Advances in Aptamers-Based Applications in Breast Cancer: Drug Delivery, Therapeutics, and Diagnostics
Abstract
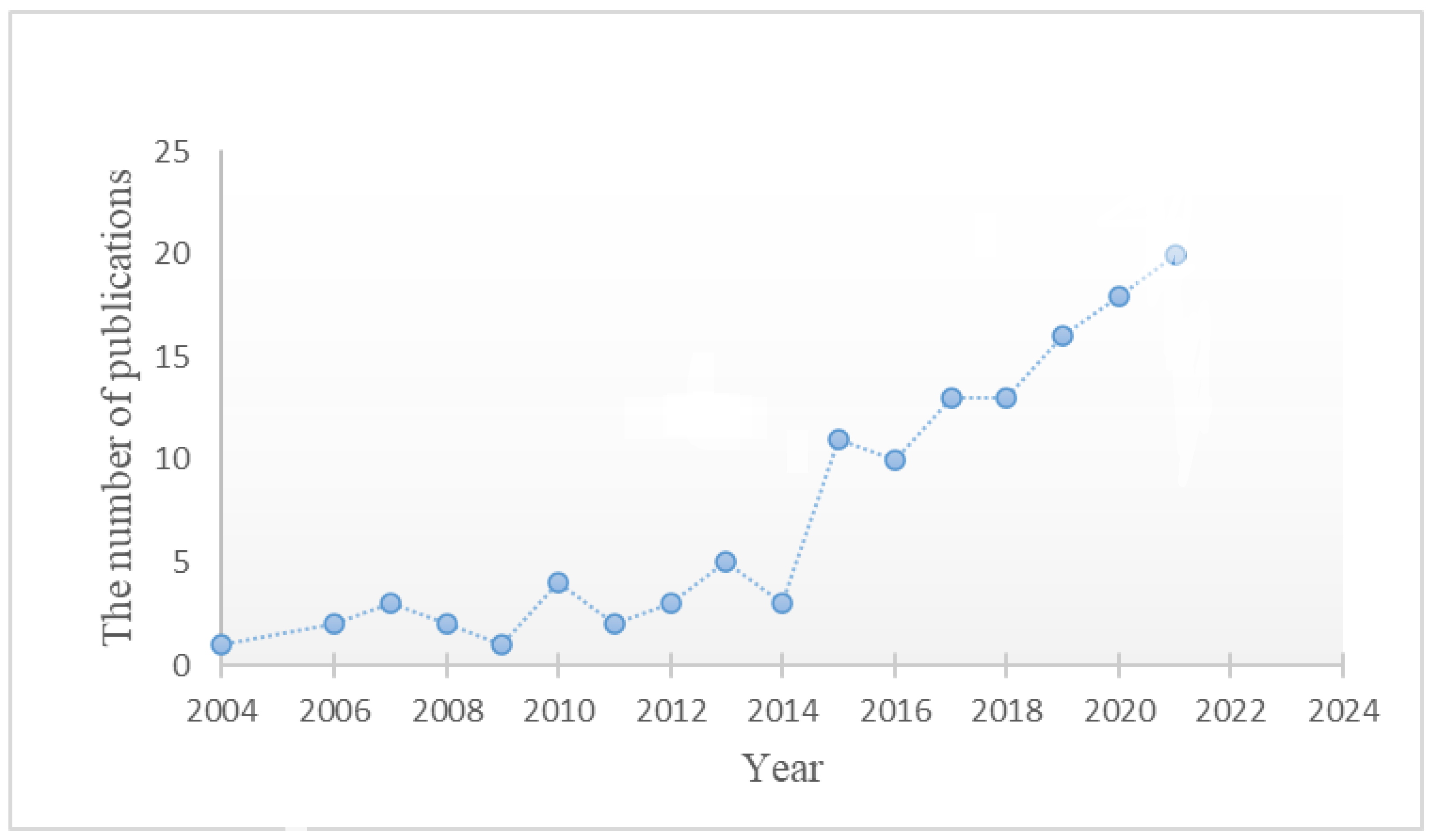
1. Introduction
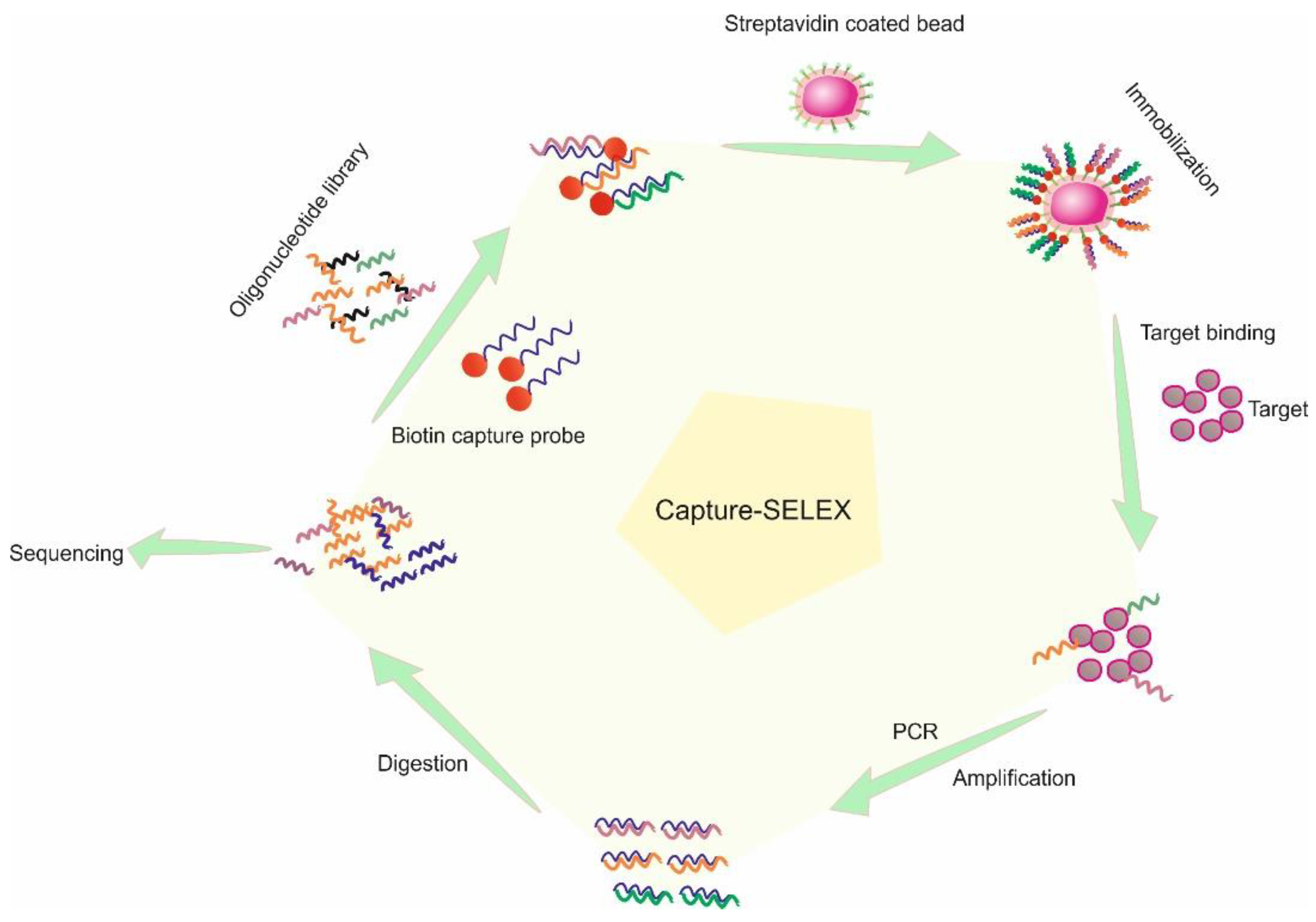
2. Affinity SELEX
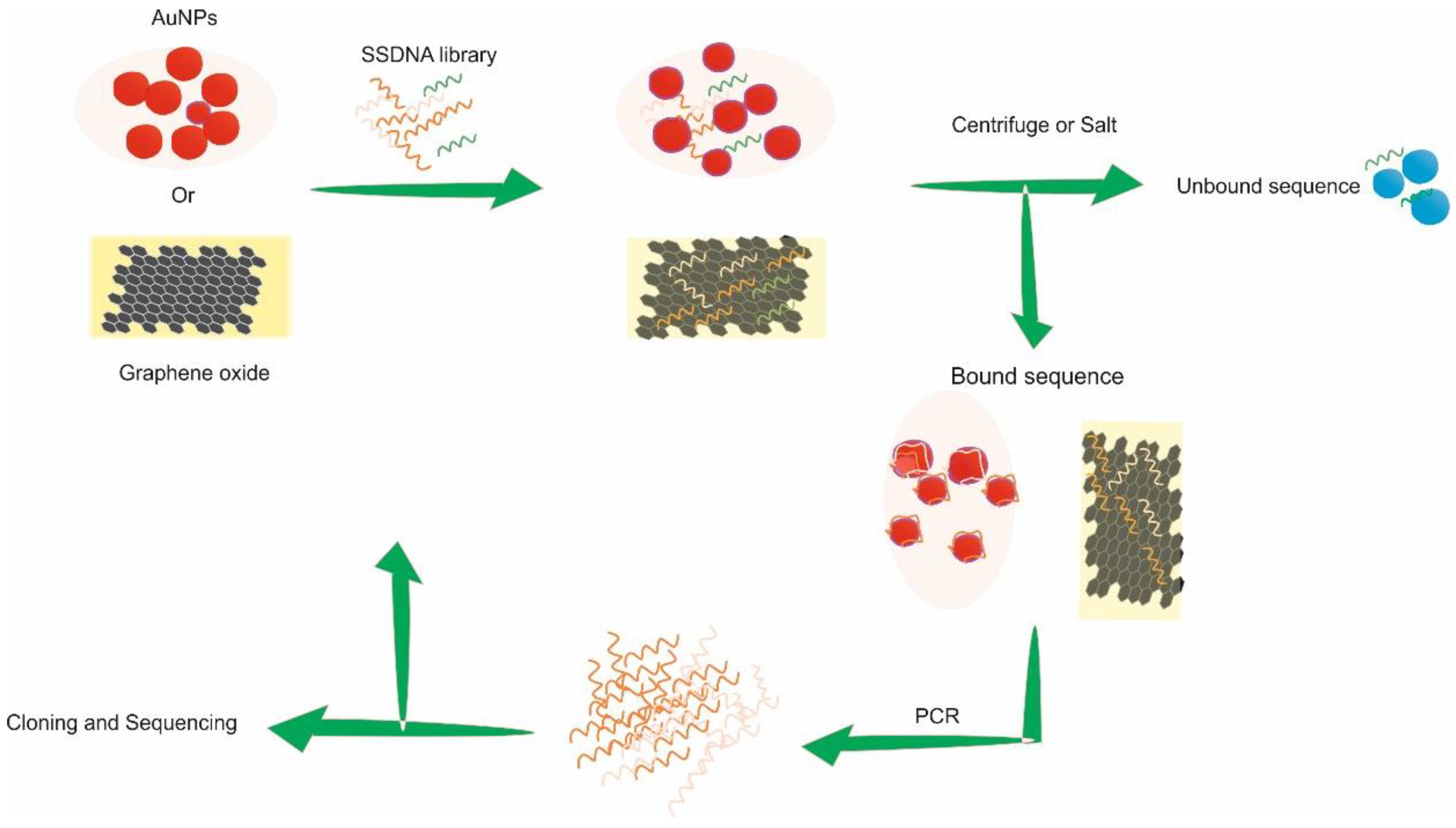
3. Label-Free SELEX
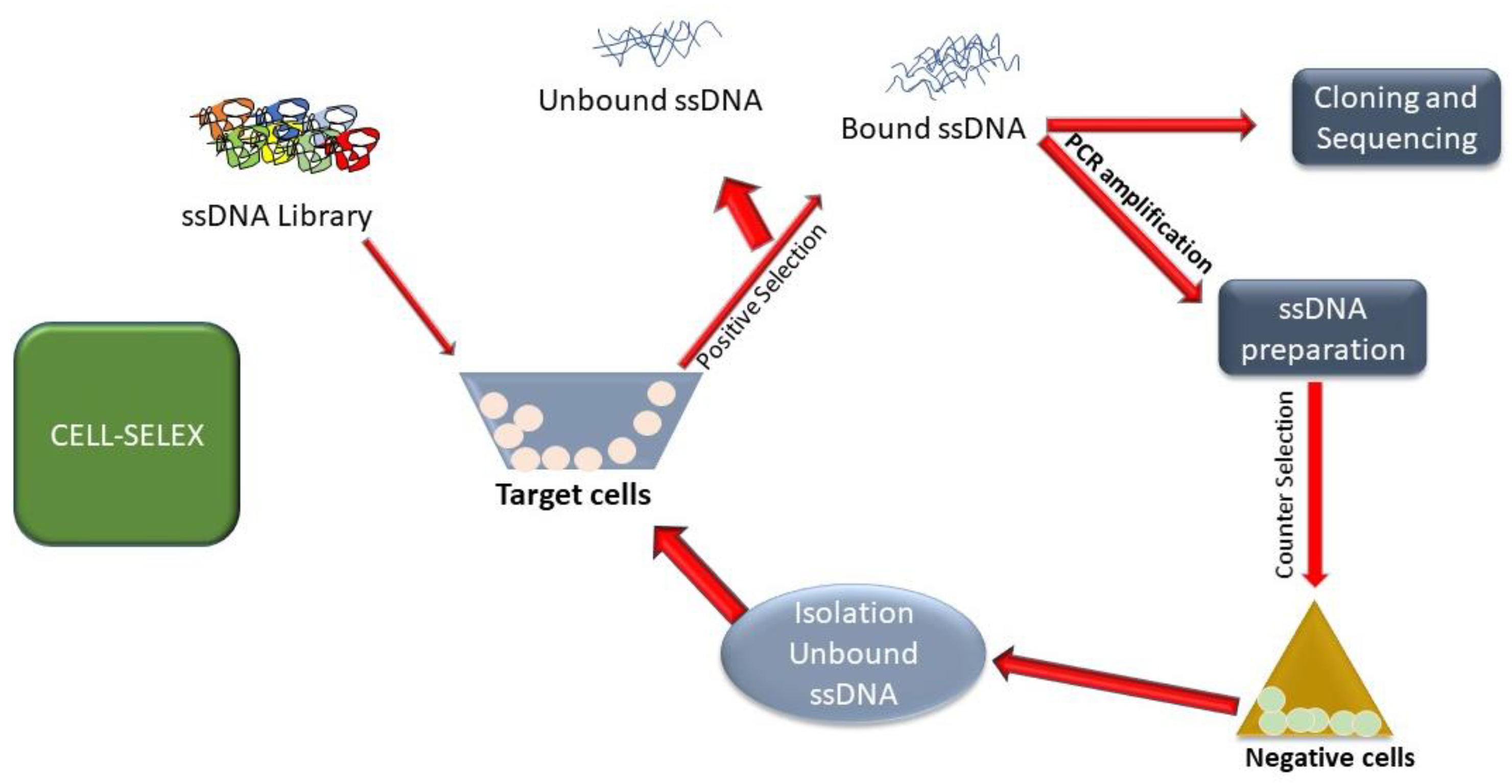
4. Cell-SELEX Applications
5. In Vivo-SELEX
6. Aptamer Utilization for Breast Cancer
| Target | Sequences | Aptamer | Kd(nM) | Purpose | Ref. |
|---|---|---|---|---|---|
| Nucleolin | GGTGGTGGTGGTTGTGGTGGTGGTGG | AS1411 | 47.3 | Diagnostic | [129] |
| SK-BR-3 cells | TGGATGGGGAGATCCGTTGAGTAAGCGGGCGTGTCTCTCTGCCGCCTTGCTATGGGG | S6 | 28.2 | Therapeutic | [151] |
| HER2 | GGGCCGTCGAACACGAGCATGGTGCGTGGACCTAGGATGACCTGAGTACTGTCC | H2 | 1.8 ± 0.5 | Diagnostic | [152] |
| Mucin-1 | GCAGTTGATCCTTTGGATACCCTGG | Muc1 | 38.3 | Diagnostic | [153] |
| ERα | ATACCAGCTTATTCAATTCGTTGCATTTAGGTG | ERaptD4 | 33 | Diagnostic | [136] |
| HER2 | AACCGCCCAAATCCCTAAGAGTCTGCACTTGT | HB5 | 18.9 | Therapeutic | [154] |
| MDA-MB-231 | GAATTCAGTCGGACAGCGAAGTAGTTTTCCTT | Xlx-1-A | 0.7 | Therapeutic | [155] |
| MCF-10AT1 cells | AGGCGGCAGTGTCAGAGTGAATAGGGGATGTA | KMF2-1a | 52 | Therapeutic | [156] |
| EpCAM | CACTACAGAGGTTGCGTCTGTCCCACGTTGTCATGGGGGGTTGGCCTG | SYL3C | 20.08 | Diagnostic | [157] |
7. Therapeutic Use of Aptamers
Therapeutic Application of Aptamers in Breast Cancer
8. Drug Delivery Pathways of Aptamer
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parkin, D.; Läärä, E.; Muir, C. Estimates of the worldwide frequency of sixteen major cancers in 1980. Int. J. Cancer 1988, 41, 184–197. [Google Scholar] [CrossRef]
- Zoon, C.K.; Starker, E.Q.; Wilson, A.M.; Emmert-Buck, M.R.; Libutti, S.K.; Tangrea, M.A. Current molecular diagnostics of breast cancer and the potential incorporation of microRNA. Expert Rev. Mol. Diagn. 2009, 9, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, R.E.; Baker, J.A.; Helvie, M.A. Breast cancer deaths averted over 3 decades. Cancer 2019, 125, 1482–1488. [Google Scholar] [CrossRef]
- Maghous, A.; Rais, F.; Ahid, S.; Benhmidou, N.; Bellahamou, K.; Loughlimi, H.; Marnouche, E.; Elmajjaoui, S.; Elkacemi, H.; Kebdani, T. Factors influencing diagnosis delay of advanced breast cancer in Moroccan women. BMC Cancer 2016, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Z.; Yang, J.; Jiang, Y.; Chen, Z.; Ali, Z.; He, N.; Wang, Z. Cell-specific biomarkers and targeted biopharmaceuticals for breast cancer treatment. Cell Prolif. 2016, 49, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, Q. Understanding the Global Cancer Statistics 2018: Implications for cancer control. Sci. China Life Sci. 2019, 64, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Zidan, H.E.; Karam, R.A.; El-Seifi, O.S.; Abd Elrahman, T.M. Circulating long non-coding RNA MALAT1 expression as molecular biomarker in Egyptian patients with breast cancer. Cancer Genet. 2018, 220, 32–37. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Fedewa, S.A.; Goding Sauer, A.; Kramer, J.L.; Smith, R.A.; Jemal, A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J. Clin. 2016, 66, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Lukong, K.E. Understanding breast cancer–The long and winding road. BBA Clin. 2017, 7, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.; Dorokhova, O.; Sunkara, J.; Schlesinger, K.; Suhrland, M.; Oktay, M.H. Estrogen, progesterone, and HER-2 receptor immunostaining in cytology: The effect of varied fixation on human breast cancer cells. Diagn. Cytopathol. 2013, 41, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Toss, A.; Cristofanilli, M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015, 17, 60. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ma, J. Association between imaging characteristics and different molecular subtypes of breast cancer. Acad. Radiol. 2017, 24, 426–434. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.; Gelber, R.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J.; Albain, K.S.; André, F.; Bergh, J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Wanchai, A.; Armer, J.M. A systematic review association of reflexology in managing symptoms and side effects of breast cancer treatment. Complement. Ther. Clin. Pract. 2020, 38, 101074. [Google Scholar] [CrossRef] [PubMed]
- Obi, E.; Tom, M.C.; Manyam, B.V.; Grobmyer, S.R.; Al-Hilli, Z.; Valente, S.; Fanning, A.; Radford, D.M.; Cherian, S.; Tendulkar, R.D. Outcomes with intraoperative radiation therapy for early-stage breast cancer. Breast J. 2020, 26, 454–457. [Google Scholar] [CrossRef]
- Gnant, M.; Harbeck, N.; Thomssen, C. St. Gallen/Vienna 2017: A brief summary of the consensus discussion about escalation and de-escalation of primary breast cancer treatment. Breast Care 2017, 12, 101–106. [Google Scholar] [CrossRef]
- Shrivastava, G.; Bakshi, H.A.; Aljabali, A.A.; Mishra, V.; Hakkim, F.L.; Charbe, N.B.; Kesharwani, P.; Chellappan, D.K.; Dua, K.; Tambuwala, M.M. Nucleic acid aptamers as a potential nucleus targeted drug delivery system. Curr. Drug Deliv. 2020, 17, 101–111. [Google Scholar] [CrossRef]
- Liang, T.; Yao, Z.; Ding, J.; Min, Q.; Jiang, L.; Zhu, J.-J. Cascaded aptamers-governed multistage drug-delivery system based on biodegradable envelope-type nanovehicle for targeted therapy of HER2-overexpressing breast cancer. ACS Appl. Mater. Interfaces 2018, 10, 34050–34059. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H. Recent developments in cell-SELEX technology for aptamer selection. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 2323–2329. [Google Scholar] [CrossRef]
- Chen, Z.; Ali, Z.; Li, S.; Liu, B.; He, N. Aptamers generated from cell-systematic evolution of ligands through exponential enrichment and their applications. J. Nanosci. Nanotechnol. 2016, 16, 9346–9358. [Google Scholar] [CrossRef]
- Xi, Z.; Zheng, B.; Wang, C. Synthesis, surface modification, and biolabeling with aptamer of Fe3O4@ SiO2 magnetic nanoparticles. Nanosci. Nanotechnol. Lett. 2016, 8, 1061–1066. [Google Scholar] [CrossRef]
- Amano, R.; Takada, K.; Tanaka, Y.; Nakamura, Y.; Kawai, G.; Kozu, T.; Sakamoto, T. Kinetic and thermodynamic analyses of interaction between a high-affinity RNA aptamer and its target protein. Biochemistry 2016, 55, 6221–6229. [Google Scholar] [CrossRef]
- Kanakaraj, I.; Chen, W.-H.; Poongavanam, M.; Dhamane, S.; Stagg, L.J.; Ladbury, J.E.; Kourentzi, K.; Strych, U.; Willson, R.C. Biophysical characterization of VEGF–aHt DNA aptamer interactions. Int. J. Biol. Macromol. 2013, 57, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Pica, A.; Russo Krauss, I.; Parente, V.; Tateishi-Karimata, H.; Nagatoishi, S.; Tsumoto, K.; Sugimoto, N.; Sica, F. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017, 45, 461–469. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ennifar, E.; Nakamura, Y. Thermodynamic study of aptamers binding to their target proteins. Biochimie 2018, 145, 91–97. [Google Scholar] [CrossRef]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Inside Cover: Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells (Angew. Chem. Int. Ed. 8/2019). Angew. Chem. Int. Ed. 2019, 58, 2158. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Zhao, Z.; Hu, Z.; Zhou, S.; Yang, N.; Huang, Y.; Zhang, Z.; Su, J.; Lan, D. A smart detection system based on specific magnetic and rolling cycle amplification signal-amplified dual-aptamers to accurately monitor minimal residual diseases in patients with T-ALL. J. Biomed. Nanotechnol. 2016, 12, 2151–2160. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Wang, L.; Zheng, Q.; Wu, G. Bi-specific aptamers on nanostructured substrates fail to capture cancer cells. J. Nanosci. Nanotechnol. 2016, 16, 8640–8647. [Google Scholar] [CrossRef]
- Shahdordizadeh, M.; Taghdisi, S.M.; Ansari, N.; Langroodi, F.A.; Abnous, K.; Ramezani, M. Aptamer based biosensors for detection of Staphylococcus aureus. Sens. Actuators B Chem. 2017, 241, 619–635. [Google Scholar] [CrossRef]
- Wang, K.; He, M.-Q.; Zhai, F.-H.; He, R.-H.; Yu, Y.-L. A novel electrochemical biosensor based on polyadenine modified aptamer for label-free and ultrasensitive detection of human breast cancer cells. Talanta 2017, 166, 87–92. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, J.; Li, J.; Liao, X.; Chen, F. Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Anal. Biochem. 2020, 598, 113620. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, N. Systematic Evolution of Novel 2′ F-PY RNA Aptamers Targeting the Membrane Protein l-Arginine/Agmatine Antiporter Purified in Mild Detergent; Middle East Technical University: Ankara, Turkey, 2021. [Google Scholar]
- Lee, J.-H.; Canny, M.D.; De Erkenez, A.; Krilleke, D.; Ng, Y.-S.; Shima, D.T.; Pardi, A.; Jucker, F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. USA 2005, 102, 18902–18907. [Google Scholar] [CrossRef] [PubMed]
- Gijs, M.; Penner, G.; Blackler, G.B.; Impens, N.R.; Baatout, S.; Luxen, A.; Aerts, A.M. Improved aptamers for the diagnosis and potential treatment of HER2-positive cancer. Pharmaceuticals 2016, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Shahdordizadeh, M.; Yazdian-Robati, R.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Aptamer application in targeted delivery systems for diagnosis and treatment of breast cancer. J. Mater. Chem. B 2016, 4, 7766–7778. [Google Scholar] [CrossRef]
- Han, J.; Gao, L.; Wang, J.; Wang, J. Application and development of aptamer in cancer: From clinical diagnosis to cancer therapy. J. Cancer 2020, 11, 6902. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Gholikhani, T.; Brito, B.J.; Livingston, F.; Kumar, S. The Potential Use of Aptamers in The Process of Drug Development. Pharm. Sci. 2021, 27, 503–510. [Google Scholar] [CrossRef]
- Nelissen, F.H.; Peeters, W.J.; Roelofs, T.P.; Nagelkerke, A.; Span, P.N.; Heus, H.A. Improving breast cancer treatment specificity using aptamers obtained by 3D cell-SELEX. Pharmaceuticals 2021, 14, 349. [Google Scholar] [CrossRef]
- Lin, C.-S.; Tsai, Y.-C.; Hsu, K.-F.; Lee, G.-B. Optimization of aptamer selection on an automated microfluidic system with cancer tissues. Lab. A Chip 2021, 21, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-C.; Lin, C.-S.; Lin, C.-N.; Hsu, K.-F.; Lee, G.-B. Screening aptamers targeting the cell membranes of clinical cancer tissues on an integrated microfluidic system. Sens. Actuators B Chem. 2021, 330, 129334. [Google Scholar] [CrossRef]
- Zhong, J.; Ding, J.; Deng, L.; Xiang, Y.; Liu, D.; Zhang, Y.; Chen, X.; Yang, Q. Selection of DNA aptamers recognizing EpCAM-positive prostate cancer by cell-SELEX for in vitro and in vivo MR imaging. Drug Des. Dev. Ther. 2021, 15, 3985. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.; Guthrie, J.W.; Zhang, H.; Li, X.-F.; Le, X.C. Selection and analytical applications of aptamers. TrAC Trends Anal. Chem. 2006, 25, 681–691. [Google Scholar] [CrossRef]
- Hamula, C.L.; Zhang, H.; Li, F.; Wang, Z.; Le, X.C.; Li, X.-F. Selection and analytical applications of aptamers binding microbial pathogens. TrAC Trends Anal. Chem. 2011, 30, 1587–1597. [Google Scholar] [CrossRef]
- Heiat, M.; Ranjbar, R.; Latifi, A.M.; Rasaee, M.J. Selection of a high-affinity and in vivo bioactive ssDNA aptamer against angiotensin II peptide. Peptides 2016, 82, 101–108. [Google Scholar] [CrossRef]
- Li, S.; Xu, H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2009, 218, 327–336. [Google Scholar] [CrossRef]
- Mi, J.; Ray, P.; Liu, J.; Kuan, C.-T.; Xu, J.; Hsu, D.; Sullenger, B.A.; White, R.R.; Clary, B.M. In vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Mol. Ther. Nucleic Acids 2016, 5, e315. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Wang, J.; Xu, L.; Chen, H.; Pei, R. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd (II). Talanta 2016, 154, 498–503. [Google Scholar] [CrossRef]
- Yang, K.-A.; Pei, R.; Stojanovic, M.N. In vitro selection and amplification protocols for isolation of aptameric sensors for small molecules. Methods 2016, 106, 58–65. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Paniel, N.; Istamboulié, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Spiga, F.M.; Maietta, P.; Guiducci, C. More DNA–aptamers for small drugs: A capture–SELEX coupled with surface plasmon resonance and high-throughput sequencing. ACS Comb. Sci. 2015, 17, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.; Kalyani, N.; Anand, A.; Khan, E.; Das, S.; Bansal, V.; Kumar, A.; Sharma, T.K. GOLD SELEX: A novel SELEX approach for the development of high-affinity aptamers against small molecules without residual activity. Microchim. Acta 2020, 187, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gao, T.; Li, W.; Luo, Y.; Ullah, S.; Fang, X.; Cao, Y.; Pei, R. Ni-Nitrilotriacetic acid affinity SELEX method for selection of DNA aptamers specific to the N-cadherin protein. ACS Comb. Sci. 2020, 22, 867–872. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Xi, Z.; Huang, R.; Li, Z.; He, N.; Wang, T.; Su, E.; Deng, Y. Selection of HBsAg-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis B virus infection. ACS Appl. Mater. Interfaces 2015, 7, 11215–11223. [Google Scholar] [CrossRef]
- Qi, S.; Duan, N.; Khan, I.M.; Dong, X.; Zhang, Y.; Wu, S.; Wang, Z. Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment. Biotechnol. Adv. 2022, 55, 107902. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Q.; Yin, Y.; Yang, Y.; Cui, H.; Dong, Y. Evolution of Interferon-Gamma Aptamer with Good Affinity and Analytical Utility by a Rational In Silico Base Mutagenesis Post-SELEX Strategy. Molecules 2022, 27, 5725. [Google Scholar] [CrossRef]
- Krasitskaya, V.V.; Goncharova, N.S.; Biriukov, V.V.; Bashmakova, E.E.; Kabilov, M.R.; Baykov, I.K.; Sokolov, A.E.; Frank, L.A. The Ca2+-Regulated Photoprotein Obelin as a Tool for SELEX Monitoring and DNA Aptamer Affinity Evaluation. Photochem. Photobiol. 2020, 96, 1041–1046. [Google Scholar] [CrossRef]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.; Appaiah, P.; Sistla, S.; Bk, B.; Bhatt, P. Bio-Layer Interferometry-Based SELEX and Label-Free Detection of Patulin Using Generated Aptamer. J. Agric. Food Chem. 2022, 70, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Canoura, J.; Yu, H.; Alkhamis, O.; Roncancio, D.; Farhana, R.; Xiao, Y. Accelerating post-SELEX aptamer engineering using exonuclease digestion. J. Am. Chem. Soc. 2020, 143, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Currás, N.; Somerson, J.; Vieira, P.A.; Ploense, K.L.; Kippin, T.E.; Plaxco, K.W. Real-time measurement of small molecules directly in awake, ambulatory animals. Proc. Natl. Acad. Sci. USA 2017, 114, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.C.; Zarbl, H. Use of cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS ONE 2012, 7, e36103. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004, 76, 5387–5392. [Google Scholar] [CrossRef]
- Hamedani, N.S.; Müller, J. Capillary electrophoresis for the selection of DNA aptamers recognizing activated protein C. In Nucleic Acid Aptamers; Springer: Berlin/Heidelberg, Germany, 2016; pp. 61–75. [Google Scholar]
- Mosing, R.K.; Mendonsa, S.D.; Bowser, M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005, 77, 6107–6112. [Google Scholar] [CrossRef]
- Yang, J.; Bowser, M.T. Capillary electrophoresis–SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal. Chem. 2013, 85, 1525–1530. [Google Scholar] [CrossRef]
- Zhang, X.h.; Wang, W.; Chen, X. Selection and identification of an ssDNA aptamer to NB4 cell. J. Clin. Lab. Anal. 2021, 35, e23718. [Google Scholar] [CrossRef]
- Nabavinia, M.S.; Charbgoo, F.; Alibolandi, M.; Mosaffa, F.; Gholoobi, A.; Ramezani, M.; Abnous, K. Comparison of flow cytometry and elasa for screening of proper candidate aptamer in cell-selex pool. Appl. Biochem. Biotechnol. 2018, 184, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Tibbs, J.; Che, C.; Peinetti, A.S.; Zhao, B.; Liu, L.; Barya, P.; Cooper, L.; Rong, L. Label-Free Digital Detection of Intact Virions by Enhanced Scattering Microscopy. J. Am. Chem. Soc. 2021, 144, 1498–1502. [Google Scholar] [CrossRef] [PubMed]
- Alkhamis, O.; Yang, W.; Farhana, R.; Yu, H.; Xiao, Y. Label-free profiling of DNA aptamer-small molecule binding using T5 exonuclease. Nucleic Acids Res. 2020, 48, e120. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Smith, M.; Blackburn, J.M. Autoantibody-based diagnostic biomarkers: Technological approaches to discovery and validation. In Autoantibodies and Cytokines; IntechOpen: London, UK, 2018. [Google Scholar]
- Gould, H.J.; Sutton, B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008, 8, 205–217. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in bioanalytical applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef]
- Kong, Q.; Yue, F.; Liu, M.; Huang, J.; Yang, F.; Liu, J.; Li, J.; Li, F.; Sun, X.; Guo, Y. Non-immobilized GO-SELEX of aptamers for label-free detection of thiamethoxam in vegetables. Anal. Chim. Acta 2022, 1202, 339677. [Google Scholar] [CrossRef]
- Guan, J.; He, K.; Gunasekaran, S. Selection of ssDNA aptamer using GO-SELEX and development of DNA nanostructure-based electrochemical aptasensor for penicillin. Biosens. Bioelectron. X 2022, 12, 100220. [Google Scholar] [CrossRef]
- Lyu, C.; Khan, I.M.; Wang, Z. Capture-SELEX for aptamer selection: A short review. Talanta 2021, 229, 122274. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-S.; Kim, E.-J.; Park, T.-K.; Bae, D.-W.; Cha, S.-S.; Kim, T.-W.; Kim, Y.-P. Gold nanoparticle-assisted SELEX as a visual monitoring platform for the development of small molecule-binding DNA aptasensors. Biosens. Bioelectron. 2021, 191, 113468. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-M.; Go, M.-J.; Lee, J.; Na, D.; Yoo, S.-M. Recent advances in micro/nanomaterial-based aptamer selection strategies. Molecules 2021, 26, 5187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Gao, Y.; Yang, C.; Zhang, X.; Hu, B.; Zhao, L.; Guo, H.; Sun, M.; Wang, L.; Jiao, B. A Novel SELEX Based on Immobilizing Libraries Enables Screening of Saxitoxin Aptamers for BLI Aptasensor Applications. Toxins 2022, 14, 228. [Google Scholar] [CrossRef] [PubMed]
- Phopin, K.; Tantimongcolwat, T. Pesticide Aptasensors—State of the Art and Perspectives. Sensors 2020, 20, 6809. [Google Scholar] [CrossRef] [PubMed]
- Zakeri-Milani, P.; Shirani, A.; Nokhodchi, A.; Mussa Farkhani, S.; Mohammadi, S.; Shahbazi Mojarrad, J.; Mahmoudian, M.; Gholikhani, T.; Farshbaf, M.; Valizadeh, H.; et al. Self-assembled peptide nanoparticles for efficient delivery of methotrexate into cancer cells. Drug Dev. Ind. Pharm. 2020, 46, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Rabai, S.; Benounis, M.; Catanante, G.; Baraket, A.; Errachid, A.; Renault, N.J.; Marty, J.-L.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 113956. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317, 126459. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Wu, P.; Li, L.; Huang, Q.; Wang, J.; Zhang, D.; Li, M.; Chen, N.; Wang, X. Ultrasensitive isothermal detection of a plant pathogen by using a gold nanoparticle-enhanced microcantilever sensor. Sens. Actuators B Chem. 2021, 338, 129874. [Google Scholar] [CrossRef]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Zhang, K. Closed Loop Aptameric Directed Evolution Selection of Therapeutic Aptamers for Chronic Myeloid Leukaemia; University of Manchester: Manchester, UK, 2019. [Google Scholar]
- Zakeri-Milani, P.; Najafi-Hajivar, S.; Sarfraz, M.; Nokhodchi, A.; Mohammadi, H.; Montazersaheb, S.; Niazi, M.; Hemmatzadeh, M.; Soleymani-Goloujeh, M.; Baradaran, B.; et al. Cytotoxicity and Immunogenicity Evaluation of Synthetic Cell-penetrating Peptides for Methotrexate Delivery. Iran. J. Pharm. Res. IJPR 2021, 20, 506. [Google Scholar]
- Li, Y.; Yang, F.; Li, S.; Yuan, R.; Xiang, Y. Target-triggered tertiary amplifications for sensitive and label-free protein detection based on lighting-up RNA aptamer transcriptions. Anal. Chim. Acta 2022, 1217, 340028. [Google Scholar] [CrossRef]
- Gaspar, I.; Ephrussi, A. Strength in numbers: Quantitative single-molecule RNA detection assays. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 135–150. [Google Scholar] [CrossRef]
- Ma, T.; Chen, L.; Shi, M.; Niu, J.; Zhang, X.; Yang, X.; Zhanghao, K.; Wang, M.; Xi, P.; Jin, D. Developing novel methods to image and visualize 3D genomes. Cell Biol. Toxicol. 2018, 34, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Agnello, L.; Fedele, M.; Camorani, S.; Cerchia, L. Profiling Cancer Cells by Cell-SELEX: Use of Aptamers for Discovery of Actionable Biomarkers and Therapeutic Applications Thereof. Pharmaceutics 2021, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Y.; Xu, X.; Liu, Y.; Lin, B.; Zhang, M.; Zhang, J.; Wan, S.; Yang, C.; Tan, W. Aptamer-based detection of circulating targets for precision medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef] [PubMed]
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wu, P.; Zhong, L.; Zhao, Y. Advances of Aptamer-based Clinical Applications for the Diagnosis and Therapy of Cancer. Discov. Med. 2020, 29, 169–180. [Google Scholar] [PubMed]
- Barman, J. Targeting cancer cells using aptamers: Cell-SELEX approach and recent advancements. RSC Adv. 2015, 5, 11724–11732. [Google Scholar] [CrossRef]
- Bakhtiari, H.; Palizban, A.A.; Khanahmad, H.; Mofid, M.R. Novel Approach to Overcome Defects of Cell-SELEX in Developing Aptamers against Aspartate β-Hydroxylase. ACS Omega 2021, 6, 11005–11014. [Google Scholar] [CrossRef]
- Bing, T.; Shangguan, D.; Wang, Y. Facile discovery of cell-surface protein targets of cancer cell aptamers. Mol. Cell. Proteom. 2015, 14, 2692–2700. [Google Scholar] [CrossRef]
- Jin, C.; Qiu, L.; Li, J.; Fu, T.; Zhang, X.; Tan, W. Cancer biomarker discovery using DNA aptamers. Analyst 2016, 141, 461–466. [Google Scholar] [CrossRef]
- Lu, M.; Zhou, L.; Zheng, X.; Quan, Y.; Wang, X.; Zhou, X.; Ren, J. A novel molecular marker of breast cancer stem cells identified by cell-SELEX method. Cancer Biomark. 2015, 15, 163–170. [Google Scholar] [CrossRef]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Uemachi, H.; Kasahara, Y.; Tanaka, K.; Okuda, T.; Yoneda, Y.; Obika, S. Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity. Pharmaceutics 2021, 13, 888. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999, 27, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Uptake and intracellular transport of RNA aptamers in African trypanosomes suggest therapeutic “piggy-back” approach. Bioorg. Med. Chem. 2001, 9, 2571–2580. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef]
- Chu, T.C.; Twu, K.Y.; Ellington, A.D.; Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006, 34, e73. [Google Scholar] [CrossRef]
- Ray, P.; White, R.R. Aptamers for targeted drug delivery. Pharmaceuticals 2010, 3, 1761–1778. [Google Scholar] [CrossRef]
- Sindhu, R.K.; Gupta, R.; Wadhera, G.; Kumar, P. Modern Herbal Nanogels: Formulation, Delivery Methods, and Applications. Gels 2022, 8, 97. [Google Scholar]
- van der Koog, L.; Gandek, T.B.; Nagelkerke, A. Liposomes and extracellular vesicles as drug delivery systems: A comparison of composition, pharmacokinetics, and functionalization. Adv. Healthc. Mater. 2022, 11, 2100639. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; Villanueva, H.; Casares, N.; Lasarte, J.J.; Bendandi, M.; Inoges, S.; de Cerio, A.L.-D.; Pastor, F. MRP1-CD28 bi-specific oligonucleotide aptamers: Target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget 2016, 7, 23182. [Google Scholar] [CrossRef]
- Rong, Y.; Chen, H.; Zhou, X.-F.; Yin, C.-Q.; Wang, B.-C.; Peng, C.-W.; Liu, S.-P.; Wang, F.-B. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget 2016, 7, 8282. [Google Scholar] [PubMed]
- Dua, P.; Kang, H.S.; Hong, S.-M.; Tsao, M.-S.; Kim, S.; Lee, D.-k. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res. 2013, 73, 1934–1945. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Qiao, L.; Zhou, S.-F.; Xiang, D.; Wang, T.; Li, Y.; Lim, L.Y.; Kong, L.; Li, L.; Duan, W. RNA aptamers targeting cancer stem cell marker CD133. Cancer Lett. 2013, 330, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Kanwar, J.R.; kumar Athalya, P.; Janakiraman, N.; Khetan, V.; Kanwar, R.K.; Eluchuri, S.; Krishnakumar, S. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J. Biomed. Sci. 2015, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dastjerdi, K.; Tabar, G.H.; Dehghani, H.; Haghparast, A. Generation of an enriched pool of DNA aptamers for an HER2-overexpressing cell line selected by Cell SELEX. Biotechnol. Appl. Biochem. 2011, 58, 226–230. [Google Scholar] [CrossRef]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef]
- Mallikaratchy, P.; Tang, Z.; Kwame, S.; Meng, L.; Shangguan, D.; Tan, W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteom. 2007, 6, 2230–2238. [Google Scholar] [CrossRef]
- Cerchia, L.; Esposito, C.L.; Camorani, S.; Rienzo, A.; Stasio, L.; Insabato, L.; Affuso, A.; De Franciscis, V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012, 20, 2291–2303. [Google Scholar] [CrossRef]
- Boltz, A.; Piater, B.; Toleikis, L.; Guenther, R.; Kolmar, H.; Hock, B. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem. 2011, 286, 21896–21905. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Chu, C.; Dai, W.; Liu, R.; Jiang, Y. Identification of a Specific RNA Aptamer Targeting Non-Small Cell Lung Cancer by In Vivo SELEX; AACR: Philadelphia, PA, USA, 2016. [Google Scholar]
- Maradani, B.S.; Parameswaran, S.; Subramanian, K. Development and characterization of DNA aptamer against Retinoblastoma by Cell-SELEX. Sci. Rep. 2022, 12, 16178. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Duan, A.; Chen, B.; Yu, X.; Wu, Y.; Zhang, X.; Li, S. Integration of an Expression Platform in the SELEX Cycle to Select DNA Aptamer Binding to a Disease Biomarker. ACS Omega 2022, 7, 10804–10811. [Google Scholar] [CrossRef] [PubMed]
- Niazi, J.H.; Verma, S.K.; Niazi, S.; Qureshi, A. In vitro HER2 protein-induced affinity dissociation of carbon nanotube-wrapped anti-HER2 aptamers for HER2 protein detection. Analyst 2015, 140, 243–249. [Google Scholar] [CrossRef]
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Label-free capacitance based aptasensor platform for the detection of HER2/ErbB2 cancer biomarker in serum. Sens. Actuators B Chem. 2015, 220, 1145–1151. [Google Scholar] [CrossRef]
- Guo, Q.; Li, X.; Shen, C.; Zhang, S.; Qi, H.; Li, T.; Yang, M. Electrochemical immunoassay for the protein biomarker mucin 1 and for MCF-7 cancer cells based on signal enhancement by silver nanoclusters. Microchim. Acta 2015, 182, 1483–1489. [Google Scholar] [CrossRef]
- Lan, J.; Li, L.; Liu, Y.; Yan, L.; Li, C.; Chen, J.; Chen, X. Upconversion luminescence assay for the detection of the vascular endothelial growth factor, a biomarker for breast cancer. Microchim. Acta 2016, 183, 3201–3208. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Development of an electrochemical aptasensor for the detection of human osteopontin. Procedia Eng. 2014, 87, 316–319. [Google Scholar] [CrossRef]
- Sett, A.; Borthakur, B.B.; Sharma, J.D.; Kataki, A.C.; Bora, U. DNA aptamer probes for detection of estrogen receptor α positive carcinomas. Transl. Res. 2017, 183, 104–120.e102. [Google Scholar] [CrossRef]
- Wu, J.; Wang, C.; Li, X.; Song, Y.; Wang, W.; Li, C.; Hu, J.; Zhu, Z.; Li, J.; Zhang, W. Identification, characterization and application of a G-quadruplex structured DNA aptamer against cancer biomarker protein anterior gradient homolog 2. PLoS ONE 2012, 7, e46393. [Google Scholar]
- Ahirwar, R.; Vellarikkal, S.K.; Sett, A.; Sivasubbu, S.; Scaria, V.; Bora, U.; Borthakur, B.B.; Kataki, A.C.; Sharma, J.D.; Nahar, P. Aptamer-assisted detection of the altered expression of estrogen receptor alpha in human breast cancer. PLoS ONE 2016, 11, e0153001. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bai, X.; Wang, N.; Chen, X.; Li, J.; Zhang, Z.; Tang, J. Aptamer-based microcantilever biosensor for ultrasensitive detection of tumor marker nucleolin. Talanta 2016, 146, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Sun, P.; Dong, X.; Zhu, C.; Liu, X.; Zheng, D.; Liu, C. Aptamers as Recognition Elements for Electrochemical Detection of Exosomes. Chem. Res. Chin. Univ. 2022, 38, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hernández, C.D.; Rodríguez-Martínez, G.; Cortés-Ramírez, S.A.; Morales-Pacheco, M.; Cruz-Burgos, M.; Losada-García, A.; Reyes-Grajeda, J.P.; González-Ramírez, I.; González-Covarrubias, V.; Camacho-Arroyo, I. Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules 2022, 12, 1056. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, C.; Wang, Y.; Chen, G. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol. Biol. Rep. 2022, 49, 7979–7993. [Google Scholar] [CrossRef]
- Jo, H.; Ban, C. Aptamer–nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med. 2016, 48, e230. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, Y.; Yu, J.; Wang, S.; Ge, S.; Song, X. Application of ZnO/graphene and S6 aptamers for sensitive photoelectrochemical detection of SK-BR-3 breast cancer cells based on a disposable indium tin oxide device. Biosens. Bioelectron. 2014, 51, 413–420. [Google Scholar] [CrossRef]
- Liu, Q.; Jin, C.; Wang, Y.; Fang, X.; Zhang, X.; Chen, Z.; Tan, W. Aptamer-conjugated nanomaterials for specific cancer cell recognition and targeted cancer therapy. NPG Asia Mater. 2014, 6, e95. [Google Scholar] [CrossRef]
- Meng, H.-M.; Fu, T.; Zhang, X.-B.; Tan, W. Cell-SELEX-based aptamer-conjugated nanomaterials for cancer diagnosis and therapy. Natl. Sci. Rev. 2015, 2, 71–84. [Google Scholar] [CrossRef]
- Zhu, G.; Zhang, S.; Song, E.; Zheng, J.; Hu, R.; Fang, X.; Tan, W. Building fluorescent DNA nanodevices on target living cell surfaces. Angew. Chem. Int. Ed. 2013, 52, 5490–5496. [Google Scholar] [CrossRef]
- Cai, S.; Li, G.; Zhang, X.; Xia, Y.; Chen, M.; Wu, D.; Chen, Q.; Zhang, J.; Chen, J. A signal-on fluorescent aptasensor based on single-stranded DNA-sensitized luminescence of terbium (III) for label-free detection of breast cancer cells. Talanta 2015, 138, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wang, Y.; Wen, X.; Li, C.; Lei, L.; Guo, Q.; Sun, G.; Yu, L.; Nie, H. A DNA Aptamer Targeting Cellular Fibronectin Rather Than Plasma Fibronectin for Bioimaging and Targeted Chemotherapy of Tumors. Adv. Funct. Mater. 2022, 32, 2205002. [Google Scholar] [CrossRef]
- Jo, H.; Her, J.; Ban, C. Dual aptamer-functionalized silica nanoparticles for the highly sensitive detection of breast cancer. Biosens. Bioelectron. 2015, 71, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, J.; Ali, Z.; Wang, Z.; Mou, X.; He, N.; Wang, Z. Synthesis of aptamer-functionalized Ag nanoclusters for MCF-7 breast cancer cells imaging. Sci. China Chem. 2017, 60, 370–376. [Google Scholar] [CrossRef]
- Malicki, S.; Pucelik, B.; Żyła, E.; Benedyk-Machaczka, M.; Gałan, W.; Golda, A.; Sochaj-Gregorczyk, A.; Kamińska, M.; Encarnação, J.C.; Chruścicka, B. Imaging of Clear Cell Renal Carcinoma with Immune Checkpoint Targeting Aptamer-Based Probe. Pharmaceuticals 2022, 15, 697. [Google Scholar] [CrossRef]
- Li, J.; Zhong, X.; Cheng, F.; Zhang, J.-R.; Jiang, L.-P.; Zhu, J.-J. One-pot synthesis of aptamer-functionalized silver nanoclusters for cell-type-specific imaging. Anal. Chem. 2012, 84, 4140–4146. [Google Scholar] [CrossRef]
- Liu, Z.; Duan, J.-H.; Song, Y.-M.; Ma, J.; Wang, F.-D.; Lu, X.; Yang, X.-D. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J. Transl. Med. 2012, 10, 148. [Google Scholar] [CrossRef]
- Wu, P.; Gao, Y.; Zhang, H.; Cai, C. Aptamer-guided silver–gold bimetallic nanostructures with highly active surface-enhanced raman scattering for specific detection and near-infrared photothermal therapy of human breast cancer cells. Anal. Chem. 2012, 84, 7692–7699. [Google Scholar] [CrossRef]
- Hua, X.; Zhou, Z.; Yuan, L.; Liu, S. Selective collection and detection of MCF-7 breast cancer cells using aptamer-functionalized magnetic beads and quantum dots based nano-bio-probes. Anal. Chim. Acta 2013, 788, 135–140. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Liu, L.; Zhu, Z.; Ouyang, G.; An, Y.; Zhao, C.; Yang, C.J. In vitro selection of DNA aptamers for metastatic breast cancer cell recognition and tissue imaging. Anal. Chem. 2014, 86, 6596–6603. [Google Scholar] [CrossRef]
- Zhang, K.; Sefah, K.; Tang, L.; Zhao, Z.; Zhu, G.; Ye, M.; Sun, W.; Goodison, S.; Tan, W. A novel aptamer developed for breast cancer cell internalization. ChemMedChem 2012, 7, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, Z.; An, Y.; Zhang, W.; Zhang, H.; Liu, D.; Yu, C.; Duan, W.; Yang, C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.-F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef]
- Pourhajibagher, M.; Etemad-Moghadam, S.; Alaeddini, M.; Mousavi, M.; Bahador, A. DNA-aptamer-nanographene oxide as a targeted bio-theragnostic system in antimicrobial photodynamic therapy against Porphyromonas gingivalis. Sci. Rep. 2022, 12, 1–18. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef]
- Wei, J.; Song, R.; Sabbagh, A.; Marisetty, A.; Shukla, N.; Fang, D.; Najem, H.; Ott, M.; Long, J.; Zhai, L. Cell-directed aptamer therapeutic targeting for cancers including those within the central nervous system. OncoImmunology 2022, 11, 2062827. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.S.; Borkowski, S.; Kurreck, J.; Stephens, A.W.; Bald, R.; Hecht, M.; Friebe, M.; Dinkelborg, L.; Erdmann, V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004, 32, 5757–5765. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H. RNA aptamers: From basic science towards therapy. RNA Towards Med. 2006, 173, 305–326. [Google Scholar]
- Eulberg, D.; Klussmann, S. Spiegelmers: Biostable aptamers. ChemBioChem 2003, 4, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yin, Q.; Chang, Y.; Zhang, Q.; Brennan, J.D.; Li, Y. In vitro selection of circular DNA aptamers for biosensing applications. Angew. Chem. Int. Ed. 2019, 58, 8013–8017. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2020, 13, 9500–9519. [Google Scholar] [CrossRef]
- Uludag, H.; Ubeda, A.; Ansari, A. At the intersection of biomaterials and gene therapy: Progress in non-viral delivery of nucleic acids. Front. Bioeng. Biotechnol. 2019, 7, 131. [Google Scholar] [CrossRef]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham Jr, E.T.; Feinsod, M.; Guyer, D.R. Pegaptanib for neovascular age-related macular degeneration. New Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef]
- De Smidt, P.C.; Doan, T.L.; Falco, S.d.; Berkel, T.J.v. Association of antisense oligonucleotides with lipoproteins prolongs the plasma half-life and modifies the tissue distribution. Nucleic Acids Res. 1991, 19, 4695–4700. [Google Scholar] [CrossRef]
- Camorani, S.; Crescenzi, E.; Gramanzini, M.; Fedele, M.; Zannetti, A.; Cerchia, L. Aptamer-mediated impairment of EGFR-integrin αvβ3 complex inhibits vasculogenic mimicry and growth of triple-negative breast cancers. Sci. Rep. 2017, 7, 46659. [Google Scholar] [CrossRef]
- Chen, K.; Liu, J.; Tong, G.; Liu, B.; Wang, G.; Liu, H. Adipo8, a high-affinity DNA aptamer, can differentiate among adipocytes and inhibit intracellular lipid accumulation in vitro. Sci. China Chem. 2015, 58, 1612–1620. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, J.S. The application of aptamer in apoptosis. Biochimie 2017, 132, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bala, J.; Bhaskar, A.; Varshney, A.; Singh, A.K.; Dey, S.; Yadava, P. In vitro selected RNA aptamer recognizing glutathione induces ROS mediated apoptosis in the human breast cancer cell line MCF 7. RNA Biol. 2011, 8, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Yuhan, J.; Zhu, L.; Zhu, L.; Huang, K.; He, X.; Xu, W. Cell-specific aptamers as potential drugs in therapeutic applications: A review of current progress. J. Control. Release 2022, 346, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef]
- Ibarra, L.E.; Camorani, S.; Agnello, L.; Pedone, E.; Pirone, L.; Chesta, C.A.; Palacios, R.E.; Fedele, M.; Cerchia, L. Selective photo-assisted eradication of Triple-Negative breast cancer cells through aptamer decoration of doped conjugated polymer nanoparticles. Pharmaceutics 2022, 14, 626. [Google Scholar] [CrossRef]
- Varshney, A.; Bala, J.; Santosh, B.; Bhaskar, A.; Kumar, S.; Yadava, P.K. Identification of an RNA aptamer binding hTERT-derived peptide and inhibiting telomerase activity in MCF7 cells. Mol. Cell. Biochem. 2017, 427, 157–167. [Google Scholar] [CrossRef]
- Bayat, P.; Abnous, K.; Balarastaghi, S.; Taghdisi, S.M.; Saeedi, M.; Yazdian-Robati, R.; Mahmoudi, M. Aptamer AS1411-functionalized gold nanoparticle-melittin complex for targeting MCF-7 breast cancer cell line. Nanomed. J. 2022, 9, 164–169. [Google Scholar]
- Chen, Y.; Shi, S. Advances and prospects of dynamic DNA nanostructures in biomedical applications. RSC Adv. 2022, 12, 30310–30320. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, Z.; Wan, Q.; Liu, X.; Qi, J.; Zu, Y. Targeted immunotherapy of triple-negative breast cancer by aptamer-engineered NK cells. Biomaterials 2022, 280, 121259. [Google Scholar] [CrossRef]
- Dai, B.; Hu, Y.; Duan, J.; Yang, X.-D. Aptamer-guided DNA tetrahedron as a novel targeted drug delivery system for MUC1-expressing breast cancer cells in vitro. Oncotarget 2016, 7, 38257. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Zeng, X.; Wu, J.; Zhu, X.; Yu, X.; Zhang, X.; Zhang, J.; Liu, G.; Mei, L. Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics 2016, 6, 470. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Ramesh, K.; Kumar, D.N.; Dehari, D.; Singh, S.; Kumar, D.; Agrawal, A.K. Polymeric micelles: A novel drug delivery system for the treatment of breast cancer. J. Drug Deliv. Sci. Technol. 2022, 77, 103886. [Google Scholar] [CrossRef]
- Beqa, L.; Fan, Z.; Singh, A.K.; Senapati, D.; Ray, P.C. Gold nano-popcorn attached SWCNT hybrid nanomaterial for targeted diagnosis and photothermal therapy of human breast cancer cells. ACS Appl. Mater. Interfaces 2011, 3, 3316–3324. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Xiang, J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9123. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Lo, Y.; Shiu, S.C.-C.; Kinghorn, A.B.; Tanner, J.A. Aptamer-Enabled Nanomaterials for Therapeutics, Drug Targeting and Imaging. Cells 2022, 11, 159. [Google Scholar] [CrossRef]
- Stern, J.M.; Stanfield, J.; Kabbani, W.; Hsieh, J.-T.; Cadeddu, J.A. Selective prostate cancer thermal ablation with laser activated gold nanoshells. J. Urol. 2008, 179, 748–753. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Thompson, L.B.; Boulos, S.P.; Sisco, P.N.; Murphy, C.J. Gold nanorods: Their potential for photothermal therapeutics and drug delivery, tempered by the complexity of their biological interactions. Adv. Drug Deliv. Rev. 2012, 64, 190–199. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Lee, A.S.W.; Yap, L.W.; Jans, D.A.; Wagstaff, K.M.; Cheng, W. Tumor cell-specific photothermal killing by SELEX-derived DNA aptamer-targeted gold nanorods. Nanoscale 2016, 8, 187–196. [Google Scholar] [CrossRef]
- Malik, M.T.; O’Toole, M.G.; Casson, L.K.; Thomas, S.D.; Bardi, G.T.; Reyes-Reyes, E.M.; Ng, C.K.; Kang, K.A.; Bates, P.J. AS1411-conjugated gold nanospheres and their potential for breast cancer therapy. Oncotarget 2015, 6, 22270. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, M.; Guo, K.; Wang, Z.; Zhang, C.; Shubhra, Q.T. Systemic Co-delivery of drugs by a pH-and photosensitive smart nanocarrier to treat cancer by chemo-photothermal-starvation combination therapy. Smart Mater. Med. 2022, 3, 390–403. [Google Scholar] [CrossRef]
- Herrmann, A.; Priceman, S.J.; Kujawski, M.; Xin, H.; Cherryholmes, G.A.; Zhang, W.; Zhang, C.; Lahtz, C.; Kowolik, C.; Forman, S.J. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Investig. 2014, 124, 2977–2987. [Google Scholar] [CrossRef]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O.; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, S.H.; Hwang, Y.; Yoo, H.; Jung, H.; Kim, S.H.; Mok, H. Multivalent Aptamer–RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells. Macromol. Biosci. 2017, 17, 1600343. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gantier, M.P.; Xiang, D.; Bean, A.G.; Bruce, M.; Zhou, S.-F.; Khasraw, M.; Ward, A.; Wang, L.; Wei, M.Q. EpCAM aptamer-mediated survivin silencing sensitized cancer stem cells to doxorubicin in a breast cancer model. Theranostics 2015, 5, 1456. [Google Scholar] [CrossRef]
| Subtype | ER | PR | Ki-67 | HER2 |
|---|---|---|---|---|
| Luminal A | + | +/− | <14% | |
| Luminal B HER2-negative | + | +/− | ≥14% | − |
| Luminal B HER2-positive | +/− | +/− | + | |
| Luminal B Triple negative | − | − | − |
| Target | Aptamer | SELEX Type | Cancer | Purpose | Ref. |
|---|---|---|---|---|---|
| MRP1 | RNA | Novel combinatorial peptide-cell SELEX | Melanoma | Therapeutic | [114] |
| CD44/CD24 | DNA | Cell-SELEX | Breast cancer | Diagnostic and Therapeutic | [104] |
| Cytokeratin 19 | DNA | Cell-SELEX | Metastatic hepatocellular carcinoma | Diagnostic and Therapeutic | [115] |
| Alkaline PhosphatasePlacental-Like2 (ALPPL-2) | RNA | Cell-SELEX | Pancreatic cancer | Diagnostic and Therapeutic | [116] |
| CD133 | RNA | Cell-SELEX | Cancer stem cell targeting | Diagnostic and Therapeutic | [117] |
| EpCAM | RNA | Cell-SELEX | Molecular imaging agents for cancer theranostics | Diagnostic and Therapeutic | [118] |
| HER2 | DNA | Cell-SELEX | HER2 positive breast cancer | Diagnostic and Therapeutic | [119] |
| PTK7 | DNA | Cell-SELEX | Acute lymphoblastic leukemia | Diagnostic and Therapeutic | [120] |
| Immunoglobin Heavy Mu Chain (IGHM) | DNA | Cell-SELEX | Burkitt lymphoma | Diagnostic and Therapeutic | [121] |
| AXL | RNA | Cell-SELEX | Human glioma cell cancer | Therapeutic | [122] |
| CD16-(FcRIII_) | DNA | Cell-SELEX | Cancer immunotherapy | Therapeutic | [123] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gholikhani, T.; Kumar, S.; Valizadeh, H.; Mahdinloo, S.; Adibkia, K.; Zakeri-Milani, P.; Barzegar-Jalali, M.; Jimenez, B. Advances in Aptamers-Based Applications in Breast Cancer: Drug Delivery, Therapeutics, and Diagnostics. Int. J. Mol. Sci. 2022, 23, 14475. https://doi.org/10.3390/ijms232214475
Gholikhani T, Kumar S, Valizadeh H, Mahdinloo S, Adibkia K, Zakeri-Milani P, Barzegar-Jalali M, Jimenez B. Advances in Aptamers-Based Applications in Breast Cancer: Drug Delivery, Therapeutics, and Diagnostics. International Journal of Molecular Sciences. 2022; 23(22):14475. https://doi.org/10.3390/ijms232214475
Chicago/Turabian StyleGholikhani, Tooba, Shalen Kumar, Hadi Valizadeh, Somayeh Mahdinloo, Khosro Adibkia, Parvin Zakeri-Milani, Mohammad Barzegar-Jalali, and Balam Jimenez. 2022. "Advances in Aptamers-Based Applications in Breast Cancer: Drug Delivery, Therapeutics, and Diagnostics" International Journal of Molecular Sciences 23, no. 22: 14475. https://doi.org/10.3390/ijms232214475
APA StyleGholikhani, T., Kumar, S., Valizadeh, H., Mahdinloo, S., Adibkia, K., Zakeri-Milani, P., Barzegar-Jalali, M., & Jimenez, B. (2022). Advances in Aptamers-Based Applications in Breast Cancer: Drug Delivery, Therapeutics, and Diagnostics. International Journal of Molecular Sciences, 23(22), 14475. https://doi.org/10.3390/ijms232214475




