. Sci. 2022, 23, 14301. https://doi.org/10.3390/ijms232214301
Velasquez-Vasconez PA, Hunt BJ, Dias RO, Souza TP, Bass C, Silva-Filho MC. Adaptation of Helicoverpa armigera to Soybean Peptidase Inhibitors Is Associated with the Transgenerational Upregulation of Serine Peptidases. International Journal of Molecular Sciences. 2022; 23(22):14301. https://doi.org/10.3390/ijms232214301
Chicago/Turabian StyleVelasquez-Vasconez, Pedro A., Benjamin J. Hunt, Renata O. Dias, Thaís P. Souza, Chris Bass, and Marcio C. Silva-Filho. 2022. "Adaptation of Helicoverpa armigera to Soybean Peptidase Inhibitors Is Associated with the Transgenerational Upregulation of Serine Peptidases" International Journal of Molecular Sciences 23, no. 22: 14301. https://doi.org/10.3390/ijms232214301
APA StyleVelasquez-Vasconez, P. A., Hunt, B. J., Dias, R. O., Souza, T. P., Bass, C., & Silva-Filho, M. C. (2022). Adaptation of Helicoverpa armigera to Soybean Peptidase Inhibitors Is Associated with the Transgenerational Upregulation of Serine Peptidases. International Journal of Molecular Sciences, 23(22), 14301. https://doi.org/10.3390/ijms232214301







