The Role of the Gallbladder, the Intestinal Barrier and the Gut Microbiota in the Development of Food Allergies and Other Disorders
Abstract
:1. Introduction
2. The Human Gut and Gallbladder
3. Microorganisms Colonizing the Gallbladder, Bile and Intestinal Lumen
4. Gallbladder and Infectious Diseases
5. Gut Microorganisms and Allergies
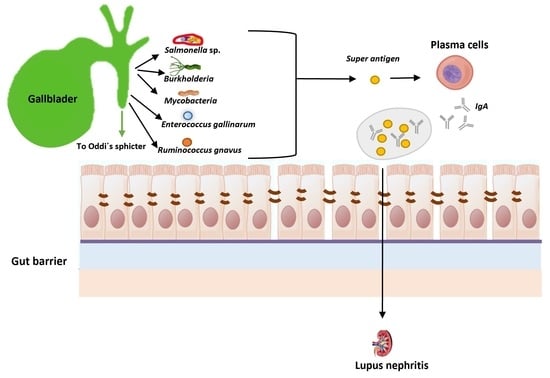
5.1. Systemic Lupus Erythematosus
5.2. Leaky Gut Syndrome
5.3. Polyamines and Food Allergies
5.4. The Gut Microbiota Can Prevent Food Allergies and Help Maintain Oral Tolerance
6. Omic Approaches Applied to the Study of Gallbladder Metagenome, Proteome, Transcriptome and Metabolome
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calo-Mata, P.; Ageitos, J.M.; Böhme, K.; Barros-Velázquez, J. Intestinal Microbiota: First Barrier Against Gut-Affecting Pathogens. In New Weapons to Control Bacterial Growth; Villa, T., Vinas, M., Eds.; Springer: Cham, Switzerland, 2016; pp. 281–314. [Google Scholar]
- Villa, T.G.; Sánchez-Pérez, A. The Gut Microbiome Affects Human Mood and Behavior. In Developmental Biology in Prokaryotes and Lower Eukaryotes; Villa, T.G., de Miguel Bouzas, T., Eds.; Springer: Cham, Switzerland, 2016; pp. 541–565. [Google Scholar]
- Villa, T.G.; Feijoo-Siota, L.; Rama, J.L.R.; Sánchez-Pérez, A.; de Miguel-Bouzas, T. Fecal Matter Implantation as a Way to Fight Diarrhea-Causing Microorganisms. In New Weapons to Control Bacterial Growth; Villa, T., Vinas, M., Eds.; Springer: Cham, Switzerland, 2016; pp. 315–352. [Google Scholar]
- Hernandez-Sanabria, E.; Vázquez-Castellanos, J.F.; Raes, J. In vitro ecology: A discovery engine for microbiome therapies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 711–712. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Nance, C.L.; Deniskin, R.; Diaz, V.C.; Paul, M.; Anvari, S.; Anagnostou, A. The Role of the Microbiome in Food Allergy: A Review. Children 2020, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Berin, M.C. Dysbiosis in food allergy and implications for microbial therapeutics. J. Clin. Investig. 2021, 131, e144994. [Google Scholar] [CrossRef]
- Gasbarrini, G.B.; Mangiola, F.; Gerardi, V.; Ianiro, G.; Corazza, G.R.; Gasbarrini, A. Coeliac disease: An old or a new disease? History of a pathology. Intern. Emerg. Med. 2014, 9, 249–256. [Google Scholar] [CrossRef]
- Katzka, D.A. Eosinophilic Esophagitis: Leaky Gullet or Leaky Gut? Am. J. Gastroenterol. 2017, 112, 1072–1073. [Google Scholar] [CrossRef]
- Mehr, S.; Kakakios, A.; Frith, K.; Kemp, A.S. Food protein-induced enterocolitis syndrome: 16-year experience. Pediatrics 2009, 123, e459–e464. [Google Scholar] [CrossRef]
- Michelet, M.; Schluckebier, D.; Petit, L.M.; Caubet, J.C. Food protein-induced enterocolitis syndrome-a review of the literature with focus on clinical management. J. Asthma Allergy 2017, 10, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Nowak-Węgrzyn, A.; Jarocka-Cyrta, E.; Castro, M.A. Food Protein-Induced Enterocolitis Syndrome. J. Investig. Allergol. Clin. Immunol. 2017, 27, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Lake, A.M. Food-Induced Eosinophilic Proctocolitis. J. Ped. Gastroenterol. Nutr. 2000, 30, 558–560. [Google Scholar] [CrossRef]
- Boné, J.; Claver, A.; Guallar, I.; Plaza, A.M. Allergic proctocolitis, food-induced enterocolitis: Immune mechanisms, diagnosis and treatment. Allerg. Immunopathol. 2009, 37, 36–42. [Google Scholar] [CrossRef]
- Bengmark, S. Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut 1998, 42, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Gerritsen, J.; Smidt, H.; Rijkers, G.T.; de Vos, W.M. Intestinal microbiota in human health and disease: The impact of probiotics. Genes Nutr. 2011, 6, 209–240. [Google Scholar] [CrossRef] [Green Version]
- Fernández, L.; Langa, S.; Martín, V.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The microbiota of human milk in healthy women. Cell. Mol. Biol. 2013, 59, 31–42. [Google Scholar]
- Molinero, N.; Ruiz, L.; Milani, C.; Gutiérrez-Díaz, I.; Sánchez, B.; Mangifesta, M.; Segura, J.; Cambero, I.; Campelo, A.B.; García-Bernardo, C.M.; et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 2019, 7, 100. [Google Scholar] [CrossRef] [Green Version]
- Nichols, H.J.; Stimmel, C.O. Protection against typhoid-like infections by vaccination. An experimental study. J. Exp. Med. 1923, 38, 283–290. [Google Scholar] [CrossRef]
- Saunders, W.E. On Cholera. Indian Med. Gazette 1887, 22, 360–365. [Google Scholar]
- Lee, J.Y.; Keane, M.G.; Pereira, S. Diagnosis and treatment of gallstone disease. Practitioner 2015, 259, 15–19. [Google Scholar] [PubMed]
- Kim, H.; Kwack, K.; Kim, D.-Y.; Ji, G.E. Oral probiotic bacterial administration suppressed allergic responses in an oval-bumin-induced allergy mouse model. FEMS. Immunol. Med. Microbiol. 2005, 45, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tan, W.-H.; Wu, J.-C.; Huang, Z.-X.; Shang, Y.-Y.; Liang, B.; Chen, J.-H.; Pang, R.; Xie, X.-Q.; Zhang, J.-M.; et al. Microbiologic risk factors of recurrent choledocholithiasis post-endoscopic sphincter-otomy. World J. Gastroenterol. 2022, 28, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Ivy, A.C.; Oldberg, E.A. hormone mechanism for gall-bladder contraction and evacuation. Am. J. Physiol. 1928, 86, 599–613. [Google Scholar] [CrossRef]
- Rehfeld, J.F. Cholecystokinin and the hormone concept. Endocr. Connect. 2021, 10, R139–R150. [Google Scholar] [CrossRef]
- Crawford, R.W.; Rosales-Reyes, R.; de Ramirez-Aguilar, M.L.; Chapa-Azuela, O.; Alpuche-Aranda, C.; Gunn, J.S. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. USA 2010, 107, 4353–4358. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and acute infection of the gall bladder by Salmonella Typhi: Un-derstanding the carrier state. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [Green Version]
- Shukla, V.K.; Tiwari, S.C.; Roy, S.K. Biliary bile acids in cholelithiasis and carcinoma of the gall bladder on JSTOR. Eur. J. Cancer Prev. 1993, 2, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Chauhan, V.S.; Nath, G.; Kumar, A.; Shukla, V.K. Role of bile bacteria in gallbladder carcinoma. Hepatogastroenterology 2007, 54, 1622–1625. [Google Scholar] [PubMed]
- Wu, T.; Zhang, Z.; Liu, B.; Hou, D.; Liang, Y.; Zhang, J.; Shi, P. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genom. 2013, 14, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Yan, Q.; Luo, F.; Shang, D.; Wu, D.; Zhang, H.; Shang, X.; Kang, X.; Abdo, M.; Liu, B.; et al. Acute cholecystitis associated with infection of Enterobacteriaceae from gut microbiota. Clin. Microbiol. Infect. 2015, 21, 851–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Fu, S.W.; Lu, L.; Zhao, H. A Preliminary Study of Biliary Microbiota in Patients with Bile Duct Stones or Distal Cholangiocarcinoma. Biomed. Res. Int. 2019, 2019, 1092563. [Google Scholar] [CrossRef] [Green Version]
- Saab, M.; Mestivier, D.; Sohrabi, M.; Rodriguez, C.; Khonsari, M.R.; Faraji, A.; Sobhani, I. Characterization of biliary microbiota dysbiosis in extrahepatic cholangiocarcinoma. PLoS ONE 2021, 16, e0247798. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Hagey, L.R.; Krasowski, M.D. Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 2010, 51, 226–246. [Google Scholar] [CrossRef] [Green Version]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [Green Version]
- Hamer, H.M.; De Preter, V.; Windey, K.; Verbeke, K. Functional analysis of colonic bacterial metabolism: Relevant to health? Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1–G9. [Google Scholar]
- Hill, M.J. Bile flow and colon cancer. Mutat. Res. 1990, 238, 313–320. [Google Scholar] [CrossRef]
- Degirolamo, C.; Modica, S.; Palasciano, G.; Moschetta, A. Bile acids and colon cancer: Solving the puzzle with nuclear re-ceptors. Trends Mol. Med. 2011, 17, 564–572. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremers, C.M.; Knoefler, D.; Vitvitsky, V.; Banerjee, R.; Jakoba, U. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, E1610–E1619. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, D.; Schuhmacher, D.A.; Barker, J.L.; Klose, K.E. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 2000, 68, 1491–1497. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Linke, K.; Jatzek, A.; Jakob, U. Severe oxidative stress causes inactivation of DnaK and activation of the re-dox-regulated chaperone Hsp33. Mol. Cell. 2005, 17, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Wholey, W.Y.; Jakob, U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol. Microbiol. 2012, 83, 981–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, D.E.; Siddique, S.S.; Weinstock, J.V. Innate immunity in disease. Clin. Gastroenterol. Hepatol. 2014, 12, 749–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kortman, G.A.; Boleij, A.; Swinkels, D.W.; Tjalsma, H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS ONE 2012, 7, e29968. [Google Scholar] [CrossRef]
- Sistrunk, J.R.; Nickerson, K.; Chanin, R.B.; Rasko, D.A.; Faherty, C.S. Survival of the Fittest: How Bacterial Pathogens Utilize Bile to Enhance Infection. Clin. Microbiol. Rev. 2016, 29, 819–836. [Google Scholar] [CrossRef] [Green Version]
- Miller, V.R.; Banwart, G.J. Effect of various concentrations of brilliant green and bile salts on Salmonellae and other microorganisms. Appl. Microbiol. 1965, 13, 77–80. [Google Scholar] [CrossRef]
- Bernstein, C.; Bernstein, H.; Payne, C.M.; Beard, S.E.; Schneider, J. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 1999, 39, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Kus, J.V.; Gebremedhin, A.; Dang, V.; Tran, S.L.; Serbanescu, A.; Foster, D.B. Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2011, 193, 4509–4515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provenzano, D.; Lauriano, C.M.; Klose, K.E. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 2001, 183, 3652–3662. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Oliver, J.D.; Wong, H.C. Adaptation of Vibrio vulnificus and an rpoS mutant to bile salts. Int. J. Food Microbiol. 2010, 140, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Chaudhuri, S.; Saha, G.; Gupta, S.; Chowdhury, R. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 2004, 186, 6809–6814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerda-Maira, F.A.; Ringelberg, C.S.; Taylor, R.K. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 2008, 190, 7441–7452. [Google Scholar] [CrossRef] [Green Version]
- Bina, J.E.; Provenzano, D.; Wang, C.; Bina, X.R.; Mekalanos, J.J. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 2006, 186, 171–181. [Google Scholar] [CrossRef]
- Herrera, C.M.; Crofts, A.A.; Henderson, J.C.; Pingali, S.C.; Davies, B.W.; Trent, M.S. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio 2014, 5, e02283-14. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, T.; Ogawa, W.; Tsuchiya, T.; Kuroda, T. Overexpression of vmeTUV encoding a multidrug efflux transporter of Vibrio parahaemolyticus causes bile acid resistance. Gene 2014, 541, 19–25. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS. Microbiol. Rev. 2005, 29, 625–651. [Google Scholar]
- Lahiri, A.; Ananthalakshmi, T.K.; Nagarajan, A.G.; Ray, S.; Chakravortty, D. TolA mediates the differential detergent re-sistance pattern between the Salmonella enterica subsp. enterica serovars Typhi and Typhimurium. Microbiology 2011, 157, 1402–1415. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.I.; Ramos-Morales, F.; Casadesus, J. Bile-induced DNA damage in Salmonella enterica. Genetics 2004, 168, 1787–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, A.I.; Ramos-Morales, F.; Casadesus, J. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 2006, 174, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaido, E.; Yamaguchi, A.; Nishino, K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhi-murium by RamA in response to environmental signals. J. Biol. Chem. 2008, 283, 24245–24253. [Google Scholar] [CrossRef]
- Nishino, K.; Latifi, T.; Groisman, A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef]
- Lin, J.; Sahin, O.; Michel, L.O.; Zhang, Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 2003, 71, 4250–4259. [Google Scholar] [CrossRef] [Green Version]
- Manfredi, R.; Nanetti, A.; Ferri, M.; Chiodo, F. Pseudomonas spp. complications in patients with HIV disease: An eight- year clinical and microbiological survey. Eur. J. Epidemiol. 2000, 16, 111–118. [Google Scholar] [CrossRef]
- Magner, W.; Hutcheson, J.M. Cholecystitis, bacteriological and experimental study. Can. Med. Assoc. J. 1932, 27, 469–477. [Google Scholar]
- Hill, M.J. Action of Bile Salts on Bacterial Cell Walls. Nature 1967, 214, 1152–1154. [Google Scholar] [CrossRef]
- Köhler, W. The present state of species within the genera Streptococcus and Enterococcus. Int. J. Med. Microbiol. 2007, 297, 133–150. [Google Scholar] [CrossRef]
- Demarne, Y.; Corring, T.; Pihet, A.; Sacquet, E. Fat absorption in germ-free and conventional rats artificially deprived of bile secretion. Gut 1982, 23, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Graber, C.D.; O’Neal, R.M.; Rabin, E.R. Effect of high fat diets on intestinal microflora and serum cholesterol in rats. J. Bacteriol. 1965, 89, 47–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drasar, B.S.; Crowther, J.S.; Goddard, P.; Hawksworth, G.; Hill, M.J.; Peach, S.; Williams, R.E.O. The relation between diet and the gut microflora in man. Proc. Nutr. Soc. 1973, 32, 49–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.J. The effect of some factors on the faecal concentration of acid steroids, neutral steroids and urobilins. J. Pathol. 1971, 104, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Salter, D.N.; Fulford, R.J. The influence of the gut microflora on the digestion of dietary and endogenous proteins: Studies of the amino acid composition of the excreta of germ-free and conventional chicks. Br. J. Nutr. 1974, 32, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R. Dietary modulation of the human gut microflora using prebiotics. Br. J. Nutr. 1998, 80, S209–S212. [Google Scholar] [CrossRef] [Green Version]
- Parola, P.; Anani, H.; Eldin, C.; Dubourg, G.; Lagier, J.C.; Levasseur, A.; Casalta, J.P.; Raoult, D.; Fournier, P.E. Case Report: Vibrio cholerae Biliary Tract Infections in Two North Africans in France. Am. J. Trop. Med. Hyg. 2020, 102, 1306–1308. [Google Scholar] [CrossRef]
- Robson, M. diseases of the gall-bladder and bile-ducts, and on gall-stones. Glasgow Med. J. 1909, 71, 133–149. [Google Scholar]
- Parker, C.T. I: The Complications of Cholelithiasis. Ann. Surg. 1893, 17, 639–651. [Google Scholar]
- McMaster, P.D.; Elman, R. Studies on urobilin physiology and pathology. VI. The relation of biliary infections to the genesis and excretion of urobilin. J. Exp. Med. 1926, 43, 753–783. [Google Scholar] [CrossRef] [Green Version]
- Moffitt, H.C. Clinical Features of Gallbladder and Gall Duct Affections. Cal. State J. Med. 1905, 3, 77–280. [Google Scholar]
- De Takats, G.; Mackenzie, W.D. Acute Pancreatic Necrosis and Its Sequelæ. A Critical Study of Thirty Cases. Ann. Surg. 1932, 96, 418–440. [Google Scholar] [CrossRef] [PubMed]
- Bree, R.L. Further observations on the usefulness of the sonographic Murphy’s sign in the evaluation of suspected acute cholecystitis. J. Clin. Ultrasound 1995, 23, 169–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, G.M. Gas Bacillus Infection. Cal. State J. Med. 1915, 13, 260–265. [Google Scholar]
- Lipshutz, B. Acute cholecystitis. Ann. Surg. 1935, 101, 902–911. [Google Scholar] [CrossRef]
- Eusterman, G.B. The Essential Factors in the Diagnosis of Chronicand Duodenal Ulcers. J. Am. Med. Assoc. 1915, 18, 1500–1503. [Google Scholar] [CrossRef] [Green Version]
- Crispin, E.L. Toxic gastric hemorrhage. Cal. State J. Med. 1917, 15, 308–312. [Google Scholar]
- Judd, E.S. Surgery of the Gallbladder and Biliary Ducts. Can. Med. Assoc. J. 1921, 11, 929–933. [Google Scholar] [CrossRef] [Green Version]
- Weiner, S.; Gramatica, L.; Voegle, L.D.; Hauman, R.L.; Anderson, M.C. Role of the lymphatic system in the pathogenesis of inflammatory disease in the biliary tract and páncreas. Am. J. Surg. 1970, 199, 55–61. [Google Scholar] [CrossRef]
- Glenn, F. Surgery of the gallbladder and biliary tract. Ann. Surg. 1936, 103, 77–85. [Google Scholar] [CrossRef]
- Han, P.; Gu, J.-Q.; Li, L.-S.; Wang, X.-Y.; Wang, H.-T.; Wang, Y.; Chang, C.; Sun, J.-L. The Association Between Intestinal Bacteria and Allergic Diseases—Cause or Consequence? Front. Cell. Infect. Microbiol. 2021, 11, 650893. [Google Scholar]
- Ramachandran, M.; Aronson, J.K. John Bostock’s first description of hayfever. J. R. Soc. Med. 2011, 104, 237–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizaka, K.; Ishizaka, T.; Hornbrook, M.M. Physicochemical properties of reaginic antibody V. Correlation of reaginic activity with ϒE-globulin antibody. J. Immunol. 1966, 97, 840–853. [Google Scholar] [PubMed]
- Watanabe, N.; Bruschi, F.; Korenaga, M. IgE: A question of protective immunity in Trichinella spiralis infection. Trends Parasitol. 2005, 21, 175–178. [Google Scholar] [CrossRef]
- Dworetzky, M.; Cohen, S.G.; Ed Frankland, A.W. The allergy archives. J. Allerg. Clin. Immunol. 2003, 111, 1142–1150. [Google Scholar] [CrossRef]
- Rank, M.A.; Li, J.T. Allergen immunotherapy. Mayo Clin. Proc. 2007, 82, 1119–1123. [Google Scholar] [CrossRef] [Green Version]
- Abramson, M.J.; Puy, R.M.; Weiner, J.M. Injection allergen immunotherapy for asthma. Cochrane Database Syst. Rev. 2010, 8, CD001186. [Google Scholar] [CrossRef]
- Fyhrquist, N. The Human Microbiota and Its Relationship with Allergies. Gastroenterol. Clin. North Am. 2019, 48, 377–387. [Google Scholar]
- Ipci, K.; Altıntoprak, N.; Muluk, N.B.; Senturk, M.; Cingi, C. The possible mechanisms of the human microbiome in allergic diseases. Eur. Arch. Otorhinolaryngol. 2017, 274, 617–626. [Google Scholar] [CrossRef]
- Pan, Q.; Guo, F.; Huang, Y.; Li, A.; Chen, S.; Chen, J.; Liu, H.-F.; Pan, Q. Gut Microbiota Dysbiosis in Systemic Lupus Ery-thematosus: Novel Insights into Mechanisms and Promising Therapeutic Strategies. Front. Immunol. 2021, 12, 799788. [Google Scholar] [CrossRef]
- Murphy, G.; Isenberg, D. Effect of gender on clinical presentation in systemic lupus erythematosus. Rheumatology 2013, 52, 2108–2115. [Google Scholar] [CrossRef] [Green Version]
- Lisnevskaia, L.; Murphy, G.; Isenberg, D. Systemic lupus erythematosus. Lancet 2014, 384, 1878–1888. [Google Scholar] [CrossRef]
- Neuman, H.; Koren, O. The gut microbiota: A possible factor influencing systemic lupus erythematosus. Curr. Opin. Rheumatol. 2017, 29, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Liu, T.; Zhao, M.; Dang, X.; Feng, S.; Ding, X.; Xu, Z.; Huang, X.; Lin, Q.; Xiang, W.; et al. Correlation Analysis between Gut Microbiota and Metabolites in Children with Systemic Lupus Erythematosus. Immunol. Res. 2021, 2021, 5579608. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; López, P.; Suárez, A.; Sánchez, B.; Margolles, A. The role of gut microbiota in lupus: What we know in 2018? Expert. Rev. Clin. Immunol. 2018, 14, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, R.; Jacob, A.; Wlaschin, J.; McGregor, M.; Quigg, R.J.; Alexander, J.J. Lupus: The microbiome angle. Immunobiology 2018, 223, 460–465. [Google Scholar] [CrossRef]
- Zhang, W.; Reichlin, M.A. Possible Link Between Infection with Burkholderia Bacteria and Systemic Lupus Erythematosus Based on Epitope Mimicry. Clin. Dev. Immunol. 2008, 2008, 683489. [Google Scholar] [CrossRef] [Green Version]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell. Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef] [Green Version]
- Van Itallie, C.M.; Anderson, J.M. Claudin interactions in and out of the tight junction. Tissue Barriers 2013, 1, e25247. [Google Scholar] [CrossRef] [Green Version]
- Furuse, M.; Hirase, T.; Itoh, M.; Nagafuchi, A.; Yonemura, S.; Tsukita, S.; Tsukita, S. Occludin: A novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993, 123, 1777–1788. [Google Scholar] [CrossRef]
- Fasano, A.; Baudry, B.; Pumplin, D.; Wasserman, S.S.; Tall, B.D.; Ketley, J.M.; Kaper, J.B. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. USA 1991, 88, 5242–5246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanuytsel, T.; Vermeire, S.; Cleynen, I. The role of Haptoglobin and its related protein, Zonulin, in inflammatory bowel disease. Tissue Barriers 2013, 1, e27321. [Google Scholar] [CrossRef] [PubMed]
- El Asmar, R.; Panigrahi, P.; Bamford, P.; Bamford, P.; Berti, I.; Not, T.; Coppa, G.V.; Catassi, C.; Fasano, A. Host-dependent activation of the zonulin system is involved in the impairment of the gut barrier function following bacterial colonization. Gastroenterology 2002, 123, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Leaky Gut and Autoimmune Diseases. Clin. Rev. Allerg. Immunol. 2012, 42, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Lammers, K.M.; Goldblum, S.; Shea-Donohue, T.; Netzel-Arnett, S.; Buzza, M.S.; Antalis, T.M.; Vogel, S.N.; Zhao, A.; Yang, S.; et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc. Natl. Acad. Sci. USA 2009, 106, 16799–16804. [Google Scholar] [CrossRef] [Green Version]
- Renyong, G.; Hirosi, S.; Shikogi, S. Endothelial Cell Motility is Compatible with Junctional Integrity. J. Cell. Physiol. 2006, 211, 327–375. [Google Scholar]
- Wan, H.; Gadmor, H.; Brown, L. Anchoring junctions in the oral mucosa:adherence junctions and desmosomes. In Oral Mucosa in Health and Disaease; Springer: Cham, Switzerland, 2018; pp. 31–51. [Google Scholar]
- Kingsley, C.; Kourtidis, A. Critical roles of adherence junctions in diseases of the oral mucosa. Tissue Barriers. 2022, 2084320. [Google Scholar] [CrossRef]
- Delva, E.; Tucker, D.K.; Kowalczy, K.A.P. The desmosome. Cold Spring Harbor. Perspect. Biol. 2009, 1, a002543. [Google Scholar]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Drago, S.; El Asmar, R.; De Pierro, M.; Clemente, M.G.; Tripathi, A.; Sapone, A.; Thakar, M.; Iacono, G.; Carroccio, A.; D’Agate, C.; et al. Gliadin, zonulin and gut permeability: Effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand. J. Gastroenterol. 2006, 41, 408–419. [Google Scholar] [CrossRef]
- Branski, D.; Fasano, A.; Troncone, R. Latest developments in the pathogenesis and treatment of celiac disease. J. Pediatr. 2006, 149, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Zonulin and its regulation of intestinal barrier function: The biological door to inflammation, autoimmunity, and cancer. Physiol. Rev. 2011, 91, 151–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meddings, J.B.; Jarand, J.; Urbanski, S.J.; Hardin, J.; Gall, D.G. Increased gastrointestinal permeability is an early lesion in the spontaneously diabetic BB rat. Am. J. Physiol. 1999, 276, G951–G957. [Google Scholar] [CrossRef] [PubMed]
- Watts, T.; Berti, I.; Sapone, A.; Gerarduzzi, T.; Not, T.; Zielke, R.; Fasano, A. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc. Natl. Acad. Sci. USA 2005, 102, 2916–2921. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.; Barmeyer, C.; Fromm, M.; Runkel, N.; Foss, H.D.; Bentzel, C.J.; Riecken, E.O.; Schulzke, J.D. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 1999, 116, 301–307. [Google Scholar] [CrossRef]
- Zeissig, S.; Bürgel, N.; Günzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfu (nction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef]
- Caffarelli, C.; Deriu, F.M.; Terzi, V.; Perrone, F.; De Angelis, G.; Atherton, D.J. Gastrointestinal symptoms in patients with asthma. Arch. Dis. Child. 2000, 82, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Hijazi, Z.; Molla, A.M.; Al-Habashi, H.; Muawad, W.; Molla, A.; Sharma, P. Intestinal permeability is increased in bron-chial asthma. Arch. Dis. Child. 2004, 89, 227–229. [Google Scholar] [CrossRef] [Green Version]
- Wendling, D.; Bidet, A.; Guidet, M. Evaluation de la perméabilité intestinale au cours de la spondylarthrite ankylosante par le test au 51Cr-EDTA. Rev. Esp. Reumatol. 1992, 19, 253–256. [Google Scholar]
- Gierynska, M.; Szulc-Dabrowska, L.; Struzik, J.; Mielcarska, M.B.; GregorczykZboroch, K.P. Integrity of the Intes-tinal Barrier: The Involvement of Epithelial Cells and Microbiota—A Mutual Relationship. Animals 2022, 12, 145. [Google Scholar] [CrossRef]
- Usuda, H.; Okamoto, T.; Wada, K. Leaky gut. Effect of dietary fiber and fats on microbiome and intestinal barrier. Int. J. Mol. Sci. 2021, 22, 7613. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Pottosin, I.; Lamade, E.; Tcherkez, G. What is the role of putrescine accumulated under potassium deficiency? Plant Cell Environ. 2020, 43, 1331–1347. [Google Scholar] [CrossRef] [PubMed]
- Brieger, L. Weitere Untersuchungen Über Ptomaine; Hirschwald: Berlin, Germany, 1885; p. 39. [Google Scholar]
- Qian, Z.-G.; Xia, X.-X.; Lee, S.Y. Metabolic Engineering of Escherichia coli for the Production of Putrescine: A Four Carbon Diamine. Biotechnol. Bioeng. 2009, 104, 651–662. [Google Scholar] [PubMed]
- Zhang, M.; Pickart, C.M.; Coffino, P. Determinants of proteasome recognition of ornithine decarboxylase, a ubiqui-tin-independent substrate. EMBO J. 2003, 22, 1488–1496. [Google Scholar] [CrossRef] [Green Version]
- Qian, Z.-G.; Xia, X.-X.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of cadaverine: A five carbon di-amine. Biotechnol. Bioeng. 2011, 10, 93–103. [Google Scholar] [CrossRef]
- Ikeguchi, Y.; Bewley, M.C.; Pegg, A.E. Aminopropyltransferases: Function, structure and genetics. J. Biochem. 2006, 139, 1–9. [Google Scholar] [CrossRef]
- Moinard, C.; Cynober, L.; De Bandt, J.P. Polyamines: Metabolism and implications in human diseases. Clin. Nutr. 2005, 24, 184–197. [Google Scholar] [CrossRef]
- Kossel, A. Über das Agmatin. Zeitsch. Physiolog. Chem 1910, 66, 257–261. [Google Scholar]
- Dandrifosse, G.; Peulen, O.; El Khefif, N.; Deloyer, P.; Dandrifosse, A.C.; Grandfils, C. Are milk polyamines preventive agents against food allergy? Proc. Nutr. Soc. 2000, 59, 81–86. [Google Scholar] [CrossRef]
- Larqué, E.; Sabater-Molina, M.; Zamora, S. Biological significance of dietary polyamines. Nutrition 2007, 23, 87–95. [Google Scholar] [CrossRef]
- Buts, J.P. Les facteurs trophiques du lait. Arch. Pediatr. 1998, 5, 298–306. [Google Scholar] [CrossRef]
- McCormack, S.A.; Johnson, L.R. Role of polyamines in gastrointestinal mucosal growth. Am. J. Physiol. 1991, 260, G795–G806. [Google Scholar] [CrossRef] [PubMed]
- Milovic, V. Polyamines in the gut lumen: Bioavailability and biodistribution. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Osborne, D.L.; Seidel, E.R. Gastrointestinal luminal polyamines: Cellular accumulation and enterohepatic circulation. Am. J. Physiol. 1990, 258, G576–G584. [Google Scholar] [CrossRef] [PubMed]
- Scemama, J.L.; Grabié, V.; Seidel, E.R. Characterization of univectorial polyamine transport in duodenal crypt cell line. Am. J. Physiol. 1993, 265, G851–G856. [Google Scholar] [CrossRef]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, 4548. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Buts, J.P.; De Keyser, N.; Kolanowski, J.; Sokal, E.; Van Hoof, F. Maturation of villus and crypt cell functions in rat small intestine. Role of dietary polyamines. Dig. Dis. Sci. 1993, 38, 1091–1098. [Google Scholar] [CrossRef]
- Guo, X.; Rao, J.N.; Liu, L.; Zou, T.-T.; Turner, D.J.; Bass, B.L.; Wang, J.-Y. Regulation of adherens junctions and epithelial paracellular permeability: A novel function for polyamines. Am. J. Physiol. Cell. Physiol. 2003, 285, C1174–C1187. [Google Scholar] [CrossRef] [Green Version]
- Velloso, N.A.; Dalmolin, G.D.; Gomes, G.M.; Rubin, M.A.; Canas, P.M.; Cunha, R.A.; Mello, C.F. Spermine improves recognition memory deficit in a rodent model of Huntington’s disease. Neurobiol. Learn. Mem. 2009, 92, 574–580. [Google Scholar] [CrossRef]
- Zhang, M.; Caragine, T.; Wang, H.; Cohen, P.S.; Botchkina, G.; Soda, K.; Bianchi, M.; Ulrich, P.; Cerami, A.; Sherry, B.; et al. Spermine Inhibits Proinflammatory Cytokine Synthesis in Human Mononuclear Cells: A Counterregulatory Mechanism that Restrains the Immune Response. J. Exp. Med. 1997, 185, 1759–1768. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Rao, J.N.; Bass, B.L.; Wang, J.Y. NF-kappaB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G992–G1004. [Google Scholar] [CrossRef]
- Kurihara, S.; Kato, K.; Asada, K.; Kumagai, H.; Suzuki, H. A Putrescine-Inducible Pathway Comprising PuuE-YneI in Which γ-Aminobutyrate Is Degraded into Succinate in Escherichia coli K-12. J. Bacteriol. 2010, 192, 4582–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky Gut as a Danger Signal for Autoimmune Diseases. Front. Immunol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyce, J.A.; Assa’ad, A.; Burks, A.W.; Jones, S.M.; Sampson, H.A.; Wood, R.A.; Plaut, M.; Cooper, S.F.; Fenton, M.J.; Arshad, S.H.; et al. Guidelines for the diagnosis and management of food allergy in the United States: Report of the NIAID–sponsored expert panel. J. Allergy Clin. Immunol. 2010, 126, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Paparo, L.; Nocerino, R.; Di Scala, C.; Della Gatta, G.; Maddalena, Y.; Buono, A.; Bruno, C.; Voto, L.; Ercolini, D. Gut Microbiome as Target for Innovative Strategies against Food Allergy. Front. Immunol. 2019, 10, 191. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Nocerino, R.; Terrin, G.; Leone, L.; Troncone, R. Hospital admissions for food–induced anaphylaxis in Italian children. Clin. Exp. Allergy 2012, 42, 1813–1814. [Google Scholar] [CrossRef]
- Rivas, N.M.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Garcia-Lloret, M.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Sonoyama, K.; Ogasawara, T.; Goto, H.; Yoshida, T.; Takemura, N.; Fujiwara, R.; Watanabe, J.; Ito, H.; Morita, T.; Tokunaga, Y.; et al. Comparison of gut microbiota and allergic reactions in BALB/c mice fed different cultivars of rice. Br. J. Nut. 2010, 103, 218–226. [Google Scholar] [CrossRef] [Green Version]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov. sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [Green Version]
- Van Passel, M.W.J.; Kant, R.; Zoetendal, E.G.; Plugge, C.M.; Derrien, M.; Malfatti, S.A.; Chain, P.S.G.; Woyke, T.; Palva, A.; de Vos, W.M.; et al. The Genome of Akkermansia muciniphila, a Dedicated Intestinal Mucin Degrader, and Its Use in Exploring Intestinal Metagenomes. PLoS ONE 2011, 6, e16876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vos, W.M. Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology 2017, 1635, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [Green Version]
- Evers, T.M.; Sheikhhassani, V.; Haks, M.C.; Storm, C.; Ottenhoff, T.H.; Mashaghi, A. Single-cell analysis reveals chemo-kine-mediated differential regulation of monocyte mechanics. iScience 2022, 25, 103555. [Google Scholar] [CrossRef]
- Mehrabian, M.; Sparkes, R.S.; Mohandas, T.; Fogelman, A.M.; Lusis, A.J. Localization of monocyte chemotactic protein-1 gene (SCYA2) to human chromosome 17q11.2-q21.1. Genomics 1991, 9, 200–203. [Google Scholar] [CrossRef]
- Craig, M.J.; Loberg, R.D. CCL2 (Monocyte Chemoattractant Protein-1) in cancer bone metastases. Cancer Metast. Rev. 2006, 25, 611–619. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [Green Version]
- Di Costanzo, M.; De Paulis, N.; Biasucci, G. Butyrate: A Link between Early Life Nutrition and Gut Microbiome in the Development of Food Allergy. Life 2021, 11, 384. [Google Scholar] [CrossRef]
- Luu, M.; Visekruna, A. Short-Chain Fatty Acids: Bacterial Messengers Modulating the Immunometabolism of T Cells. Eur. J. Immunol. 2019, 49, 842–848. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.P.; Bradfield, C.A. The Search for Endogenous Activators of the Aryl Hydrocarbon Receptor. Chem. Res. Toxicol. 2008, 21, 102–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos. 2015, 43, 1522–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Juan, A.; Segura, E. Modulation of Immune Responses by Nutritional Ligands of Aryl Hydrocarbon Receptor. Front. Immunol. 2021, 12, 645168. [Google Scholar] [CrossRef] [PubMed]
- Pirzadeh, M.; Khalili, N.; Rezaei, N. The interplay between aryl hydrocarbon receptor, H. pylori, tryptophan, and arginine in the pathogenesis of gastric cancer. Int. Rev. Immunol. 2022, 41, 299–312. [Google Scholar] [CrossRef]
- Muñoz-Esparza, N.C.; Latorre-Moratalla, M.L.; Comas-Basté, O.; Toro-Funes, N.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Polyamines in Food. Front. Nutr. 2019, 6, 108. [Google Scholar] [CrossRef] [Green Version]
- Van de Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogénesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut home-ostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [Green Version]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef] [Green Version]
- Kaye, D.M.; Shihata, W.A.; Jama, H.A.; Tsyganov, K.; Ziemann, M.; Kiriazis, H.; Horlock, D.; Vijay, A.; Giam, B.; Vinh, A.; et al. Deficiency of Prebiotic Fiber and Insufficient Signaling Through Gut Metabolite-Sensing Receptors Leads to Cardiovascular Disease. Circulation 2020, 141, 1393–1403. [Google Scholar] [CrossRef]
- Muralitharan, R.R.; Jama, H.A.; Xie, L.; Peh, A.; Snelson, M.; Marques, F.Z. Microbial Peer Pressure: The Role of the Gut Microbiota in Hypertension and Its Complications. Hypertension 2020, 76, 1674–1687. [Google Scholar] [CrossRef] [PubMed]
- Jose, A.; Apewokin, S.; Hussein, W.E.; Ollberding, N.J.; Elwing, J.M.; Haslam, D.B. A unique gut microbiota signature in pulmonary arterial hypertension: A pilot study. Pulm. Circ. 2022, 12, e12051. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.; Stewart, C.S.; Flint, H.J. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [PubMed] [Green Version]
- Martín, R.; Bermúdez-Humarán, L.G.; Langella, P. Searching for the Bacterial Effector: The Example of the Multi-Skilled Commensal Bacterium Faecalibacterium prausnitzii. Front. Microbiol. 2018, 9, 346. [Google Scholar] [CrossRef]
- Sjödin, K.S.; Vidman, L.; Rydén, P.; West, C.E. Emerging evidence of the role of gut microbiota in the development of al-lergic diseases. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 390–395. [Google Scholar]
- Codoñer, F.M.; Ramírez-Bosca, A.; Climent, E.; Carrión-Gutierrez, M.; Guerrero, M.; Pérez-Orquín, J.M.; Horga de la Parte, J.; Genovés, S.; Ramón, D.; Navarro-López, V.; et al. Gut microbial composition in patients with psoriasis. Sci. Rep. 2018, 8, 3812. [Google Scholar] [CrossRef]
- Demirci, M.; Tokman, H.B.; Uysal, H.K.; Demiryas, S.; Karakullukcu, A.; Saribas, S.; Cokugras, H.; Kocazeybek, B.S. Re-duced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allerg. Immunopathol. 2019, 47, 365–371. [Google Scholar] [CrossRef]
- Hu, W.; Lu, W.; Li, L.; Zhang, H.; Lee, Y.-K.; Chen, W.; Zhao, J. Both living and dead Faecalibacterium prausnitzii alleviate house dust mite-induced allergic asthma through the modulation of gut microbiota and short-chain fatty acid production. J. Sci. Food Agric. 2021, 101, 5563–5573. [Google Scholar] [CrossRef]
- Jia, W.; Whitehead, R.N.; Griffiths, L.; Dawson, C.; Waring, R.H.; Ramsden, D.B.; Hunter, J.O.; Cole, J.A. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn’s disease? FEMS Microbiol. Lett. 2010, 310, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Beh. Imm. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, A.; Wang, W.; Guo, R.; Liu, H. Faecalibacterium prausnitzii (ATCC 27766) has preventive and therapeutic effects on chronic unpredictable mild stress-induced depression-like and anxiety-like behavior in rats. Psychoneuroendocrinology 2019, 104, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, R.; George, G.; Kabeerdoss, J.; Hepsiba, J.; Chandragunasekaran, A.; Ramakrishna, B. Quantitative differ-ences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br. J. Nutr. 2010, 103, 335–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenman, L.K.; Burcelin, R.; Lahtinen, S. Establishing a causal link between gut microbes, body weight gain and glucose metabolism in humans-towards treatment with probiotics. Benefl. Microb. 2015, 7, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Hippe, B.; Zanner, J.; Aumueller, E.; Brath, H.; Haslberger, G.A. Gut Microbiota of Obese, Type 2 Diabetic Individuals is Enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after Weight Loss. Endocr. Metab. Immune Disord. Drug Targets 2016, 16, 99–106. [Google Scholar] [CrossRef]
- Björkqvist, O.; Rangel, I.; Serrander, L.; Magnusson, C.; Halfvarson, J.; Norén, T.; Bergman-Jungeström, M. Faecalibacterium prausnitzii increases following fecal microbiota transplantation in recurrent Clostridioides difficile infection. PLoS ONE 2021, 16, e0249861. [Google Scholar] [CrossRef]
- Quevrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.M.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Rossi, O.; Khan, M.T.; Schwarzer, M.; Hudcovic, T.; Srutkova, D.; Duncan, S.H.; Stolte, E.H.; Kozakova, H.; Flint, H.J.; Samsom, J.N.; et al. Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS ONE 2015, 10, e0123013. [Google Scholar] [CrossRef] [Green Version]
- Rossi, O.; van Berkel, L.A.; Chain, F.; Khan, M.T.; Taverne, N.; Sokol, H.; Duncan, S.H.; Flint, H.J.; Harmsen, H.J.M.; Langella, P.; et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci. Rep. 2016, 6, 18507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlsson, A.H.; Yakymenko, O.; Olivier, I.; Håkansson, F.; Postma, E.; Keita, A.V.; Söderholm, J.D. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand. J. Gastroenterol. 2013, 48, 1136–1144. [Google Scholar]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Invest. 2021, 131, e141935. [Google Scholar] [CrossRef] [PubMed]
- Lukić, I.; Getselter, D.; Ziv, O.; Oron, O.; Reuveni, E.; Koren, O.; Elliott, E. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Translat. Psychiatry 2019, 9, 133. [Google Scholar]
- Chua, H.-H.; Chou, Y.H.-C.; Tung, Y.-L.; Chiang, B.-L.; Liao, C.-C.; Liu, H.-H.; Ni, Y.H. Intestinal Dysbiosis Featuring Abun-dance of Ruminococcus gnavus Associates with Allergic Diseases in Infants. Gastroenterology 2018, 154, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Dot, D.; Osawa, R.; Stackebrandt, E. Phascolarctobacterium faecium gen. nov, spec. nov., a Novel Taxon of the Sporomusa Group of Bacteria. Syst. Appl. Microbiol. 1993, 16, 380–384. [Google Scholar] [CrossRef]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium faecium abundant colonization in human gas-trointestinal tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef] [Green Version]
- Kampf, C.; Mardinoglu, A.; Fagerberg, L.; Hallström, B.M.; Danielsson, A.; Nielsen, J.; Pontén, F.; Uhlen, M. Defining the human gallbladder proteome by transcriptomics and affinity proteomics. Proteomics 2014, 14, 2498–2507. [Google Scholar] [CrossRef]
- Nepal, C.; Zhu, B.; O’Rourke, C.J.; Bhatt, D.K.; Lee, D.; Song, L.; Wang, D.; Van Dyke, A.L.; Choo-Wosoba, H.; Liu, Z.; et al. Integrated molecular characterization of gallbladder cancer reveals micro-environment-associated subtype. J. Hepatol. 2021, 74, 1132–1144. [Google Scholar] [CrossRef]
- Zali, M.R.; Azodi, M.Z.; Razzaghi, Z.; Heydari, M.H. Gallbladder cancer integrated bioinformatics analysis of protein profile data. Gastroenterol. Hepatol. Bed Bench 2019, 12 (Suppl. S1), S66. [Google Scholar]
- Shen, H.; Ye, F.; Xie, L.; Yang, J.; Li, Z.; Xu, P.; Meng, F.; Li, L.; Chen, Y.; Bo, X.; et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci. Rep. 2015, 5, 17450. [Google Scholar] [CrossRef] [PubMed]
- Kose, S.H.; Grice, K.; Orsi, W.D.; Ballal, M.; Coolen, M.J.L. Metagenomics of pigmented and cholesterol gallstones: The putative role of bacteria. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Zhang, S.; Li, S.; Wang, G.; Zhang, A.; Jin, T.; Zhang, Y.; Lv, Q.; Xiao, M.; Sun, Y.; et al. Cultivation and genomic characterization of the bile bacterial species from cholecystitis patients. Front. Microbiol. 2021, 12, 739621. [Google Scholar] [CrossRef]
- Sharma, N.; Yadav, M.; Tripathi, G.; Mathew, B.; Bindal, V.; Falari, S.; Maras, J.S. Bile multi-omics analysis classifies lipid species and microbial peptides predictive of Carcinoma of Gallbladder. Hepatology 2022, 76, 920–935. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Raftery, D. (Eds.) NMR-Based Metabolomics Methods and Protocols. In Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 2037, ISBN 978-1-4939-9689-6. [Google Scholar]
- Jayalakshmi, K.; Sonkar, K.; Behari, A.; Kapoor, V.K.; Sinha, N. Lipid profiling of cancerous and benign gallbladder tissues by 1H NMR spectroscopy. NMR Biomed. 2011, 24, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Sonkar, K.; Behari, A.; Kapoor, V.K.; Sinha, N. 1H NMR metabolic profiling of human serum associated with benign and malignant gallstone diseases. Metabolomics 2013, 9, 515–528. [Google Scholar] [CrossRef]
- Sharma, R.K.; Sonkar, K.; Sinha, N.; Rebala, P.; Albani, A.E.; Behari, A.; Reddy, D.N.; Farooqui, A.; Kapoor, V.K. Gallstones: A Worldwide Multifaceted Disease and Its Correlations with Gallbladder Carcinoma. PLoS ONE 2016, 11, e0166351. [Google Scholar] [CrossRef]
- Bharti, S.K.; Behari, A.; Kapoor, V.K.; Kumari, N.; Krishnani, N.; Roy, R. Magic angle spinning NMR spectroscopic metabolic profiling of gall bladder tissues for differentiating malignant from benign disease. Metabolomics 2013, 9, 101–118. [Google Scholar] [CrossRef]
- Ko, H.; Choi, I.; Chang, K.; Jeong, G.; Gong, G.; Seo, H.; Ryu, D.; Lee, K.G.; Choi, D.; Chung, H.; et al. Amphiphilic metabolites in gallbladder bile: Potential biomarkers for gallbladder diseases. Appl. Spectrosc. Rev. 2016, 51, 706–717. [Google Scholar] [CrossRef]
- Srivastava, M.; Sharma, A.; Kapoor, V.K.; Nagana Gowda, G.A. Stones from cancerous and benign gallbladders are different: A proton nuclear magnetic resonance spectroscopy study. Hepatol. Res. 2008, 38, 997–1005. [Google Scholar] [CrossRef]
- Poland, J.C.; Schrimpe-Rutledge, A.C.; Sherrod, S.D.; Flynn, C.R.; McLean, J.A. Utilizing Untargeted Ion Mobility-Mass Spectrometry to Profile Changes in the Gut Metabolome following Biliary Diversion Surgery. Anal. Chem. 2019, 91, 14417–14423. [Google Scholar] [CrossRef] [PubMed]





| Genera | Cholelithiasis Group a | Control Group | p Value b |
|---|---|---|---|
| Acidibacter | 0.13 ± 0.33 | 3.15 ± 4.4 | 0.005 |
| Actinobacillus | 3.34 ± 12.03 | 0.00 ± 0.00 | 0.225 |
| Alistipes | 3.85 ± 2.16 | 1.72 ± 2.31 | 0.031 |
| Alloprevotella | 0.53 ± 0.43 | 0.23 ± 0.35 | 0.346 |
| Bacteroides | 10.21 ± 6.94 | 2.74 ± 3.77 | 0.001 |
| Barnesiella | 1.45 ± 0.90 | 0.73 ± 1.05 | 0.084 |
| Bifidobacterium | 3.01 ± 5.02 | 7.60 ± 17.98 | 0.599 |
| Blautia | 1.15 ± 1.22 | 0.79 ± 1.63 | 0.56 |
| Bradyrhizobium | 0.18 ± 0.44 | 6.90 ± 8.56 | 0.004 |
| Brevundimonas | 0.15 ± 0.20 | 2.80 ± 3.31 | 0.003 |
| Christensenellaceae R.7 group | 0.56 ± 0.44 | 0.08 ± 0.20 | 0.04 |
| Coprococcus 3 | 1.19 ± 0.69 | 0.35 ± 0.67 | 0.009 |
| Dialister | 1.49 ± 1.40 | 0.12 ± 0.25 | 0.002 |
| Escherichia-Shigella | 4.90 ± 10.52 | 0.30 ± 0.49 | 0.001 |
| Eubacterium comprostanoligenes group | 2.22 ± 2.53 | 0.59 ± 0.81 | 0.01 |
| Faecalibacterium | 2.22 ± 1.31 | 1.53 ± 3.66 | 0.661 |
| Haemophilus | 7.09 ± 25.28 | 0.02 ± 0.04 | 0.011 |
| Haliangium | 0.01 ± 0.04 | 0.54 ± 0.88 | 0.006 |
| Helicobacter | 10.84 ± 8.46 | 6.29 ± 9.32 | 0.232 |
| Lachonospira | 2.75 ± 5.32 | 0.54 ± 1.32 | 0.055 |
| Lachnospiraceae NK4A136 group | 1.41 ± 0.92 | 0.63 ± 0.88 | 0.039 |
| Lactococcus | 0.53 ± 1.10 | 13.14 ± 24.37 | 0.007 |
| Methylobacterium | 0.04 ± 0.04 | 1.80 ± 3.76 | 0.003 |
| Parabacteroides | 0.68 ± 0.43 | 0.14 ± 0.28 | 0.067 |
| Prevotella | 1.16 ± 1.17 | 1.17 ± 0.57 | 0.011 |
| Prevotellaceae NK3B31 group | 0.59 ± 0.56 | 0.06 ± 0.18 | 0.04 |
| Propionibacterium | 0.58 ± 0.40 | 10.77 ± 18.48 | 0.01 |
| Pseudobytyrividrio | 0.59 ± 0.36 | 1.11 ± 3.20 | 0.977 |
| Ruminococcaceae UCG-002 | 0.82 ± 0.60 | 0.17 ± 0.33 | 0.023 |
| Ruminococcaceae UCG-004 | 0.56 ± 0.34 | 0.23 ± 0.35 | 0.229 |
| Ruminococcus | 0.51 ± 0.48 | 015 ± 0.42 | 0.182 |
| Sediminibacterium | 0.55 ± 0.94 | 4.65 ± 4.75 | 0.008 |
| Sphingomonas | 0.03 ± 0.05 | 2.74 ± 4.68 | 0.001 |
| Streptococcus | 6.67 ± 22.33 | 0.89 ± 0.81 | 0.957 |
| Subdoligranulum | 1.65 ± 1.24 | 0.47 ± 0.71 | 0.013 |
| U.m. of Bacteroidales S24-7 group family | 1.48 ± 1.05 | 0.78 ± 1.11 | 0.111 |
| U.m. of Caulobacteraceae family | 1.14 ± 0.11 | 2.52 ± 3.14 | 0.009 |
| U.m. of Lachnospiraceae family | 6.05 ± 3.05 | 3.46 ± 4.33 | 0.108 |
| U.m. of Ruminococcaceae family | 1.05 ± 0.58 | 1.09 ± 2.17 | 0.986 |
| Pathogen | Function of Outer Membrane Proteins (Reference[s]) | Mechanism of Induction of Stress Response Genes | Efflux Pump (s) |
|---|---|---|---|
| Escherichia coli | Repression of ompF [54] | sulA to correct DNA damage [55] | AcrAB [56] |
| Vibrio | Regulated expression of ompU and ompT [57] | RpoS in V. vulnificus [58] | AcrAB, BreR repressor, vexAB and vexCD, VprAB, Vme pumps [59,60,61,62,63] |
| Salmonella | Repression of OmpF and OmpC, utilization of TolA and TolC [64,65] | SoxRS, OxyR [66,67] | AcrAB [68,69], AcrEF, MdtABC, MdsCBA, EmrAB, MdtK, and MacAB |
| Campylobacter | CmeABC and CmeDEF [70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, A.G.; Villa, T.G.; Sánchez-Pérez, Á.; Notario, V.; Carrera, M. The Role of the Gallbladder, the Intestinal Barrier and the Gut Microbiota in the Development of Food Allergies and Other Disorders. Int. J. Mol. Sci. 2022, 23, 14333. https://doi.org/10.3390/ijms232214333
Abril AG, Villa TG, Sánchez-Pérez Á, Notario V, Carrera M. The Role of the Gallbladder, the Intestinal Barrier and the Gut Microbiota in the Development of Food Allergies and Other Disorders. International Journal of Molecular Sciences. 2022; 23(22):14333. https://doi.org/10.3390/ijms232214333
Chicago/Turabian StyleAbril, Ana G., Tomás G. Villa, Ángeles Sánchez-Pérez, Vicente Notario, and Mónica Carrera. 2022. "The Role of the Gallbladder, the Intestinal Barrier and the Gut Microbiota in the Development of Food Allergies and Other Disorders" International Journal of Molecular Sciences 23, no. 22: 14333. https://doi.org/10.3390/ijms232214333
APA StyleAbril, A. G., Villa, T. G., Sánchez-Pérez, Á., Notario, V., & Carrera, M. (2022). The Role of the Gallbladder, the Intestinal Barrier and the Gut Microbiota in the Development of Food Allergies and Other Disorders. International Journal of Molecular Sciences, 23(22), 14333. https://doi.org/10.3390/ijms232214333