Dissection of Functional Domains of Orc1-2, the Archaeal Global DNA Damage-Responsive Regulator
Abstract
:1. Introduction
2. Results
2.1. Structural Modelling Revealed Key Residues in Orc1-2 Responsible for DNA Binding
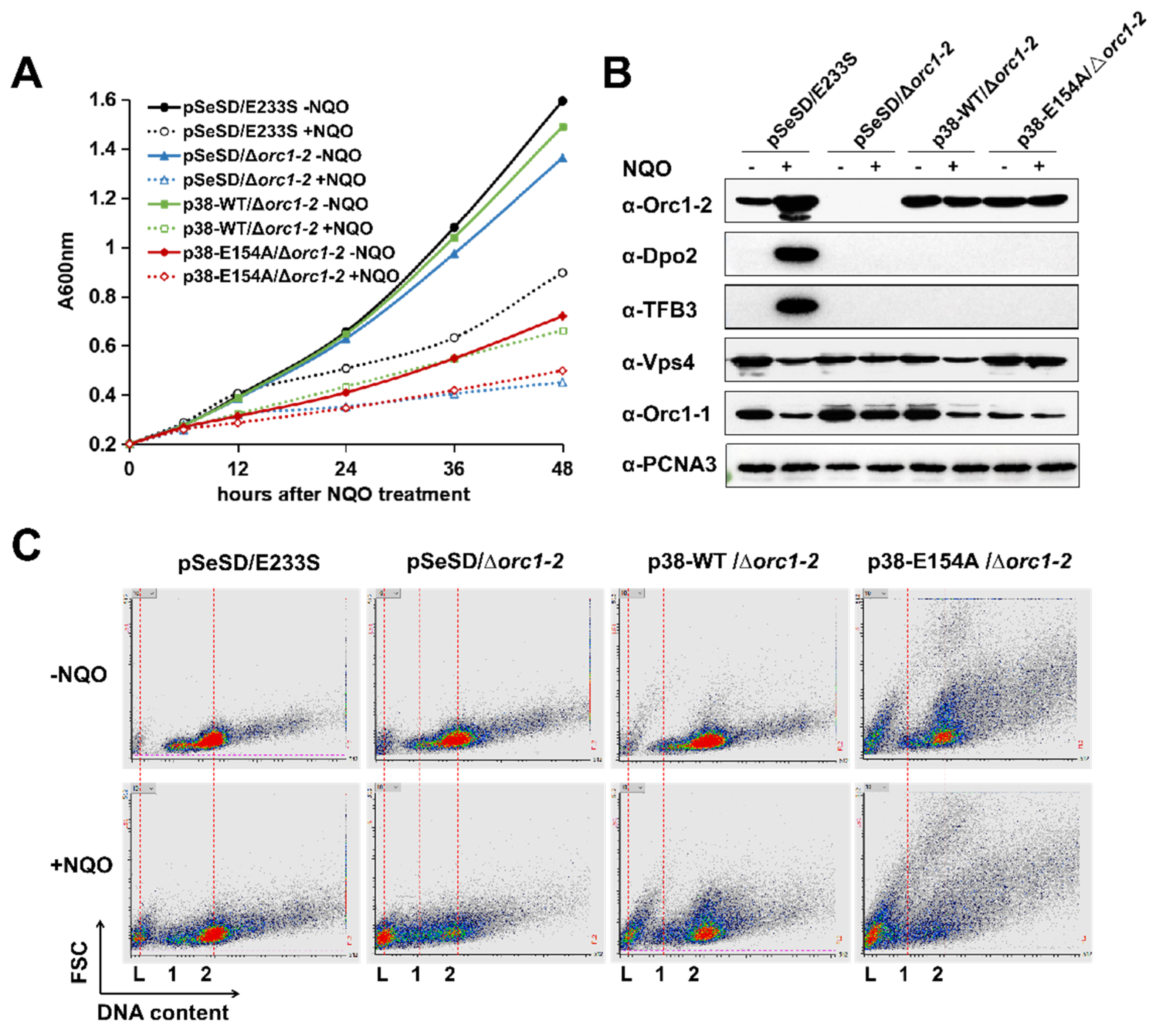
2.2. Inactivation of ATP Hydrolysis of Orc1-2 Yielded Cell Death in S. islandicus
2.3. Each Functional Domain of Orc1-2 Is Essential for DNA Damage Tolerance of S. islandicus
2.4. All Orc1-2 Functional Modules Are Important for Regulated Expression of DDR genes in S. islandicus
2.5. Identification of Orc1-2 Domains Responsible for DNA Binding
2.6. Orc1-2 Mediated Two Distinct Modes of Cell Death in S. islandicus
3. Discussion
3.1. Distinct Features of Orc1-2 Protein–DNA Interactions
3.2. Functions of the AAA+ ATPase Domain
3.3. Orc1-2-Directed DDR Regulation Is a Stepwise Process
4. Materials and Methods
4.1. Cell Growth, Transformation and NQO Treatment of Saccharolobus
4.2. Construction of S. islandicus orc1-2 Mutants
4.3. Microscopy and Flow Cytometry
4.4. Total RNA Preparation and Real-Time Quantitative PCR
4.5. Western-Blotting Analysis
4.6. Construction of Expression Plasmids of Mutated orc1-2 Genes and Purification of Their Encoded Proteins from E. coli
4.7. Electrophoretic Mobility Shift Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Feng, X.; Liu, Z.; Han, W.; Liang, Y.X.; She, Q. An Orc1/Cdc6 ortholog functions as a key regulator in the DNA damage response in Archaea. Nucleic Acids Res. 2018, 46, 6697–6711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duderstadt, K.E.; Berger, J.M. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.D. Archaeal orc1/cdc6 proteins. Subcell Biochem. 2012, 62, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.M.; Recalde, A.; Brasen, C.; Counts, J.A.; Nussbaum, P.; Bost, J.; Schocke, L.; Shen, L.; Willard, D.J.; Quax, T.E.F.; et al. The biology of thermoacidophilic archaea from the order Sulfolobales. FEMS Microbiol. Rev. 2021, 45, fuaa063. [Google Scholar] [CrossRef]
- Guo, L.; Brugger, K.; Liu, C.; Shah, S.A.; Zheng, H.; Zhu, Y.; Wang, S.; Lillestol, R.K.; Chen, L.; Frank, J.; et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011, 193, 1672–1680. [Google Scholar] [CrossRef] [Green Version]
- Samson, R.Y.; Xu, Y.; Gadelha, C.; Stone, T.A.; Faqiri, J.N.; Li, D.; Qin, N.; Pu, F.; Liang, Y.X.; She, Q.; et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 2013, 3, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Smith, C.L.; DeRyckere, D.; DeAngelis, K.; Martin, G.S.; Berger, J.M. Structure and function of Cdc6/Cdc18: Implications for origin recognition and checkpoint control. Mol. Cell 2000, 6, 637–648. [Google Scholar] [CrossRef]
- Grabowski, B.; Kelman, Z. Autophosphorylation of Archaeal Cdc6 Homologues Is Regulated by DNA. J. Bacteriol. 2001, 183, 5459–5464. [Google Scholar] [CrossRef]
- Capaldi, S.A.; Berger, J.M. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 2004, 32, 4821–4832. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, F.; Forterre, P.; Ishino, Y.; Myllykallio, H. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA 2001, 98, 11152–11157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, F.; Glatigny, A.; Mucchielli-Giorgi, M.-H.; Agier, N.; Delacroix, H.; Marisa, L.; Durosay, P.; Ishino, Y.; Aggerbeck, L.; Forterre, P. Genomewide and biochemical analyses of DNA-binding activity of Cdc6/Orc1 and Mcm proteins in Pyrococcus sp. Nucleic Acids Res. 2007, 35, 3214–3222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, N.P.; Dionne, I.; Lundgren, M.; Marsh, V.L.; Bernander, R.; Bell, S.D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 2004, 116, 25–38. [Google Scholar] [CrossRef]
- De Felice, M.; Esposito, L.; Rossi, M.; Pisani, F.M. Biochemical characterization of two Cdc6/ORC1-like proteins from the crenarchaeon Sulfolobus solfataricus. Extremophiles 2006, 10, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Dueber, E.C.; Costa, A.; Corn, J.E.; Bell, S.D.; Berger, J.M. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 2011, 39, 3621–3631. [Google Scholar] [CrossRef] [Green Version]
- Grainge, I.; Scaife, S.; Wigley, D.B. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003, 31, 4888–4898. [Google Scholar] [CrossRef]
- Singleton, M.R.; Morales, R.; Grainge, I.; Cook, N.; Isupov, M.N.; Wigley, D.B. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 2004, 343, 547–557. [Google Scholar] [CrossRef]
- Haugland, G.T.; Innselset, M.; Madern, D.; Birkeland, N.K. Characterization of the Cdc6 Homologues from the Euryarchaeon Thermoplasma acidophilum. Open Biochem. J. 2008, 2, 129–134. [Google Scholar] [CrossRef]
- Duggin, I.G.; McCallum, S.A.; Bell, S.D. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 2008, 105, 16737–16742. [Google Scholar] [CrossRef]
- Arora, J.; Goswami, K.; Saha, S. Characterization of the replication initiator Orc1/Cdc6 from the Archaeon Picrophilus torridus. J. Bacteriol. 2014, 196, 276–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludt, K.; Soppa, J. Influence of Origin Recognition Complex Proteins on the Copy Numbers of Three Chromosomes in Haloferax volcanii. J. Bacteriol. 2018, 200, e00161-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majernik, A.I.; Chong, J.P. A conserved mechanism for replication origin recognition and binding in archaea. Biochem. J. 2008, 409, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Dueber, E.L.; Corn, J.E.; Bell, S.D.; Berger, J.M. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science (New York N. Y.) 2007, 317, 1210–1213. [Google Scholar] [CrossRef]
- Gaudier, M.; Schuwirth, B.S.; Westcott, S.L.; Wigley, D.B. Structural basis of DNA replication origin recognition by an ORC protein. Science (New York N. Y.) 2007, 317, 1213–1216. [Google Scholar] [CrossRef] [Green Version]
- He, Z.G.; Feng, Y.; Wang, J.; Jiang, P.X. The regulatory function of N-terminal AAA+ ATPase domain of eukaryote-like archaeal Orc1/Cdc6 protein during DNA replication initiation. Arch. Biochem. Biophys. 2008, 471, 176–183. [Google Scholar] [CrossRef]
- She, Q.; Zhang, C.; Deng, L.; Peng, N.; Chen, Z.; Liang, Y.X. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 2009, 37, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef] [Green Version]
- Dalal, S.; Rosser, M.F.; Cyr, D.M.; Hanson, P.I. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell 2004, 15, 637–648. [Google Scholar] [CrossRef] [Green Version]
- Weibezahn, J.; Schlieker, C.; Bukau, B.; Mogk, A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003, 278, 32608–32617. [Google Scholar] [CrossRef]
- Samson, R.Y.; Abeyrathne, P.D.; Bell, S.D. Mechanism of Archaeal MCM Helicase Recruitment to DNA Replication Origins. Mol. Cell 2016, 61, 287–296. [Google Scholar] [CrossRef]
- Greci, M.D.; Bell, S.D. Archaeal DNA Replication. Annu. Rev. Microbiol. 2020, 74, 65–80. [Google Scholar] [CrossRef]
- Peng, N.; Xia, Q.; Chen, Z.; Liang, Y.X.; She, Q. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 2009, 74, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, X.; Xu, R.; Zhao, R.; Feng, W.; Liao, J.; Han, W.; She, Q. A Unique B-Family DNA Polymerase Facilitating Error-Prone DNA Damage Tolerance in Crenarchaeota. Front. Microbiol. 2020, 11, 1585. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhang, B.; Xu, R.; Gao, Z.; Liu, X.; Yuan, G.; Ishino, S.; Feng, M.; Shen, Y.; Ishino, Y.; et al. Enzymatic Switching Between Archaeal DNA Polymerases Facilitates Abasic Site Bypass. Front. Microbiol. 2021, 12, 802670. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhang, B.; Gao, Z.; Xu, R.; Liu, X.; Ishino, S.; Feng, M.; Shen, Y.; Ishino, Y.; She, Q. A Well-Conserved Archaeal B-Family Polymerase Functions as an Extender in Translesion Synthesis. mBio 2022, 13, e0265921. [Google Scholar] [CrossRef]
- Feng, X.; Sun, M.; Han, W.; Liang, Y.X.; She, Q. A transcriptional factor B paralog functions as an activator to DNA damage-responsive expression in archaea. Nucleic Acids Res. 2018, 46, 7085–7096. [Google Scholar] [CrossRef] [Green Version]
- Ajon, M.; Frols, S.; van Wolferen, M.; Stoecker, K.; Teichmann, D.; Driessen, A.J.; Grogan, D.W.; Albers, S.V.; Schleper, C. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol 2011, 82, 807–817. [Google Scholar] [CrossRef] [Green Version]
- van Wolferen, M.; Wagner, A.; van der Does, C.; Albers, S.V. The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. USA 2016, 113, 2496–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Zhu, H.; Chen, Z.; Liang, Y.X.; She, Q. Unmarked gene deletion and host–vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 2009, 13, 735–746. [Google Scholar] [CrossRef]
- Feng, X.; She, Q. Recombinant protein expression in Sulfolobus islandicus. Methods Enzymol. 2021, 659, 275–295. [Google Scholar]
- Zillig, W.; Kletzin, A.; Schleper, C.; Holz, I.; Janekovic, D.; Hain, J.; Lanzendorfer, M.; Kristjansson, J.K. Screening for Sulfolobales, their Plasmids and their Viruses in Icelandic Solfataras. Syst. Appl. Microbiol. 1994, 16, 609–628. [Google Scholar] [CrossRef]
- Han, W.; Xu, Y.; Feng, X.; Liang, Y.X.; Huang, L.; Shen, Y.; She, Q. NQO-Induced DNA-Less Cell Formation Is Associated with Chromatin Protein Degradation and Dependent on A0A1-ATPase in Sulfolobus. Front. Microbiol. 2017, 8, 1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pan, S.; Zhang, Y.; Ren, M.; Feng, M.; Peng, N.; Chen, L.; Liang, Y.X.; She, Q. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016, 44, e34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.; Feng, M.; Feng, X.; Liang, Y.X.; She, Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting featuRes. and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015, 43, 406–417. [Google Scholar] [CrossRef]
- Warrens, A.N.; Jones, M.D.; Lechler, R.I. Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest. Gene 1997, 186, 29–35. [Google Scholar] [CrossRef]
- Han, W.; Feng, X.; She, Q. Reverse Gyrase Functions in Genome Integrity Maintenance by Protecting DNA Breaks In Vivo. Int. J. Mol. Sci. 2017, 18, 1340. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Peng, N.; Deng, L.; Mei, Y.; Jiang, D.; Hu, Y.; Awayez, M.; Liang, Y.; She, Q. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012, 78, 5630–5637. [Google Scholar] [CrossRef]



| Name | Mutation | Mutant Viability | Protein Stability | NQO Sensitivity | Cell Aggregation | Cell Population (%) | |
|---|---|---|---|---|---|---|---|
| <1 | 2 | ||||||
| WT | 413 aa | Yes | ++ | S | ++ | 14.9 | 69.3 |
| Δorc1-2 | In-frame deletion retaining 35 aa at N-terminus | Yes | NV | HS | − | 72.0 | 20.0 |
| ΔC | 298 aa lacking wH domain | Yes | + | HS | − | 75.0 | 15.5 |
| ΔN | 115 aa lacking ATPase domain | Yes | ++ | HS | − | 77.8 | 8.9 |
| WA | K72A Walker A | Yes | + | HS | − | 69.4 | 22.5 |
| WB | E154A WalkerB | No | ++ | NV | NV | NV | NV |
| ISM | G127D, L128D initiator specific motif | Yes | ++ | HS | − | 73.3 | 19.7 |
| wH-m1 | R353A, R354A wH domain | Yes | ++ | S* | + | 52.4 | 41.4 |
| wH-m2 | R381A, R383A wH domain | Yes | ++ | HS | − | 73.7 | 18.7 |
| wH | quadruple substitutions wH domain | Yes | ++ | HS | − | 80.2 | 12.3 |
| WBwH | E154A, R353A, R354A, R381A and R383A pentuple substitutions | Yes | ++ | HS | − | 62.1 | 28.6 |
| Name | Description | Promotor | Orc1-2 Level | Broth Growth | NQO Sensitivity | Cell Aggregation | Giant Cells | |
|---|---|---|---|---|---|---|---|---|
| SCV | ACV | |||||||
| pSeSD/E233S | Genomic: Orc1-2 | Porc1-2 | ++ | ++ | ++ | S | ++ | No |
| pSeSD/Δorc1-2 | Orc1-2-deficient | NV | NV | ++ | ++ | HS | − | No |
| p38-WT/Δorc1-2 | Plasmid-borne: Orc1-2 | ParaS-38 | + | ++ | ++ | HS | − | No |
| p38-E154A/Δorc1-2 | Plasmid-borne: Orc1-2E154A | ParaS-38 | + | ++ | + | HS | − | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Sun, M.; Xu, R.; Shen, Y.; Huang, Q.; Feng, X.; She, Q. Dissection of Functional Domains of Orc1-2, the Archaeal Global DNA Damage-Responsive Regulator. Int. J. Mol. Sci. 2022, 23, 14609. https://doi.org/10.3390/ijms232314609
Liu X, Sun M, Xu R, Shen Y, Huang Q, Feng X, She Q. Dissection of Functional Domains of Orc1-2, the Archaeal Global DNA Damage-Responsive Regulator. International Journal of Molecular Sciences. 2022; 23(23):14609. https://doi.org/10.3390/ijms232314609
Chicago/Turabian StyleLiu, Xiaotong, Mengmeng Sun, Ruyi Xu, Yulong Shen, Qihong Huang, Xu Feng, and Qunxin She. 2022. "Dissection of Functional Domains of Orc1-2, the Archaeal Global DNA Damage-Responsive Regulator" International Journal of Molecular Sciences 23, no. 23: 14609. https://doi.org/10.3390/ijms232314609
APA StyleLiu, X., Sun, M., Xu, R., Shen, Y., Huang, Q., Feng, X., & She, Q. (2022). Dissection of Functional Domains of Orc1-2, the Archaeal Global DNA Damage-Responsive Regulator. International Journal of Molecular Sciences, 23(23), 14609. https://doi.org/10.3390/ijms232314609







