MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor
Abstract
1. Introduction
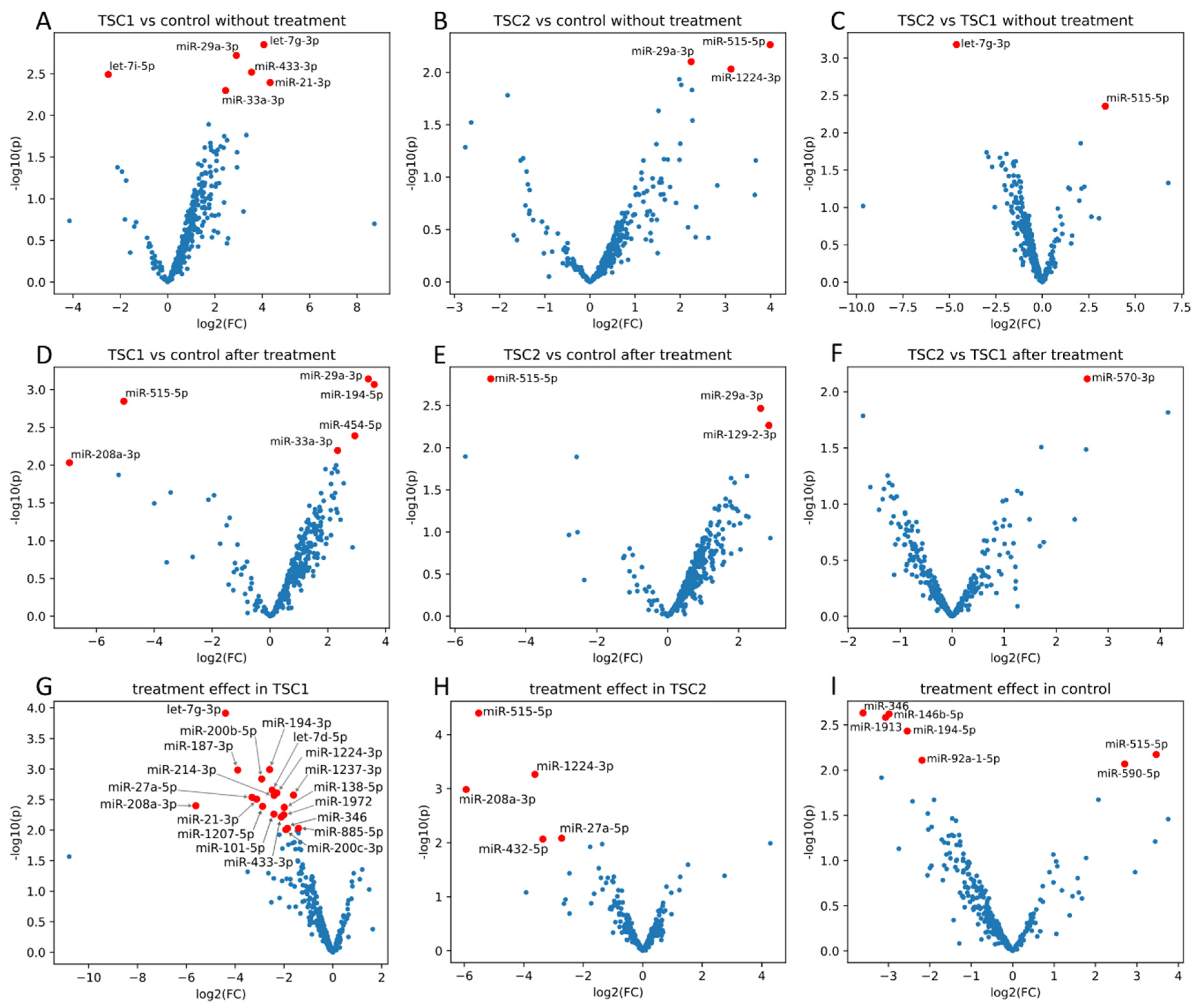
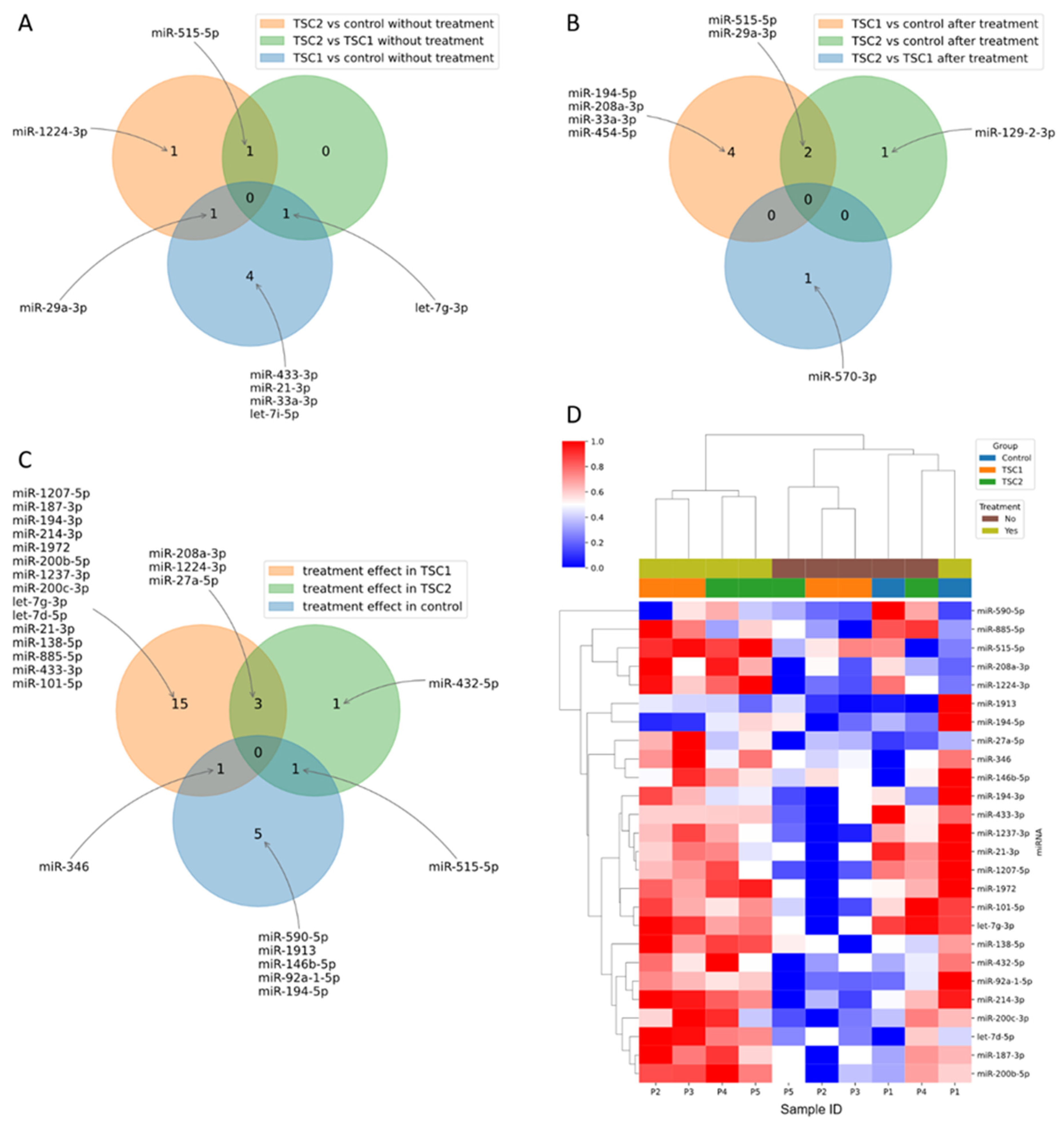
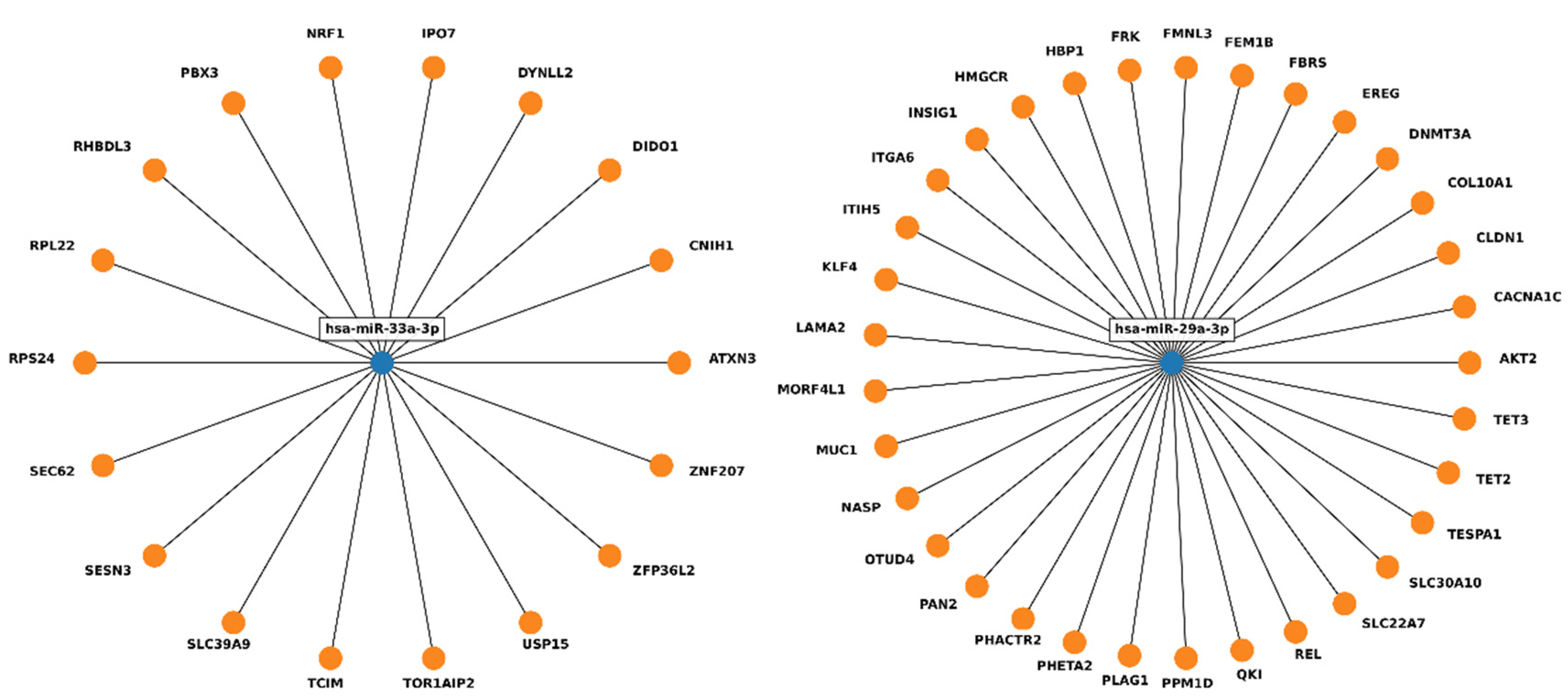
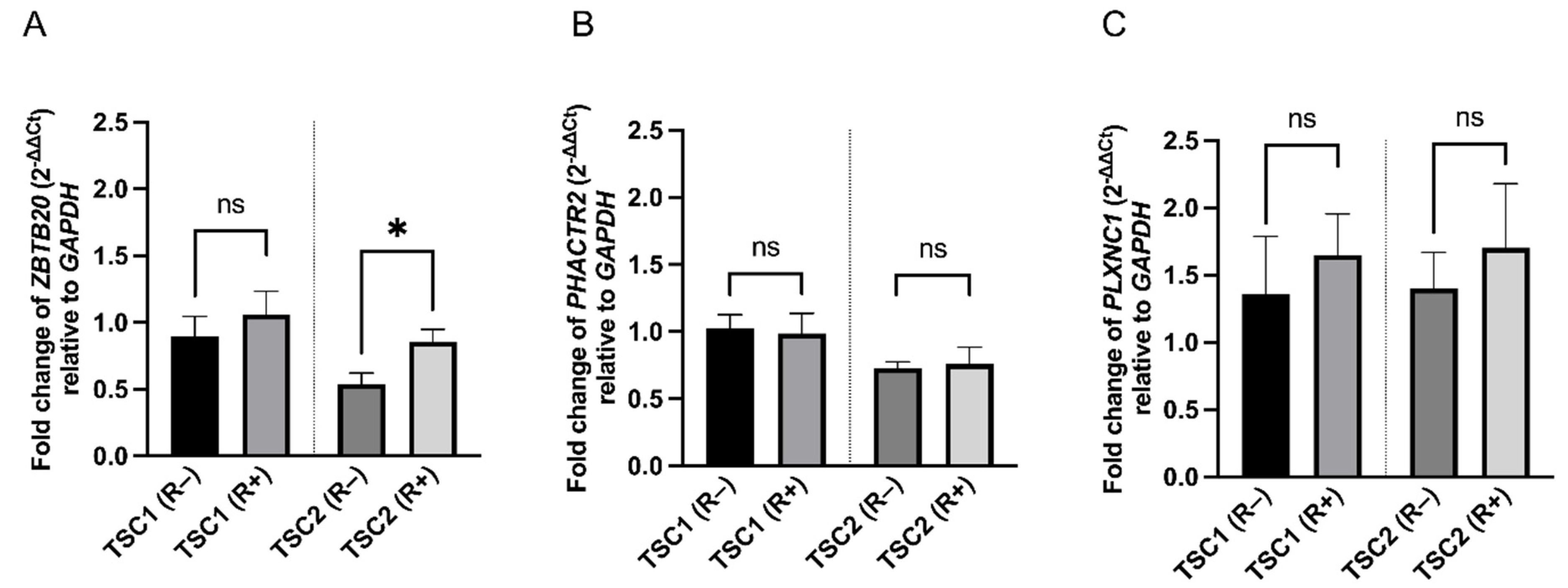
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MicroRNA Profiling
4.3. Reverse Transcriptase Reaction
4.4. Real-Time PCR (qPCR)
4.5. RNA Isolation and Gene Expression Study
4.6. Bioinformatical and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wataya-Kaneda, M. Mammalian Target of Rapamycin and Tuberous Sclerosis Complex. J. Dermatol. Sci. 2015, 79, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Manning, B.D. The TSC1-TSC2 Complex: A Molecular Switchboard Controlling Cell Growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Crino, P.B.; Nathanson, K.L.; Henske, E.P. The Tuberous Sclerosis Complex. N. Engl. J. Med. 2006, 355, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Franz, D.N.; Belousova, E.; Sparagana, S.; Bebin, E.M.; Frost, M.; Kuperman, R.; Witt, O.; Kohrman, M.H.; Flamini, J.R.; Wu, J.Y.; et al. Efficacy and Safety of Everolimus for Subependymal Giant Cell Astrocytomas Associated with Tuberous Sclerosis Complex (EXIST-1): A Multicentre, Randomised, Placebo-Controlled Phase 3 Trial. Lancet 2013, 381, 125–132. [Google Scholar] [CrossRef]
- Bissler, J.J.; Kingswood, J.C.; Radzikowska, E.; Zonnenberg, B.A.; Frost, M.; Belousova, E.; Sauter, M.; Nonomura, N.; Brakemeier, S.; De Vries, P.J.; et al. Everolimus for Angiomyolipoma Associated with Tuberous Sclerosis Complex or Sporadic Lymphangioleiomyomatosis (EXIST-2): A Multicentre, Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 2013, 381, 817–824. [Google Scholar] [CrossRef]
- Bissler, J.J.; McCormack, F.X.; Young, L.R.; Elwing, J.M.; Chuck, G.; Leonard, J.M.; Schmithorst, V.J.; Laor, T.; Brody, A.S.; Bean, J.; et al. Sirolimus for Angiomyolipoma in Tuberous Sclerosis Complex or Lymphangioleiomyomatosis. N. Engl. J. Med. 2008, 358, 140–151. [Google Scholar] [CrossRef]
- Krueger, D.A.; Care, M.M.; Holland, K.; Agricola, K.; Tudor, C.; Mangeshkar, P.; Wilson, K.A.; Byars, A.; Sahmoud, T.; Franz, D.N. Everolimus for Subependymal Giant-Cell Astrocytomas in Tuberous Sclerosis. N. Engl. J. Med. 2010, 363, 1801–1811. [Google Scholar] [CrossRef]
- Franz, D.N.; Leonard, J.; Tudor, C.; Chuck, G.; Care, M.; Sethuraman, G.; Dinopoulos, A.; Thomas, G.; Crone, K.R. Rapamycin Causes Regression of Astrocytomas in Tuberous Sclerosis Complex. Ann. Neurol. 2006, 59, 490–498. [Google Scholar] [CrossRef]
- Miller, J.M.; Wachsman, A.; Haker, K.; Majlessipour, F.; Danielpour, M.; Puliyanda, D. The Effects of Everolimus on Tuberous Sclerosis-Associated Lesions Can Be Dramatic but May Be Impermanent. Pediatr. Nephrol. 2015, 30, 173–177. [Google Scholar] [CrossRef]
- Romaker, D.; Kumar, V.; Cerqueira, D.M.; Cox, R.M.; Wessely, O. MicroRNAs Are Critical Regulators of Tuberous Sclerosis Complex and MTORC1 Activity in the Size Control of the Xenopus Kidney. Proc. Natl. Acad. Sci. USA 2014, 111, 6335–6340. [Google Scholar] [CrossRef]
- Fragkouli, A.; Doxakis, E. MiR-7 and MiR-153 Protect Neurons against MPP+-Induced Cell Death via Upregulation of MTOR Pathway. Front. Cell. Neurosci. 2014, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- AlQurashi, N.; Hashimi, S.M.; Wei, M.Q. Chemical Inhibitors and MicroRNAs (MiRNA) Targeting the Mammalian Target of Rapamycin (MTOR) Pathway: Potential for Novel Anticancer Therapeutics. Int. J. Mol. Sci. 2013, 14, 3874–3900. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, B.; Smyczyńska, U.; Grabia, S.; Fendler, W.; Dróżdż, I.; Bąbol-Pokora, K.; Kotulska, K.; Jóźwiak, S.; Borkowska, J.; Młynarski, W.; et al. MTOR Inhibitor Treatment in Patients with Tuberous Sclerosis Complex Is Associated with Specific Changes in MicroRNA Serum Profile. J. Clin. Med. 2022, 11, 3395. [Google Scholar] [CrossRef] [PubMed]
- Trelinska, J.; Fendler, W.; Dachowska, I.; Kotulska, K.; Jozwiak, S.; Antosik, K.; Gnys, P.; Borowiec, M.; Mlynarski, W. Abnormal Serum MicroRNA Profiles in Tuberous Sclerosis Are Normalized during Treatment with Everolimus: Possible Clinical Implications. Orphanet J. Rare Dis. 2016, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Trindade, A.J.; Medvetz, D.A.; Neuman, N.A.; Myachina, F.; Yu, J.; Priolo, C.; Henske, E.P. MicroRNA-21 Is Induced by Rapamycin in a Model of Tuberous Sclerosis (TSC) and Lymphangioleiomyomatosis (LAM). PLoS ONE 2013, 8, e60014. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of Patterns in Biological Sequences at the ALGGEN Server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Franz, D.N.; Belousova, E.; Sparagana, S.; Bebin, E.M.; Frost, M.D.; Kuperman, R.; Witt, O.; Kohrman, M.H.; Flamini, J.R.; Wu, J.Y.; et al. Long-Term Use of Everolimus in Patients with Tuberous Sclerosis Complex: Final Results from the EXIST-1 Study. PLoS ONE 2016, 11, e0158476. [Google Scholar] [CrossRef]
- Bissler, J.J.; Franz, D.N.; Frost, M.D.; Belousova, E.; Bebin, E.M.; Sparagana, S.; Berkowitz, N.; Ridolfi, A.; Kingswood, J.C. The Effect of Everolimus on Renal Angiomyolipoma in Pediatric Patients with Tuberous Sclerosis Being Treated for Subependymal Giant Cell Astrocytoma. Pediatr. Nephrol. 2018, 33, 101–109. [Google Scholar] [CrossRef]
- Franz, D.N.; Lawson, J.A.; Yapici, Z.; Ikeda, H.; Polster, T.; Nabbout, R.; Curatolo, P.; de Vries, P.J.; Dlugos, D.J.; Voi, M.; et al. Everolimus for Treatment-Refractory Seizures in TSC: Extension of a Randomized Controlled Trial. Neurol. Clin. Pract. 2018, 8, 412–420. [Google Scholar] [CrossRef]
- Trelinska, J.; Dachowska, I.; Kotulska, K.; Baranska, D.; Fendler, W.; Jozwiak, S.; Mlynarski, W. Factors Affecting Response to Everolimus Therapy for Subependymal Giant Cell Astrocytomas Associated with Tuberous Sclerosis. Pediatr. Blood Cancer 2015, 62, 616–621. [Google Scholar] [CrossRef]
- Calsina, B.; Jaime Castro-Vega, L.; Torres-Pérez, R.; Inglada-Pérez, L.; Currás-Freixes, M.; María Roldán-Romero, J.; Mancikova, V.; Letón, R.; Remacha, L.; Santos, M.; et al. Integrative Multi-Omics Analysis Identifies a Prognostic MiRNA Signature and a Targetable MiR-21-3p/TSC2/ MTOR Axis in Metastatic Pheochromocytoma/ Paraganglioma. Theranostics 2019, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Jin, D.; Zhuo, Z.; Zhang, B.; Chen, K. MiR-1224-5p Activates Autophagy, Cell Invasion and Inhibits Epithelial-to-Mesenchymal Transition in Osteosarcoma Cells by Directly Targeting PLK1 Through PI3K/AKT/MTOR Signaling Pathway. Onco. Targets Ther. 2020, 13, 11807–11818. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wei, J.; Tang, T.; Huang, Z. Role of MicroRNA-33a in Malignant Cells (Review). Oncol. Lett. 2020, 20, 2537–2556. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farré, D.; Nuñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of Known Transcription Reg-ulatory Elements Using Species-Tailored Searches. Bioinform. Appl. Note 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, K.; Shi, L.; Zhang, Q.; Zhang, S. CircPVT1 Promoted the Progression of Breast Cancer by Regulating MiR-29a-3p-Mediated AGR2-HIF-1α Pathway. Cancer Manag. Res. 2020, 12, 11477–11490. [Google Scholar] [CrossRef] [PubMed]
- Land, S.C.; Tee, A.R. Hypoxia-Inducible Factor 1α Is Regulated by the Mammalian Target of Rapamycin (MTOR) via an MTOR Signaling Motif. J. Biol. Chem. 2007, 282, 20534–20543. [Google Scholar] [CrossRef]
- Song, Q.; Zhang, H.; He, J.; Kong, H.; Tao, R.; Huang, Y.; Yu, H.; Zhang, Z.; Huang, Z.; Wei, L.; et al. Long Non-Coding RNA LINC00473 Acts as a MicroRNA-29a-3p Sponge to Promote Hepatocellular Carcinoma Development by Activating Robo1-Dependent PI3K/AKT/MTOR Signaling Pathway. Ther. Adv. Med. Oncol. 2020, 12, 1758835920937890. [Google Scholar] [CrossRef]
- Korneenko, T.V.; Pestov, N.B.; Ahmad, N.; Okkelman, I.A.; Dmitriev, R.I.; Shakhparonov, M.I.; Modyanov, N.N. Evolutionary Diversification of the BetaM Interactome Acquired through Co-Option of the ATP1B4 Gene in Placental Mammals. Nat. Publ. Gr. 2016, 6, 22395. [Google Scholar] [CrossRef]
- Doeppner, T.R.; Herz, J.; Bähr, M.; Tonchev, A.B.; Stoykova, A. Zbtb20 Regulates Developmental Neurogenesis in the Olfactory Bulb and Gliogenesis After Adult Brain Injury. Mol. Neurobiol. 2019, 56, 567–582. [Google Scholar] [CrossRef]
- Nielsen, J.V.; Nielsen, F.H.; Ismail, R.; Noraberg, J.; Jensen, N.A. Hippocampus-like Corticoneurogenesis Induced by Two Isoforms of the BTB-Zinc Finger Gene Zbtb20 in Mice. Development 2007, 134, 1133–1140. [Google Scholar] [CrossRef]
- Chabrat, A.; Brisson, G.; Doucet-Beaupré, H.; Salesse, C.; Profes, M.S.; Dovonou, A.; Akitegetse, C.; Charest, J.; Lemstra, S.; Côté, D.; et al. Transcriptional Repression of Plxnc1 by Lmx1a and Lmx1b Directs Topographic Dopaminergic Circuit Formation. Nat. Commun. [CrossRef] [PubMed]
- König, K.; Marth, L.; Roissant, J.; Granja, T.; Jennewein, C.; Devanathan, V.; Schneider, M.; Köhler, D.; Zarbock, A.; Rosenberger, P. The Plexin C1 Receptor Promotes Acute Inflammation. Eur. J. Immunol. 2014, 44, 2648–2658. [Google Scholar] [CrossRef]
- Walzer, T.; Galibert, L.; De Smedt, T. Dendritic Cell Function in Mice Lacking Plexin C1. Int. Immunology. [CrossRef] [PubMed]
- Kim, J.Y.; Choi, S.Y.; Moon, Y.; Kim, H.J.; Chin, J.H.; Kim, H.; Sun, W. Different Expression Patterns of Phactr Family Members in Normal and Injured Mouse Brain. Neuroscience 2012, 221, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Wider, C.; Lincoln, S.J.; Heckman, M.G.; Diehl, N.N.; Jeremy, T.; Haugarvoll, K.; Aasly, J.O.; Gibson, J.M.; Lynch, T.; Rajput, M.L.; et al. Phactr2 and Parkinson’s Disease. Neurosci Lett. 2010, 453, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.A.; Hubbard, A.E.; Cutler, A.; Barcellos, L.F. An Application of Random Forests to a Genome-Wide Association Dataset: Methodological Considerations and New Findings. BMC Genet. 2010, 11, 49. [Google Scholar] [CrossRef]
- Yuan, F.; Miao, Z.; Chen, W.; Wu, F.; Wei, C.; Yong, J.; Xiao, C. Long Non-Coding RNA PHACTR2-AS1 Promotes Tongue Squamous Cell Carcinoma Metastasis by Regulating Snail. J. Biochem. 2020, 168, 651–657. [Google Scholar] [CrossRef]
- Chu, W.; Zhang, X.; Qi, L.; Fu, Y.; Wang, P.; Zhao, W.; Du, J.; Zhang, J.; Zhan, J.; Wang, Y.; et al. The EZH2-PHACTR2-AS1-Ribosome Axis Induces Genomic Instability and Promotes Growth and Metastasis in Breast Cancer. Cancer Res. 2020, 80, 2737–2750. [Google Scholar] [CrossRef]
- Yang, T.; Zhu, L.; Zhai, Y.; Zhao, Q.; Peng, J.; Zhang, H.; Yang, Z.; Zhang, L.; Ding, W.; Zhao, Y. TSC1 Controls IL-1β Expression in Macrophages via MTORC1-Dependent C/EBPβ Pathway. Cell. Mol. Immunol. 2016, 13, 640–650. [Google Scholar] [CrossRef]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. Corrigendum: PI3Kγ Is a Molecular Switch That Controls Immune Suppression. Nature 2017, 542, 124. [Google Scholar] [CrossRef]
- Huang, E.; Huang, H.; Guan, T.; Liu, C.; Qu, D.; Xu, Y.; Yang, J.; Yan, L.; Xiong, Y.; Liang, T.; et al. Involvement of C/EBPβ-Related Signaling Pathway in Methamphetamine-Induced Neuronal Autophagy and Apoptosis. Toxicol. Lett. 2019, 312, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Inoki, K.; Karbowniczek, M.; Petroulakis, E.; Sonenberg, N.; Henske, E.P.; Guan, K.-L. Constitutive MTOR Activation in TSC Mutants Sensitizes Cells to Energy Starvation and Genomic Damage via P53. EMBO J. 2007, 26, 4812–4823. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, H.; Levine, A.J.; Jin, S. The Coordinate Regulation of the P53 and MTOR Pathways in Cells. Proc. Natl. Acad. Sci. USA 2005, 102, 8204–8209. [Google Scholar] [CrossRef] [PubMed]
- Liszewska, E.; Majchrowicz, L.; Krogulec, E.; Kotulska, K.; Kaczmarek, L.; Kalita, K.; Dobrzyń, A.; Jaworski, J. Establishment of Two HiPSC Lines (IIMCBi001-A and IIMCBi002-A) from Dermal Fibroblasts of Healthy Donors and Characterization of Their Cell Cycle. Stem Cell Res. 2021, 52, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Robak, P.; Dró, I.; Jarych, D.; Mikulski, D.; Siemieniuk-Ry, M.; Misiewicz, M.; Stawiski, K.; Fendler, W.; Szemraj, J.; Smolewski, P.; et al. The Value of Serum MicroRNA Expression Signature in Predicting Refractoriness to Bortezomib-Based Therapy in Multiple Myeloma Patients. Cancers 2020, 12, 2569. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Grabia, S.; Smyczynska, U.; Pagacz, K.; Fendler, W. NormiRazor: Tool Applying GPU-Accelerated Computing for Determination of Internal References in MicroRNA Transcription Studies. BMC Bioinformatics 2020, 21, 1–16. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. MiRWalk: An Online Resource for Prediction of MicroRNA Binding Sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. MiRTarBase 2020: Updates to the Experimentally Validated MicroRNA-Target Interaction Database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawlik, B.; Grabia, S.; Smyczyńska, U.; Fendler, W.; Dróżdż, I.; Liszewska, E.; Jaworski, J.; Kotulska, K.; Jóźwiak, S.; Młynarski, W.; et al. MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor. Int. J. Mol. Sci. 2022, 23, 14493. https://doi.org/10.3390/ijms232214493
Pawlik B, Grabia S, Smyczyńska U, Fendler W, Dróżdż I, Liszewska E, Jaworski J, Kotulska K, Jóźwiak S, Młynarski W, et al. MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor. International Journal of Molecular Sciences. 2022; 23(22):14493. https://doi.org/10.3390/ijms232214493
Chicago/Turabian StylePawlik, Bartłomiej, Szymon Grabia, Urszula Smyczyńska, Wojciech Fendler, Izabela Dróżdż, Ewa Liszewska, Jacek Jaworski, Katarzyna Kotulska, Sergiusz Jóźwiak, Wojciech Młynarski, and et al. 2022. "MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor" International Journal of Molecular Sciences 23, no. 22: 14493. https://doi.org/10.3390/ijms232214493
APA StylePawlik, B., Grabia, S., Smyczyńska, U., Fendler, W., Dróżdż, I., Liszewska, E., Jaworski, J., Kotulska, K., Jóźwiak, S., Młynarski, W., & Trelińska, J. (2022). MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor. International Journal of Molecular Sciences, 23(22), 14493. https://doi.org/10.3390/ijms232214493









