Similarities and Differences between the Orai1 Variants: Orai1α and Orai1β
Abstract
:1. Introduction
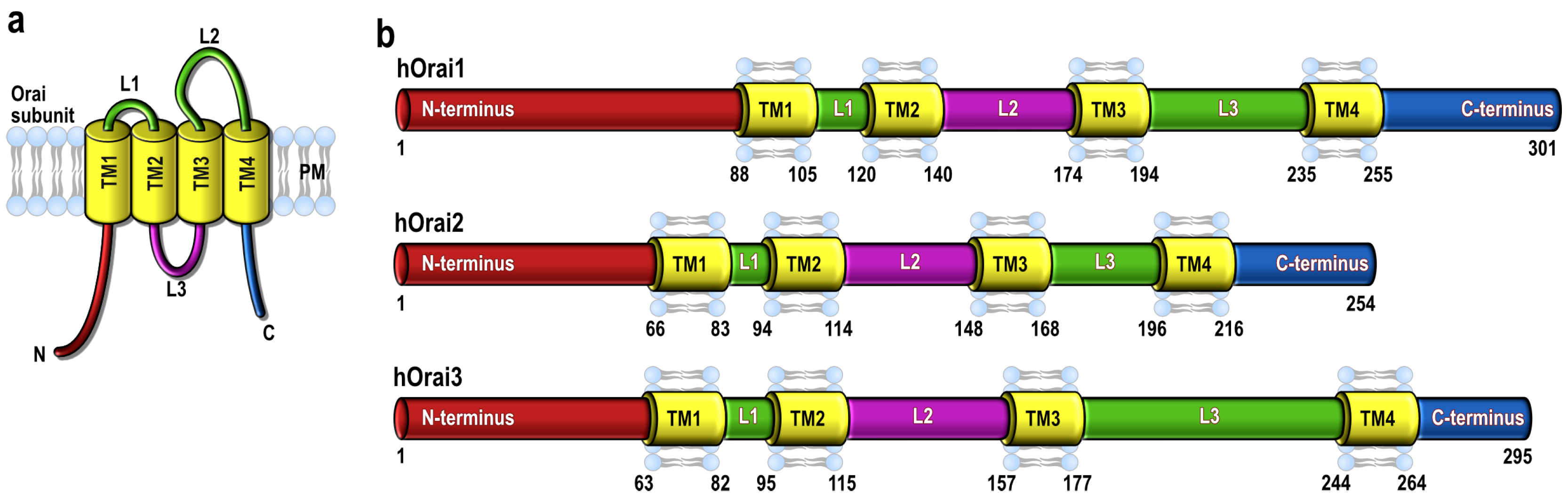
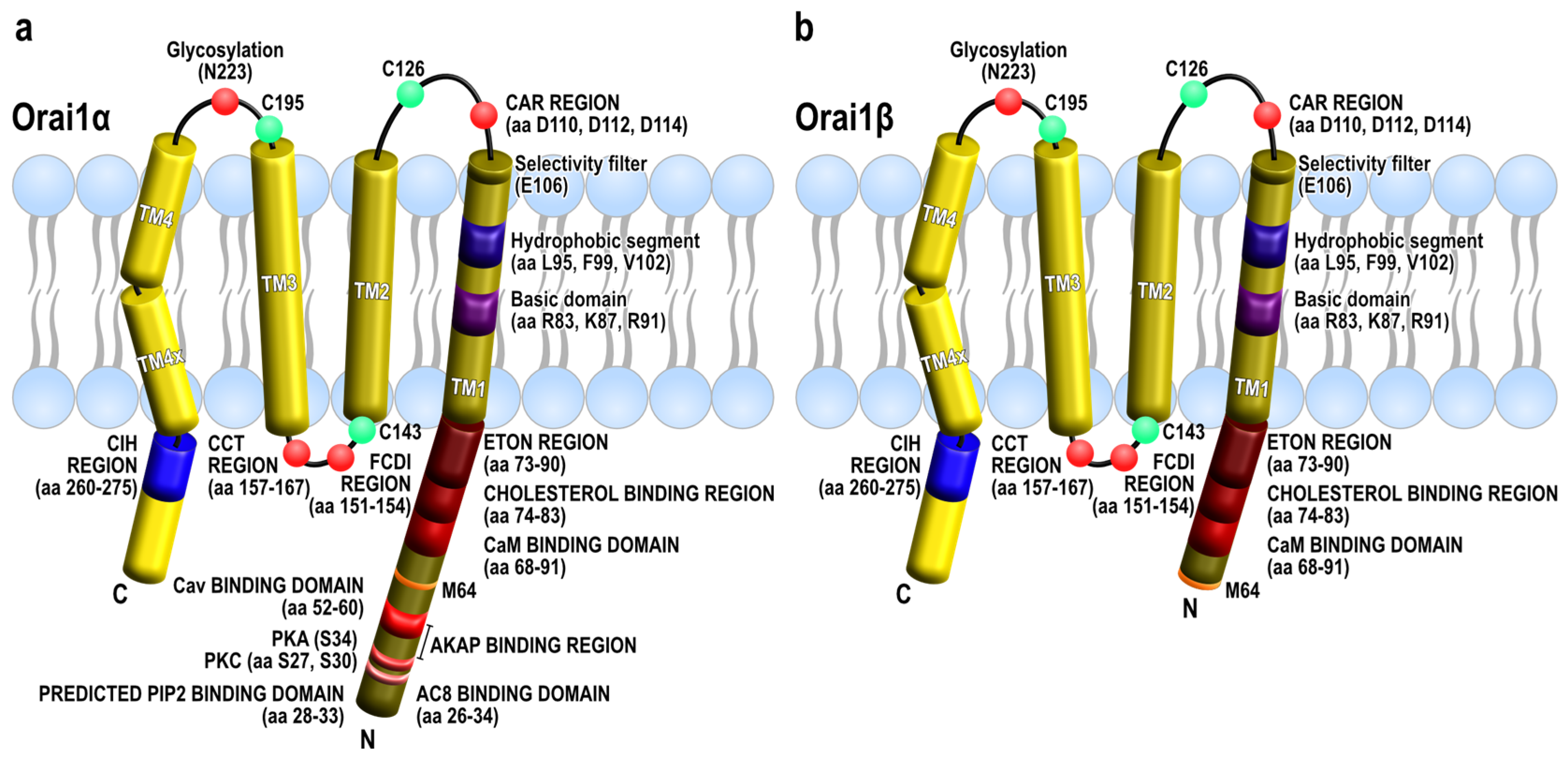
2. Orai1 Variants: Orai1α and Orai1β
3. Functional Properties of Orai1α and Orai1β
4. Orai1 Variants and NFAT Activation
5. Regulation of Orai1α by AC8 in Breast Cancer Cells
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Berridge, M.J. Unlocking the secrets of cell signaling. Annu. Rev. Physiol. 2005, 67, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Putney, J.W., Jr. A model for receptor-regulated calcium entry. Cell Calcium 1986, 7, 1–12. [Google Scholar] [CrossRef]
- Desai, P.N.; Zhang, X.; Wu, S.; Janoshazi, A.; Bolimuntha, S.; Putney, J.W.; Trebak, M. Multiple types of calcium channels arising from alternative translation initiation of the Orai1 message. Sci. Signal. 2015, 8, ra74. [Google Scholar] [CrossRef] [Green Version]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef]
- Vig, M.; Peinelt, C.; Beck, A.; Koomoa, D.L.; Rabah, D.; Koblan-Huberson, M.; Kraft, S.; Turner, H.; Fleig, A.; Penner, R.; et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 2006, 312, 1220–1223. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.L.; Yeromin, A.V.; Zhang, X.H.; Yu, Y.; Safrina, O.; Penna, A.; Roos, J.; Stauderman, K.A.; Cahalan, M.D. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA 2006, 103, 9357–9362. [Google Scholar] [CrossRef] [Green Version]
- Lacruz, R.S.; Feske, S. Diseases caused by mutations in ORAI1 and STIM1. Ann. N. Y. Acad. Sci. 2015, 1356, 45–79. [Google Scholar] [CrossRef] [Green Version]
- Concepcion, A.R.; Vaeth, M.; Wagner, L.E., 2nd; Eckstein, M.; Hecht, L.; Yang, J.; Crottes, D.; Seidl, M.; Shin, H.P.; Weidinger, C.; et al. Store-operated Ca2+ entry regulates Ca2+-activated chloride channels and eccrine sweat gland function. J. Clin. Investig. 2016, 126, 4303–4318. [Google Scholar] [CrossRef]
- Vaeth, M.; Eckstein, M.; Shaw, P.J.; Kozhaya, L.; Yang, J.; Berberich-Siebelt, F.; Clancy, R.; Unutmaz, D.; Feske, S. Store-Operated Ca(2+) Entry in Follicular T Cells Controls Humoral Immune Responses and Autoimmunity. Immunity 2016, 44, 1350–1364. [Google Scholar] [CrossRef] [Green Version]
- Feske, S. CRAC channels and disease—From human CRAC channelopathies and animal models to novel drugs. Cell Calcium 2019, 80, 112–116. [Google Scholar] [CrossRef]
- DeHaven, W.I.; Smyth, J.T.; Boyles, R.R.; Putney, J.W., Jr. Calcium inhibition and calcium potentiation of Orai1, Orai2, and Orai3 calcium release-activated calcium channels. J. Biol. Chem. 2007, 282, 17548–17556. [Google Scholar] [CrossRef] [Green Version]
- Lis, A.; Peinelt, C.; Beck, A.; Parvez, S.; Monteilh-Zoller, M.; Fleig, A.; Penner, R. CRACM1, CRACM2, and CRACM3 are store-operated Ca2+ channels with distinct functional properties. Curr. Biol. 2007, 17, 794–800. [Google Scholar] [CrossRef] [Green Version]
- Emrich, S.M.; Yoast, R.E.; Xin, P.; Arige, V.; Wagner, L.E.; Hempel, N.; Gill, D.L.; Sneyd, J.; Yule, D.I.; Trebak, M. Omnitemporal choreographies of all five STIM/Orai and IP3Rs underlie the complexity of mammalian Ca2+ signaling. Cell Rep. 2021, 34, 108760. [Google Scholar] [CrossRef]
- Yoast, R.E.; Emrich, S.M.; Zhang, X.; Xin, P.; Johnson, M.T.; Fike, A.J.; Walter, V.; Hempel, N.; Yule, D.I.; Sneyd, J.; et al. The native ORAI channel trio underlies the diversity of Ca2+ signaling events. Nat. Commun. 2020, 11, 2444. [Google Scholar] [CrossRef]
- Yoast, R.E.; Emrich, S.M.; Trebak, M. The anatomy of native CRAC channel(s). Curr. Opin. Physiol. 2020, 17, 89–95. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Cantonero, C.; Jardin, I.; Regodon, S.; Redondo, P.C.; Gordillo, J.; Smani, T.; Salido, G.M.; Rosado, J.A. Orai2 Modulates Store-Operated Ca2+ Entry and Cell Cycle Progression in Breast Cancer Cells. Cancers 2021, 14, 114. [Google Scholar] [CrossRef]
- Fahrner, M.; Schindl, R.; Romanin, C. Studies of structure-function and subunit composition of Orai/STIM channel. In Calcium Entry Channels in Non-Excitable Cells; Kozak, J.A., Putney, J.W., Jr., Eds.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2018; pp. 25–50. [Google Scholar]
- Mignen, O.; Thompson, J.L.; Shuttleworth, T.J. Both Orai1 and Orai3 are essential components of the arachidonate-regulated Ca2+-selective (ARC) channels. J. Physiol. 2008, 586, 185–195. [Google Scholar] [CrossRef]
- Shuttleworth, T.J. Orai3—The ‘exceptional’ Orai? J. Physiol. 2012, 590, 241–257. [Google Scholar] [CrossRef]
- Hou, X.; Pedi, L.; Diver, M.M.; Long, S.B. Crystal structure of the calcium release-activated calcium channel Orai. Science 2012, 338, 1308–1313. [Google Scholar] [CrossRef] [Green Version]
- Demuro, A.; Penna, A.; Safrina, O.; Yeromin, A.V.; Amcheslavsky, A.; Cahalan, M.D.; Parker, I. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc. Natl. Acad. Sci. USA 2011, 108, 17832–17837. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, Y.; Ogura, T.; Mio, K.; Kato, K.; Kaneko, T.; Kiyonaka, S.; Mori, Y.; Sato, C. Tetrameric Orai1 is a teardrop-shaped molecule with a long, tapered cytoplasmic domain. J. Biol. Chem. 2009, 284, 13676–13685. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Demuro, A.; Yeromin, A.V.; Zhang, S.L.; Safrina, O.; Parker, I.; Cahalan, M.D. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 2008, 456, 116–120. [Google Scholar] [CrossRef] [Green Version]
- Derler, I.; Plenk, P.; Fahrner, M.; Muik, M.; Jardin, I.; Schindl, R.; Gruber, H.J.; Groschner, K.; Romanin, C. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J. Biol. Chem. 2013, 288, 29025–29034. [Google Scholar] [CrossRef] [Green Version]
- Frischauf, I.; Zayats, V.; Deix, M.; Hochreiter, A.; Jardin, I.; Muik, M.; Lackner, B.; Svobodova, B.; Pammer, T.; Litvinukova, M.; et al. A calcium-accumulating region, CAR, in the channel Orai1 enhances Ca2+ permeation and SOCE-induced gene transcription. Sci. Signal. 2015, 8, ra131. [Google Scholar] [CrossRef] [Green Version]
- Srikanth, S.; Jung, H.J.; Ribalet, B.; Gwack, Y. The intracellular loop of Orai1 plays a central role in fast inactivation of Ca2+ release-activated Ca2+ channels. J. Biol. Chem. 2010, 285, 5066–5075. [Google Scholar] [CrossRef] [Green Version]
- Hodeify, R.; Nandakumar, M.; Own, M.; Courjaret, R.J.; Graumann, J.; Hubrack, S.Z.; Machaca, K. The CCT chaperonin is a novel regulator of Ca(2+) signaling through modulation of Orai1 trafficking. Sci. Adv. 2018, 4, eaau1935. [Google Scholar] [CrossRef] [Green Version]
- Dorr, K.; Kilch, T.; Kappel, S.; Alansary, D.; Schwar, G.; Niemeyer, B.A.; Peinelt, C. Cell type-specific glycosylation of Orai1 modulates store-operated Ca2+ entry. Sci. Signal. 2016, 9, ra25. [Google Scholar] [CrossRef]
- Bogeski, I.; Kummerow, C.; Al-Ansary, D.; Schwarz, E.C.; Koehler, R.; Kozai, D.; Takahashi, N.; Peinelt, C.; Griesemer, D.; Bozem, M.; et al. Differential redox regulation of ORAI ion channels: A mechanism to tune cellular calcium signaling. Sci. Signal. 2010, 3, ra24. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, X.; Loktionova, N.A.; Wang, X.; Nwokonko, R.M.; Wang, X.; Wang, Y.; Rothberg, B.S.; Trebak, M.; Gill, D.L. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat. Commun. 2016, 7, 13725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muik, M.; Frischauf, I.; Derler, I.; Fahrner, M.; Bergsmann, J.; Eder, P.; Schindl, R.; Hesch, C.; Polzinger, B.; Fritsch, R.; et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 2008, 283, 8014–8022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.Y.; Hoover, P.J.; Mullins, F.M.; Bachhawat, P.; Covington, E.D.; Raunser, S.; Walz, T.; Garcia, K.C.; Dolmetsch, R.E.; Lewis, R.S. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 2009, 136, 876–890. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.P.; Zeng, W.; Dorwart, M.R.; Choi, Y.J.; Worley, P.F.; Muallem, S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 2009, 11, 337–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muik, M.; Fahrner, M.; Derler, I.; Schindl, R.; Bergsmann, J.; Frischauf, I.; Groschner, K.; Romanin, C. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J. Biol. Chem. 2009, 284, 8421–8426. [Google Scholar] [CrossRef] [Green Version]
- Stathopulos, P.B.; Schindl, R.; Fahrner, M.; Zheng, L.; Gasmi-Seabrook, G.M.; Muik, M.; Romanin, C.; Ikura, M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 2013, 4, 2963. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Sun, L.; Machaca, K. Orai1 internalization and STIM1 clustering inhibition modulate SOCE inactivation during meiosis. Proc. Natl. Acad. Sci. USA 2009, 106, 17401–17406. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Sun, L.; Machaca, K. Constitutive recycling of the store-operated Ca2+ channel Orai1 and its internalization during meiosis. J. Cell Biol. 2010, 191, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Mercer, J.C.; Dehaven, W.I.; Smyth, J.T.; Wedel, B.; Boyles, R.R.; Bird, G.S.; Putney, J.W., Jr. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J. Biol. Chem. 2006, 281, 24979–24990. [Google Scholar] [CrossRef] [Green Version]
- Rothberg, B.S.; Wang, Y.; Gill, D.L. Orai channel pore properties and gating by STIM: Implications from the Orai crystal structure. Sci. Signal. 2013, 6, pe9. [Google Scholar] [CrossRef]
- Palty, R.; Isacoff, E.Y. Cooperative Binding of Stromal Interaction Molecule 1 (STIM1) to the N and C Termini of Calcium Release-activated Calcium Modulator 1 (Orai1). J. Biol. Chem. 2016, 291, 334–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humer, C.; Romanin, C.; Hoglinger, C. Highlighting the Multifaceted Role of Orai1 N-Terminal- and Loop Regions for Proper CRAC Channel Functions. Cells 2022, 11, 371. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, M.; Pandey, S.K.; Muik, M.; Traxler, L.; Butorac, C.; Stadlbauer, M.; Zayats, V.; Krizova, A.; Plenk, P.; Frischauf, I.; et al. Communication between N terminus and loop2 tunes Orai activation. J. Biol. Chem. 2018, 293, 1271–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derler, I.; Butorac, C.; Krizova, A.; Stadlbauer, M.; Muik, M.; Fahrner, M.; Frischauf, I.; Romanin, C. Authentic CRAC channel activity requires STIM1 and the conserved portion of the Orai N terminus. J. Biol. Chem. 2018, 293, 1259–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakriya, M. The molecular physiology of CRAC channels. Immunol. Rev. 2009, 231, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zheng, X.; Mueller, G.A.; Sobhany, M.; DeRose, E.F.; Zhang, Y.; London, R.E.; Birnbaumer, L. Crystal structure of calmodulin binding domain of orai1 in complex with Ca2+ calmodulin displays a unique binding mode. J. Biol. Chem. 2012, 287, 43030–43041. [Google Scholar] [CrossRef] [Green Version]
- Traxler, L.; Rathner, P.; Fahrner, M.; Stadlbauer, M.; Faschinger, F.; Charnavets, T.; Muller, N.; Romanin, C.; Hinterdorfer, P.; Gruber, H.J. Detailed Evidence for an Unparalleled Interaction Mode between Calmodulin and Orai Proteins. Angew. Chem. Int. Ed. Engl. 2017, 56, 15755–15759. [Google Scholar] [CrossRef] [Green Version]
- Maganti, L.; Dutta, S.; Ghosh, M.; Chakrabarti, J. Allostery in Orai1 binding to calmodulin revealed from conformational thermodynamics. J. Biomol. Struct. Dyn. 2019, 37, 493–502. [Google Scholar] [CrossRef]
- Palty, R.; Raveh, A.; Kaminsky, I.; Meller, R.; Reuveny, E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 2012, 149, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Albarran, L.; Lopez, J.J.; Amor, N.B.; Martin-Cano, F.E.; Berna-Erro, A.; Smani, T.; Salido, G.M.; Rosado, J.A. Dynamic interaction of SARAF with STIM1 and Orai1 to modulate store-operated calcium entry. Sci. Rep. 2016, 6, 24452. [Google Scholar] [CrossRef] [Green Version]
- Jardin, I.; Nieto-Felipe, J.; Alvarado, S.; Diez-Bello, R.; Lopez, J.J.; Salido, G.M.; Smani, T.; Rosado, J.A. SARAF and EFHB Modulate Store-Operated Ca2+ Entry and Are Required for Cell Proliferation, Migration and Viability in Breast Cancer Cells. Cancers 2021, 13, 4160. [Google Scholar] [CrossRef] [PubMed]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soboloff, J.; Spassova, M.A.; Tang, X.D.; Hewavitharana, T.; Xu, W.; Gill, D.L. Orai1 and STIM reconstitute store-operated calcium channel function. J. Biol. Chem. 2006, 281, 20661–20665. [Google Scholar] [CrossRef] [Green Version]
- Ji, W.; Xu, P.; Li, Z.; Lu, J.; Liu, L.; Zhan, Y.; Chen, Y.; Hille, B.; Xu, T.; Chen, L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. USA 2008, 105, 13668–13673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignen, O.; Thompson, J.L.; Shuttleworth, T.J. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J. Physiol. 2008, 586, 419–425. [Google Scholar] [CrossRef]
- Scrimgeour, N.; Litjens, T.; Ma, L.; Barritt, G.J.; Rychkov, G.Y. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J. Physiol. 2009, 587, 2903–2918. [Google Scholar] [CrossRef]
- Hoover, P.J.; Lewis, R.S. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA 2011, 108, 13299–13304. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, L.; Deng, Y.; Ji, W.; Du, W.; Xu, P.; Chen, L.; Xu, T. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 2011, 21, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Fahrner, M.; Muik, M.; Schindl, R.; Butorac, C.; Stathopulos, P.; Zheng, L.; Jardin, I.; Ikura, M.; Romanin, C. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1). J. Biol. Chem. 2014, 289, 33231–33244. [Google Scholar] [CrossRef] [Green Version]
- Perni, S.; Dynes, J.L.; Yeromin, A.V.; Cahalan, M.D.; Franzini-Armstrong, C. Nanoscale patterning of STIM1 and Orai1 during store-operated Ca2+ entry. Proc. Natl. Acad. Sci. USA 2015, 112, E5533–E5542. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, X.; Wang, X.; Loktionova, N.A.; Cai, X.; Nwokonko, R.M.; Vrana, E.; Wang, Y.; Rothberg, B.S.; Gill, D.L. STIM1 dimers undergo unimolecular coupling to activate Orai1 channels. Nat. Commun. 2015, 6, 8395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, M.; Lewis, R.S. Numbers count: How STIM and Orai stoichiometry affect store-operated calcium entry. Cell Calcium 2019, 79, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, J.H., Jr.; Zhou, Y.; Nwokonko, R.M.; Jennette, M.R.; Kazzaz, S.A.; Stenson, J.M.; Whitsell, A.L.; Wang, Y.; Trebak, M.; Gill, D.L. Orai channel C-terminal peptides are key modulators of STIM-Orai coupling and calcium signal generation. Cell Rep. 2021, 35, 109322. [Google Scholar] [CrossRef]
- Maus, M.; Jairaman, A.; Stathopulos, P.B.; Muik, M.; Fahrner, M.; Weidinger, C.; Benson, M.; Fuchs, S.; Ehl, S.; Romanin, C.; et al. Missense mutation in immunodeficient patients shows the multifunctional roles of coiled-coil domain 3 (CC3) in STIM1 activation. Proc. Natl. Acad. Sci. USA 2015, 112, 6206–6211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Zhou, Y.; Nwokonko, R.M.; Loktionova, N.A.; Wang, X.; Xin, P.; Trebak, M.; Wang, Y.; Gill, D.L. The Orai1 Store-operated Calcium Channel Functions as a Hexamer. J. Biol. Chem. 2016, 291, 25764–25775. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Nwokonko, R.M.; Baraniak, J.H., Jr.; Trebak, M.; Lee, K.P.K.; Gill, D.L. The remote allosteric control of Orai channel gating. PLoS Biol. 2019, 17, e3000413. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Jardin, I.; Lopez, J.J.; Salido, G.M.; Rosado, J.A. Orai1 mediates the interaction between STIM1 and hTRPC1 and regulates the mode of activation of hTRPC1-forming Ca2+ channels. J. Biol. Chem. 2008, 283, 25296–25304. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Yuan, J.P.; Kim, M.S.; Choi, Y.J.; Huang, G.N.; Worley, P.F.; Muallem, S. STIM1 gates TRPC channels, but not Orai1, by electrostatic interaction. Mol. Cell 2008, 32, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Pani, B.; Ong, H.L.; Brazer, S.C.; Liu, X.; Rauser, K.; Singh, B.B.; Ambudkar, I.S. Activation of TRPC1 by STIM1 in ER-PM microdomains involves release of the channel from its scaffold caveolin-1. Proc. Natl. Acad. Sci. USA 2009, 106, 20087–20092. [Google Scholar] [CrossRef]
- Ong, H.L.; Jang, S.I.; Ambudkar, I.S. Distinct contributions of Orai1 and TRPC1 to agonist-induced [Ca2+]i signals determine specificity of Ca2+-dependent gene expression. PLoS ONE 2012, 7, e47146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Berna-Erro, A.; Camello, P.J.; Cantonero, C.; Smani, T.; Salido, G.M.; Rosado, J.A. Orai1alpha, but not Orai1beta, co-localizes with TRPC1 and is required for its plasma membrane location and activation in HeLa cells. Cell. Mol. Life Sci. 2022, 79, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pathak, T.; Yoast, R.; Emrich, S.; Xin, P.; Nwokonko, R.M.; Johnson, M.; Wu, S.; Delierneux, C.; Gueguinou, M.; et al. A calcium/cAMP signaling loop at the ORAI1 mouth drives channel inactivation to shape NFAT induction. Nat. Commun. 2019, 10, 1971. [Google Scholar] [CrossRef] [Green Version]
- Kar, P.; Lin, Y.P.; Bhardwaj, R.; Tucker, C.J.; Bird, G.S.; Hediger, M.A.; Monico, C.; Amin, N.; Parekh, A.B. The N terminus of Orai1 couples to the AKAP79 signaling complex to drive NFAT1 activation by local Ca2+ entry. Proc. Natl. Acad. Sci. USA 2021, 118, e2012908118. [Google Scholar] [CrossRef]
- Paria, B.C.; Malik, A.B.; Kwiatek, A.M.; Rahman, A.; May, M.J.; Ghosh, S.; Tiruppathi, C. Tumor necrosis factor-alpha induces nuclear factor-kappaB-dependent TRPC1 expression in endothelial cells. J. Biol. Chem. 2003, 278, 37195–37203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Watanabe, H.; Murakami, M.; Ohba, T.; Radovanovic, M.; Ono, K.; Iijima, T.; Ito, H. Involvement of transient receptor potential canonical 1 (TRPC1) in angiotensin II-induced vascular smooth muscle cell hypertrophy. Atherosclerosis 2007, 195, 287–296. [Google Scholar] [CrossRef]
- Schaar, A.; Sukumaran, P.; Sun, Y.; Dhasarathy, A.; Singh, B.B. TRPC1-STIM1 activation modulates transforming growth factor beta-induced epithelial-to-mesenchymal transition. Oncotarget 2016, 7, 80554–80567. [Google Scholar] [CrossRef] [Green Version]
- Mignen, O.; Thompson, J.L.; Shuttleworth, T.J. The molecular architecture of the arachidonate-regulated Ca2+-selective ARC channel is a pentameric assembly of Orai1 and Orai3 subunits. J. Physiol. 2009, 587, 4181–4197. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, W.; Gonzalez-Cobos, J.C.; Jardin, I.; Romanin, C.; Matrougui, K.; Trebak, M. Complex role of STIM1 in the activation of store-independent Orai1/3 channels. J. Gen. Physiol. 2014, 143, 345–359. [Google Scholar] [CrossRef] [Green Version]
- Faddy, H.M.; Smart, C.E.; Xu, R.; Lee, G.Y.; Kenny, P.A.; Feng, M.; Rao, R.; Brown, M.A.; Bissell, M.J.; Roberts-Thomson, S.J.; et al. Localization of plasma membrane and secretory calcium pumps in the mammary gland. Biochem. Biophys. Res. Commun. 2008, 369, 977–981. [Google Scholar] [CrossRef]
- Feng, M.; Grice, D.M.; Faddy, H.M.; Nguyen, N.; Leitch, S.; Wang, Y.; Muend, S.; Kenny, P.A.; Sukumar, S.; Roberts-Thomson, S.J.; et al. Store-independent activation of Orai1 by SPCA2 in mammary tumors. Cell 2010, 143, 84–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantome, A.; Potier-Cartereau, M.; Clarysse, L.; Fromont, G.; Marionneau-Lambot, S.; Gueguinou, M.; Pages, J.C.; Collin, C.; Oullier, T.; Girault, A.; et al. Pivotal role of the lipid Raft SK3-Orai1 complex in human cancer cell migration and bone metastases. Cancer Res. 2013, 73, 4852–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantonero, C.; Sanchez-Collado, J.; Gonzalez-Nunez, M.A.; Salido, G.M.; Lopez, J.J.; Jardin, I.; Rosado, J.A. Store-independent Orai1-mediated Ca(2+) entry and cancer. Cell Calcium 2019, 80, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gueguinou, M.; Trebak, M. Store-independent Orai channels regulated by STIM. In Calcium Entry Channels in Non-Excitable Cells; Kozak, J.A., Putney, J.W., Jr., Eds.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2018; pp. 197–214. [Google Scholar]
- Cai, X. Molecular evolution and structural analysis of the Ca2+ release-activated Ca2+ channel subunit, Orai. J. Mol. Biol. 2007, 368, 1284–1291. [Google Scholar] [CrossRef]
- Fukushima, M.; Tomita, T.; Janoshazi, A.; Putney, J.W. Alternative translation initiation gives rise to two isoforms of Orai1 with distinct plasma membrane mobilities. J. Cell Sci. 2012, 125, 4354–4361. [Google Scholar] [CrossRef] [Green Version]
- Putney, J.W. Alternative forms of the store-operated calcium entry mediators, STIM1 and Orai1. Curr. Top. Membr. 2013, 71, 109–123. [Google Scholar] [CrossRef]
- Willoughby, D.; Everett, K.L.; Halls, M.L.; Pacheco, J.; Skroblin, P.; Vaca, L.; Klussmann, E.; Cooper, D.M. Direct binding between Orai1 and AC8 mediates dynamic interplay between Ca2+ and cAMP signaling. Sci. Signal. 2012, 5, ra29. [Google Scholar] [CrossRef]
- Kawasaki, T.; Ueyama, T.; Lange, I.; Feske, S.; Saito, N. Protein kinase C-induced phosphorylation of Orai1 regulates the intracellular Ca2+ level via the store-operated Ca2+ channel. J. Biol. Chem. 2010, 285, 25720–25730. [Google Scholar] [CrossRef] [Green Version]
- Calloway, N.; Owens, T.; Corwith, K.; Rodgers, W.; Holowka, D.; Baird, B. Stimulated association of STIM1 and Orai1 is regulated by the balance of PtdIns(4,5)P2 between distinct membrane pools. J. Cell Sci. 2011, 124, 2602–2610. [Google Scholar] [CrossRef] [Green Version]
- Maleth, J.; Choi, S.; Muallem, S.; Ahuja, M. Translocation between PI(4,5)P2-poor and PI(4,5)P2-rich microdomains during store depletion determines STIM1 conformation and Orai1 gating. Nat. Commun. 2014, 5, 5843. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Camello, P.J.; Falcon, D.; Regodon, S.; Salido, G.M.; Smani, T.; Rosado, J.A. Adenylyl Cyclase Type 8 Overexpression Impairs Phosphorylation-Dependent Orai1 Inactivation and Promotes Migration in MDA-MB-231 Breast Cancer Cells. Cancers 2019, 11, 1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.T.; Liu, X.; Ong, H.L.; Swaim, W.; Ambudkar, I.S. Local Ca2+ entry via Orai1 regulates plasma membrane recruitment of TRPC1 and controls cytosolic Ca2+ signals required for specific cell functions. PLoS Biol. 2011, 9, e1001025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignen, O.; Shuttleworth, T.J. I(ARC), a novel arachidonate-regulated, noncapacitative Ca(2+) entry channel. J. Biol. Chem. 2000, 275, 9114–9119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignen, O.; Thompson, J.L.; Shuttleworth, T.J. STIM1 regulates Ca2+ entry via arachidonate-regulated Ca2+-selective (ARC) channels without store depletion or translocation to the plasma membrane. J. Physiol. 2007, 579, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, N.; Kaminuma, O. Isoform-Selective NFAT Inhibitor: Potential Usefulness and Development. Int. J. Mol. Sci. 2021, 22, 2725. [Google Scholar] [CrossRef]
- Muller, M.R.; Rao, A. NFAT, immunity and cancer: A transcription factor comes of age. Nat. Rev. Immunol. 2010, 10, 645–656. [Google Scholar] [CrossRef]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Eder, P.; Chang, B.; Molkentin, J.D. TRPC channels are necessary mediators of pathologic cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2010, 107, 7000–7005. [Google Scholar] [CrossRef] [Green Version]
- Graef, I.A.; Mermelstein, P.G.; Stankunas, K.; Neilson, J.R.; Deisseroth, K.; Tsien, R.W.; Crabtree, G.R. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 1999, 401, 703–708. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Correll, R.N.; Gao, H.; Zhang, H.; Yang, B.; Berretta, R.M.; Rizzo, V.; Molkentin, J.D.; Houser, S.R. A caveolae-targeted L-type Ca2+ channel antagonist inhibits hypertrophic signaling without reducing cardiac contractility. Circ. Res. 2012, 110, 669–674. [Google Scholar] [CrossRef]
- Murphy, J.G.; Crosby, K.C.; Dittmer, P.J.; Sather, W.A.; Dell’Acqua, M.L. AKAP79/150 recruits the transcription factor NFAT to regulate signaling to the nucleus by neuronal L-type Ca2+ channels. Mol. Biol. Cell 2019, 30, 1743–1756. [Google Scholar] [CrossRef]
- Chiang, C.S.; Huang, C.H.; Chieng, H.; Chang, Y.T.; Chang, D.; Chen, J.J.; Chen, Y.C.; Chen, Y.H.; Shin, H.S.; Campbell, K.P.; et al. The Ca(v)3.2 T-type Ca(2+) channel is required for pressure overload-induced cardiac hypertrophy in mice. Circ. Res. 2009, 104, 522–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markandeya, Y.S.; Phelan, L.J.; Woon, M.T.; Keefe, A.M.; Reynolds, C.R.; August, B.K.; Hacker, T.A.; Roth, D.M.; Patel, H.H.; Balijepalli, R.C. Caveolin-3 Overexpression Attenuates Cardiac Hypertrophy via Inhibition of T-type Ca2+ Current Modulated by Protein Kinase Calpha in Cardiomyocytes. J. Biol. Chem. 2015, 290, 22085–22100. [Google Scholar] [CrossRef] [Green Version]
- Kosiorek, M.; Zylinska, L.; Zablocki, K.; Pikula, S. Calcineurin/NFAT signaling represses genes Vamp1 and Vamp2 via PMCA-dependent mechanism during dopamine secretion by Pheochromocytoma cells. PLoS ONE 2014, 9, e92176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, P.; Hawkins, A.; Stiber, J.; Shelton, J.M.; Hutcheson, K.; Bassel-Duby, R.; Shin, D.M.; Yan, Z.; Williams, R.S. TRPC3 channels confer cellular memory of recent neuromuscular activity. Proc. Natl. Acad. Sci. USA 2004, 101, 9387–9392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onohara, N.; Nishida, M.; Inoue, R.; Kobayashi, H.; Sumimoto, H.; Sato, Y.; Mori, Y.; Nagao, T.; Kurose, H. TRPC3 and TRPC6 are essential for angiotensin II-induced cardiac hypertrophy. EMBO J. 2006, 25, 5305–5316. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Zhu, H.; Yan, Q.; Shen, X.; Lu, X.; Wang, J.; Li, J.; Chen, L. TRPV2-induced Ca2+-calcineurin-NFAT signaling regulates differentiation of osteoclast in multiple myeloma. Cell Commun. Signal. 2018, 16, 68. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Zhang, H.; Cao, X.; Yin, Y.; Zhang, B. Activation of TRPV1 mediates thymic stromal lymphopoietin release via the Ca2+/NFAT pathway in airway epithelial cells. FEBS Lett. 2014, 588, 3047–3054. [Google Scholar] [CrossRef] [Green Version]
- Weber, K.S.; Hildner, K.; Murphy, K.M.; Allen, P.M. Trpm4 differentially regulates Th1 and Th2 function by altering calcium signaling and NFAT localization. J. Immunol. 2010, 185, 2836–2846. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.M.; Huang, C.M.; Hsieh, M.S.; Lin, C.S.; Lee, W.H.; Yeh, C.T.; Liu, S.C. TRPM7 via calcineurin/NFAT pathway mediates metastasis and chemotherapeutic resistance in head and neck squamous cell carcinoma. Aging 2022, 14, 5250–5270. [Google Scholar] [CrossRef]
- Li, S.; Sun, X.; Wu, H.; Yu, P.; Wang, X.; Jiang, Z.; Gao, E.; Chen, J.; Li, D.; Qiu, C.; et al. TRPA1 Promotes Cardiac Myofibroblast Transdifferentiation after Myocardial Infarction Injury via the Calcineurin-NFAT-DYRK1A Signaling Pathway. Oxid. Med. Cell. Longev. 2019, 2019, 6408352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolmetsch, R.E.; Lewis, R.S.; Goodnow, C.C.; Healy, J.I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 1997, 386, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Tomida, T.; Hirose, K.; Takizawa, A.; Shibasaki, F.; Iino, M. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 2003, 22, 3825–3832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Llopis, J.; Whitney, M.; Zlokarnik, G.; Tsien, R.Y. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature 1998, 392, 936–941. [Google Scholar] [CrossRef]
- Kar, P.; Parekh, A.B. Distinct spatial Ca2+ signatures selectively activate different NFAT transcription factor isoforms. Mol. Cell 2015, 58, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Shibasaki, F.; Price, E.R.; Milan, D.; McKeon, F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 1996, 382, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Kar, P.; Samanta, K.; Kramer, H.; Morris, O.; Bakowski, D.; Parekh, A.B. Dynamic assembly of a membrane signaling complex enables selective activation of NFAT by Orai1. Curr. Biol. 2014, 24, 1361–1368. [Google Scholar] [CrossRef] [Green Version]
- Jans, R.; Mottram, L.; Johnson, D.L.; Brown, A.M.; Sikkink, S.; Ross, K.; Reynolds, N.J. Lysophosphatidic acid promotes cell migration through STIM1- and Orai1-mediated Ca2+(i) mobilization and NFAT2 activation. J. Investig. Dermatol. 2013, 133, 793–802. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.Y.; Putney, J.W. Orai1-mediated calcium entry plays a critical role in osteoclast differentiation and function by regulating activation of the transcription factor NFATc1. FASEB J. 2012, 26, 1484–1492. [Google Scholar] [CrossRef] [Green Version]
- Sahu, I.; Pelzl, L.; Sukkar, B.; Fakhri, H.; Al-Maghout, T.; Cao, H.; Hauser, S.; Gutti, R.; Gawaz, M.; Lang, F. NFAT5-sensitive Orai1 expression and store-operated Ca2+ entry in megakaryocytes. FASEB J. 2017, 31, 3439–3448. [Google Scholar] [CrossRef]
- Vaeth, M.; Yang, J.; Yamashita, M.; Zee, I.; Eckstein, M.; Knosp, C.; Kaufmann, U.; Karoly Jani, P.; Lacruz, R.S.; Flockerzi, V.; et al. ORAI2 modulates store-operated calcium entry and T cell-mediated immunity. Nat. Commun. 2017, 8, 14714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsvilovskyy, V.; Solis-Lopez, A.; Schumacher, D.; Medert, R.; Roers, A.; Kriebs, U.; Freichel, M. Deletion of Orai2 augments endogenous CRAC currents and degranulation in mast cells leading to enhanced anaphylaxis. Cell Calcium 2018, 71, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Yissachar, N.; Sharar Fischler, T.; Cohen, A.A.; Reich-Zeliger, S.; Russ, D.; Shifrut, E.; Porat, Z.; Friedman, N. Dynamic response diversity of NFAT isoforms in individual living cells. Mol. Cell 2013, 49, 322–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, P.; Mirams, G.R.; Christian, H.C.; Parekh, A.B. Control of NFAT Isoform Activation and NFAT-Dependent Gene Expression through Two Coincident and Spatially Segregated Intracellular Ca2+ Signals. Mol. Cell 2016, 64, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Parekh, A.B. Regulation of CRAC channels by Ca2+-dependent inactivation. Cell Calcium 2017, 63, 20–23. [Google Scholar] [CrossRef]
- Khannpnavar, B.; Mehta, V.; Qi, C.; Korkhov, V. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol. 2020, 63, 34–41. [Google Scholar] [CrossRef]
- Dessauer, C.W.; Watts, V.J.; Ostrom, R.S.; Conti, M.; Dove, S.; Seifert, R. International Union of Basic and Clinical Pharmacology. CI. Structures and Small Molecule Modulators of Mammalian Adenylyl Cyclases. Pharmacol. Rev. 2017, 69, 93–139. [Google Scholar] [CrossRef]
- Fagan, K.A.; Graf, R.A.; Tolman, S.; Schaack, J.; Cooper, D.M. Regulation of a Ca2+-sensitive adenylyl cyclase in an excitable cell. Role of voltage-gated versus capacitative Ca2+ entry. J. Biol. Chem. 2000, 275, 40187–40194. [Google Scholar] [CrossRef] [Green Version]
- Watson, E.L.; Wu, Z.; Jacobson, K.L.; Storm, D.R.; Singh, J.C.; Ott, S.M. Capacitative Ca2+ entry is involved in cAMP synthesis in mouse parotid acini. Am J. Physiol. 1998, 274, C557–C565. [Google Scholar] [CrossRef]
- Parker, T.; Wang, K.W.; Manning, D.; Dart, C. Soluble adenylyl cyclase links Ca2+ entry to Ca2+/cAMP-response element binding protein (CREB) activation in vascular smooth muscle. Sci. Rep. 2019, 9, 7317. [Google Scholar] [CrossRef]
- Lefkimmiatis, K.; Srikanthan, M.; Maiellaro, I.; Moyer, M.P.; Curci, S.; Hofer, A.M. Store-operated cyclic AMP signalling mediated by STIM1. Nat. Cell Biol. 2009, 11, 433–442. [Google Scholar] [CrossRef]
- Spirli, C.; Mariotti, V.; Villani, A.; Fabris, L.; Fiorotto, R.; Strazzabosco, M. Adenylyl cyclase 5 links changes in calcium homeostasis to cAMP-dependent cyst growth in polycystic liver disease. J. Hepatol. 2017, 66, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Motiani, R.K.; Tanwar, J.; Raja, D.A.; Vashisht, A.; Khanna, S.; Sharma, S.; Srivastava, S.; Sivasubbu, S.; Natarajan, V.T.; Gokhale, R.S. STIM1 activation of adenylyl cyclase 6 connects Ca2+ and cAMP signaling during melanogenesis. EMBO J. 2018, 37, e97597. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Salido, G.M.; Rosado, J.A. Cross-Talk between the Adenylyl Cyclase/cAMP Pathway and Ca(2+) Homeostasis. Rev. Physiol. Biochem. Pharmacol. 2021, 179, 73–116. [Google Scholar] [CrossRef] [PubMed]
- Fagan, K.A.; Mahey, R.; Cooper, D.M. Functional co-localization of transfected Ca(2+)-stimulable adenylyl cyclases with capacitative Ca2+ entry sites. J. Biol. Chem. 1996, 271, 12438–12444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaccolo, M.; Zerio, A.; Lobo, M.J. Subcellular Organization of the cAMP Signaling Pathway. Pharmacol. Rev. 2021, 73, 278–309. [Google Scholar] [CrossRef]
- Postler, T.S. A most versatile kinase: The catalytic subunit of PKA in T-cell biology. Int. Rev. Cell Mol. Biol. 2021, 361, 301–318. [Google Scholar] [CrossRef]
- Willoughby, D.; Cooper, D.M. Organization and Ca2+ regulation of adenylyl cyclases in cAMP microdomains. Physiol. Rev. 2007, 87, 965–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, A.C.; Willoughby, D.; Ciruela, A.; Ayling, L.J.; Pagano, M.; Wachten, S.; Tengholm, A.; Cooper, D.M. Capacitative Ca2+ entry via Orai1 and stromal interacting molecule 1 (STIM1) regulates adenylyl cyclase type 8. Mol. Pharmacol. 2009, 75, 830–842. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.E.; Gu, C.; Fagan, K.A.; Hu, B.; Cooper, D.M. Residence of adenylyl cyclase type 8 in caveolae is necessary but not sufficient for regulation by capacitative Ca2+ entry. J. Biol. Chem. 2002, 277, 6025–6031. [Google Scholar] [CrossRef]
- Delint-Ramirez, I.; Willoughby, D.; Hammond, G.R.; Ayling, L.J.; Cooper, D.M. Palmitoylation targets AKAP79 protein to lipid rafts and promotes its regulation of calcium-sensitive adenylyl cyclase type 8. J. Biol. Chem. 2011, 286, 32962–32975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugnier, C. The Complexity and Multiplicity of the Specific cAMP Phosphodiesterase Family: PDE4, Open New Adapted Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 10616. [Google Scholar] [CrossRef] [PubMed]
- Kar, P.; Barak, P.; Zerio, A.; Lin, Y.P.; Parekh, A.J.; Watts, V.J.; Cooper, D.M.F.; Zaccolo, M.; Kramer, H.; Parekh, A.B. AKAP79 Orchestrates a Cyclic AMP Signalosome Adjacent to Orai1 Ca2+ Channels. Function 2021, 2, zqab036. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.C.; Zhang, P.; Poon, E.; Kong, C.W.; Boheler, K.R.; Huang, Y.; Li, R.A.; Yao, X. Nitric Oxide-cGMP-PKG Pathway Acts on Orai1 to Inhibit the Hypertrophy of Human Embryonic Stem Cell-Derived Cardiomyocytes. Stem. Cells 2015, 33, 2973–2984. [Google Scholar] [CrossRef]
- Jardin, I.; Albarran, L.; Salido, G.M.; Lopez, J.J.; Sage, S.O.; Rosado, J.A. Fine-tuning of store-operated calcium entry by fast and slow Ca2+-dependent inactivation: Involvement of SARAF. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 463–469. [Google Scholar] [CrossRef]
- Rosado, J.A.; Porras, T.; Conde, M.; Sage, S.O. Cyclic nucleotides modulate store-mediated calcium entry through the activation of protein-tyrosine phosphatases and altered actin polymerization in human platelets. J. Biol. Chem. 2001, 276, 15666–15675. [Google Scholar] [CrossRef] [Green Version]
- Heemskerk, J.W.; Feijge, M.A.; Sage, S.O.; Walter, U. Indirect regulation of Ca2+ entry by cAMP-dependent and cGMP-dependent protein kinases and phospholipase C in rat platelets. Eur. J. Biochem. 1994, 223, 543–551. [Google Scholar] [CrossRef]
- Cuinas, A.; Garcia-Morales, V.; Vina, D.; Gil-Longo, J.; Campos-Toimil, M. Activation of PKA and Epac proteins by cyclic AMP depletes intracellular calcium stores and reduces calcium availability for vasoconstriction. Life Sci. 2016, 155, 102–109. [Google Scholar] [CrossRef]
- Ahmed, M.B.; Alghamdi, A.A.A.; Islam, S.U.; Lee, J.S.; Lee, Y.S. cAMP Signaling in Cancer: A PKA-CREB and EPAC-Centric Approach. Cells 2022, 11, 2020. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, J.J.; Huang, X.Y. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef]
- Peretti, M.; Badaoui, M.; Girault, A.; Van Gulick, L.; Mabille, M.P.; Tebbakha, R.; Sevestre, H.; Morjani, H.; Ouadid-Ahidouch, H. Original association of ion transporters mediates the ECM-induced breast cancer cell survival: Kv10.1-Orai1-SPCA2 partnership. Sci. Rep. 2019, 9, 1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, I.; Milevskiy, M.J.G.; Chalmers, S.B.; Yapa, K.; Robitaille, M.; Henry, C.; Baillie, G.J.; Thompson, E.W.; Roberts-Thomson, S.J.; Monteith, G.R. ORAI1 and ORAI3 in Breast Cancer Molecular Subtypes and the Identification of ORAI3 as a Hypoxia Sensitive Gene and a Regulator of Hypoxia Responses. Cancers 2019, 11, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAndrew, D.; Grice, D.M.; Peters, A.A.; Davis, F.M.; Stewart, T.; Rice, M.; Smart, C.E.; Brown, M.A.; Kenny, P.A.; Roberts-Thomson, S.J.; et al. ORAI1-mediated calcium influx in lactation and in breast cancer. Mol. Cancer Ther. 2011, 10, 448–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motiani, R.K.; Abdullaev, I.F.; Trebak, M. A novel native store-operated calcium channel encoded by Orai3: Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. J. Biol. Chem. 2010, 285, 19173–19183. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Moyano, M.; Diaz, I.; Dionisio, N.; Zhang, X.; Avila-Medina, J.; Calderon-Sanchez, E.; Trebak, M.; Rosado, J.A.; Ordonez, A.; Smani, T. Urotensin-II promotes vascular smooth muscle cell proliferation through store-operated calcium entry and EGFR transactivation. Cardiovasc. Res. 2013, 100, 297–306. [Google Scholar] [CrossRef]
- Pulver, R.A.; Rose-Curtis, P.; Roe, M.W.; Wellman, G.C.; Lounsbury, K.M. Store-operated Ca2+ entry activates the CREB transcription factor in vascular smooth muscle. Circ. Res. 2004, 94, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Watanabe, H.; Murakami, M.; Ono, K.; Munehisa, Y.; Koyama, T.; Nobori, K.; Iijima, T.; Ito, H. Functional role of stromal interaction molecule 1 (STIM1) in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2007, 361, 934–940. [Google Scholar] [CrossRef]
- Wei, F.; Qiu, C.S.; Kim, S.J.; Muglia, L.; Maas, J.W.; Pineda, V.V.; Xu, H.M.; Chen, Z.F.; Storm, D.R.; Muglia, L.J.; et al. Genetic elimination of behavioral sensitization in mice lacking calmodulin-stimulated adenylyl cyclases. Neuron 2002, 36, 713–726. [Google Scholar] [CrossRef] [Green Version]
- DiRocco, D.P.; Scheiner, Z.S.; Sindreu, C.B.; Chan, G.C.; Storm, D.R. A role for calmodulin-stimulated adenylyl cyclases in cocaine sensitization. J. Neurosci. 2009, 29, 2393–2403. [Google Scholar] [CrossRef] [Green Version]
- Maus, M.; Cuk, M.; Patel, B.; Lian, J.; Ouimet, M.; Kaufmann, U.; Yang, J.; Horvath, R.; Hornig-Do, H.T.; Chrzanowska-Lightowlers, Z.M.; et al. Store-Operated Ca2+ Entry Controls Induction of Lipolysis and the Transcriptional Reprogramming to Lipid Metabolism. Cell Metab. 2017, 25, 698–712. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jardin, I.; Berna-Erro, A.; Nieto-Felipe, J.; Macias, A.; Sanchez-Collado, J.; Lopez, J.J.; Salido, G.M.; Rosado, J.A. Similarities and Differences between the Orai1 Variants: Orai1α and Orai1β. Int. J. Mol. Sci. 2022, 23, 14568. https://doi.org/10.3390/ijms232314568
Jardin I, Berna-Erro A, Nieto-Felipe J, Macias A, Sanchez-Collado J, Lopez JJ, Salido GM, Rosado JA. Similarities and Differences between the Orai1 Variants: Orai1α and Orai1β. International Journal of Molecular Sciences. 2022; 23(23):14568. https://doi.org/10.3390/ijms232314568
Chicago/Turabian StyleJardin, Isaac, Alejandro Berna-Erro, Joel Nieto-Felipe, Alvaro Macias, Jose Sanchez-Collado, Jose J. Lopez, Gines M. Salido, and Juan A. Rosado. 2022. "Similarities and Differences between the Orai1 Variants: Orai1α and Orai1β" International Journal of Molecular Sciences 23, no. 23: 14568. https://doi.org/10.3390/ijms232314568
APA StyleJardin, I., Berna-Erro, A., Nieto-Felipe, J., Macias, A., Sanchez-Collado, J., Lopez, J. J., Salido, G. M., & Rosado, J. A. (2022). Similarities and Differences between the Orai1 Variants: Orai1α and Orai1β. International Journal of Molecular Sciences, 23(23), 14568. https://doi.org/10.3390/ijms232314568







