miRNA Pathway Alteration in Response to Non-Coding RNA Delivery in Viral Vector-Based Gene Therapy
Abstract
:1. Introduction
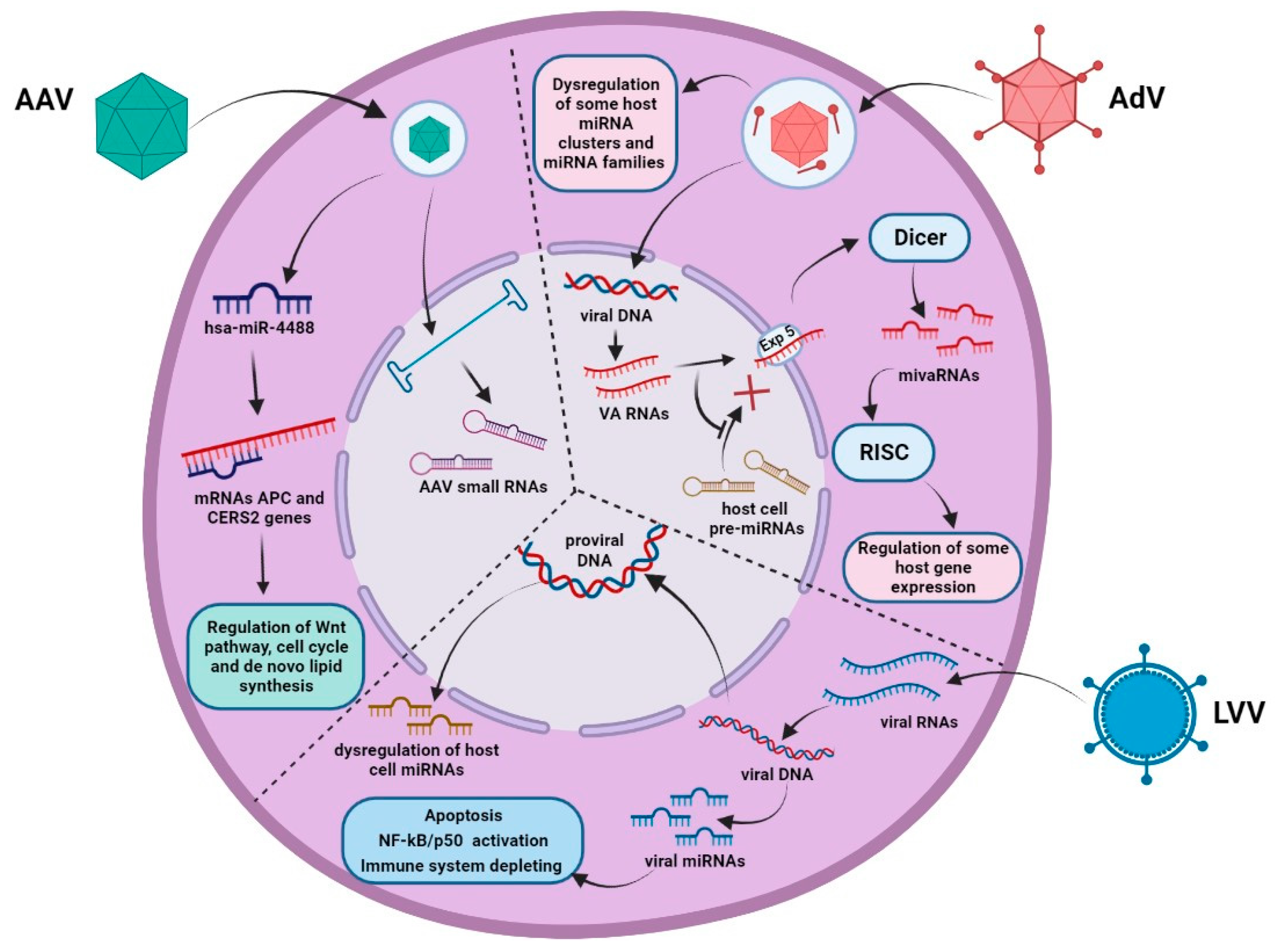
2. miRNA Pathway Dysregulation in Response to the Entry of Genotherapeutic Viruses
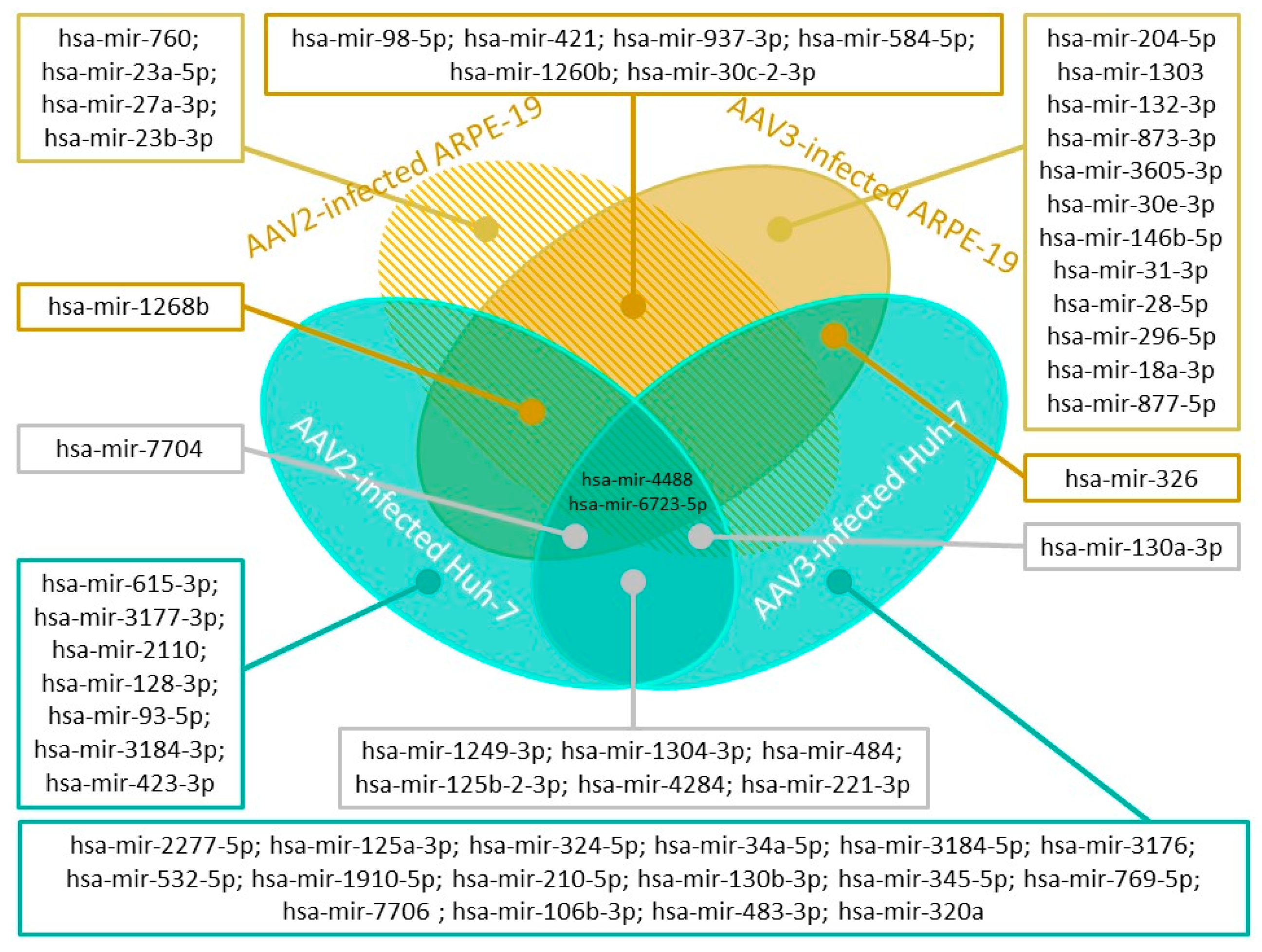
2.1. Adeno-Associated Virus
2.2. Adenovirus
2.3. Lentivirus
3. Examples of miRNA Pathway Dysregulation in Response to Non-Coding RNA Delivery
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Friedmann, T.; Roblin, R. Gene Therapy for Human Genetic Disease? Science (1979) 1972, 175, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, K.; Cai, Y. An Overview of Development in Gene Therapeutics in China. Gene Ther. 2020, 27, 338–348. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Library of Medicine. ClinicalTrials.gov. Available online: https://clinicaltrials.gov (accessed on 30 October 2022).
- European Medicines Agency. Available online: https://www.ema.europa.eu/en (accessed on 30 October 2022).
- Finer, M.; Glorioso, J. A Brief Account of Viral Vectors and Their Promise for Gene Therapy. Gene Ther. 2017, 24, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier, A.F.; Fraefel, C.; Seyffert, M. The Interplay between Adeno-Associated Virus and Its Helper Viruses. Viruses 2020, 12, 662. [Google Scholar] [CrossRef]
- Saraiva, J.; Nobre, R.J.; Pereira de Almeida, L. Gene Therapy for the CNS Using AAVs: The Impact of Systemic Delivery by AAV9. J. Control. Release 2016, 241, 94–109. [Google Scholar] [CrossRef]
- Dudek, A.M.; Zabaleta, N.; Zinn, E.; Pillay, S.; Zengel, J.; Porter, C.; Franceschini, J.S.; Estelien, R.; Carette, J.E.; Zhou, G.L.; et al. GPR108 Is a Highly Conserved AAV Entry Factor. Mol. Ther. 2020, 28, 367–381. [Google Scholar] [CrossRef]
- Kattenhorn, L.M.; Tipper, C.H.; Stoica, L.; Geraghty, D.S.; Wright, T.L.; Clark, K.R.; Wadsworth, S.C. Adeno-Associated Virus Gene Therapy for Liver Disease. Hum. Gene Ther. 2016, 27, 947–961. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhong, L.; Nahid, M.A.; Gao, G. The Potential of Adeno-Associated Viral Vectors for Gene Delivery to Muscle Tissue. Expert Opin. Drug Deliv. 2014, 11, 345–364. [Google Scholar] [CrossRef]
- Su, H.; Yeghiazarians, Y.; Lee, A.; Huang, Y.; Arakawa-Hoyt, J.; Ye, J.; Orcino, G.; Grossman, W.; Kan, Y.W. AAV Serotype 1 Mediates More Efficient Gene Transfer to Pig Myocardium than AAV Serotype 2 and Plasmid. J. Gene Med. 2008, 10, 33–41. [Google Scholar] [CrossRef]
- Greenberg, B.; Yaroshinsky, A.; Zsebo, K.M.; Butler, J.; Felker, G.M.; Voors, A.A.; Rudy, J.J.; Wagner, K.; Hajjar, R.J. Design of a Phase 2b Trial of Intracoronary Administration of AAV1/SERCA2a in Patients With Advanced Heart Failure. JACC Heart Fail. 2014, 2, 84–92. [Google Scholar] [CrossRef]
- Hadri, L.; Kratlian, R.G.; Benard, L.; Maron, B.A.; Dorfmüller, P.; Ladage, D.; Guignabert, C.; Ishikawa, K.; Aguero, J.; Ibanez, B.; et al. Therapeutic Efficacy of AAV1.SERCA2a in Monocrotaline-Induced Pulmonary Arterial Hypertension. Circulation 2013, 128, 512–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zincarelli, C.; Soltys, S.; Rengo, G.; Rabinowitz, J.E. Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol. Ther. 2008, 16, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Kotchey, N.M.; Adachi, K.; Zahid, M.; Inagaki, K.; Charan, R.; Parker, R.S.; Nakai, H. A Potential Role of Distinctively Delayed Blood Clearance of Recombinant Adeno-Associated Virus Serotype 9 in Robust Cardiac Transduction. Mol. Ther. 2011, 19, 1079–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samulski, R.J.; Muzyczka, N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu. Rev. Virol. 2014, 1, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020, 28, 723–746. [Google Scholar] [CrossRef]
- Wagner, H.J.; Weber, W.; Fussenegger, M. Synthetic Biology: Emerging Concepts to Design and Advance Adeno-Associated Viral Vectors for Gene Therapy. Adv. Sci. 2021, 8, 2004018. [Google Scholar] [CrossRef]
- Ronzitti, G.; Gross, D.-A.; Mingozzi, F. Human Immune Responses to Adeno-Associated Virus (AAV) Vectors. Front. Immunol. 2020, 11, 670. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral Vector Platforms within the Gene Therapy Landscape. Signal Transduct Target Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Ghebremedhin, B. Human Adenovirus: Viral Pathogen with Increasing Importance. Eur. J. Microbiol. Immunol. (Bp.) 2014, 4, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zeng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-Mediated Gene Delivery: Potential Applications for Gene and Cell-Based Therapies in the New Era of Personalized Medicine. Genes Dis. 2017, 4, 43–63. [Google Scholar] [CrossRef]
- Allen, R.J.; Byrnes, A.P. Interaction of Adenovirus with Antibodies, Complement, and Coagulation Factors. FEBS Lett 2019, 593, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Ricobaraza, A.; Gonzalez-Aparicio, M.; Mora-Jimenez, L.; Lumbreras, S.; Hernandez-Alcoceba, R. High-Capacity Adenoviral Vectors: Expanding the Scope of Gene Therapy. Int. J. Mol. Sci. 2020, 21, 3643. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, S.A.; Lorincz, R.; Boucher, P.; Curiel, D.T. Adenoviral Vector Vaccine Platforms in the SARS-CoV-2 Pandemic. NPJ Vaccines 2021, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Lasaro, M.O.; Ertl, H.C. New Insights on Adenovirus as Vaccine Vectors. Mol. Ther. 2009, 17, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Wang, H.; Ichino, M.; Ura, T.; Mizuki, N.; Okuda, K. Biodistribution and Immunity of Adenovirus 5/35 and Modified Vaccinia Ankara Vector Vaccines against Human Immunodeficiency Virus 1 Clade C. Gene Ther. 2022, 29, 636–642. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Kim, K.T.; Lee, S.-J.; Hong, S.-H.; Moon, J.Y.; Yoon, E.K.; Kim, S.; Kim, E.O.; Kang, S.H.; Kim, S.K.; et al. Image-Aided Suicide Gene Therapy Utilizing Multifunctional HTERT-Targeting Adenovirus for Clinical Translation in Hepatocellular Carcinoma. Theranostics 2016, 6, 357–368. [Google Scholar] [CrossRef]
- Seitz, R. Human Immunodeficiency Virus (HIV). Transfus. Med. Hemotherapy 2016, 43, 203–222. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, N.P.; Vink, C.A. The Impact of Lentiviral Vector Genome Size and Producer Cell Genomic to Gag-Pol MRNA Ratios on Packaging Efficiency and Titre. Mol. Ther. Methods Clin. Dev. 2021, 21, 574–584. [Google Scholar] [CrossRef]
- Pauwels, K.; Gijsbers, R.; Toelen, J.; Schambach, A.; Willard-Gallo, K.; Verheust, C.; Debyser, Z.; Herman, P. State-of-the-Art Lentiviral Vectors for Research Use: Risk Assessment and Biosafety Recommendations. Curr. Gene Ther. 2009, 9, 459–474. [Google Scholar] [CrossRef] [Green Version]
- Stein, S.; Ott, M.G.; Schultze-Strasser, S.; Jauch, A.; Burwinkel, B.; Kinner, A.; Schmidt, M.; Krämer, A.; Schwäble, J.; Glimm, H.; et al. Genomic Instability and Myelodysplasia with Monosomy 7 Consequent to EVI1 Activation after Gene Therapy for Chronic Granulomatous Disease. Nat. Med. 2010, 16, 198–204. [Google Scholar] [CrossRef]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion Independence and HMGA2 Activation after Gene Therapy of Human β-Thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavazzana-Calvo, M.; Hacein-Bey, S.; de Saint Basile, G.; Gross, F.; Yvon, E.; Nusbaum, P.; Selz, F.; Hue, C.; Certain, S.; Casanova, J.L.; et al. Gene Therapy of Human Severe Combined Immunodeficiency (SCID)-X1 Disease. Science (1979) 2000, 288, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; et al. LMO2-Associated Clonal T Cell Proliferation in Two Patients after Gene Therapy for SCID-X1. Science (1979) 2003, 302, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Howe, S.J.; Mansour, M.R.; Schwarzwaelder, K.; Bartholomae, C.; Hubank, M.; Kempski, H.; Brugman, M.H.; Pike-Overzet, K.; Chatters, S.J.; de Ridder, D.; et al. Insertional Mutagenesis Combined with Acquired Somatic Mutations Causes Leukemogenesis Following Gene Therapy of SCID-X1 Patients. J. Clin. Investig. 2008, 118, 3143–3150. [Google Scholar] [CrossRef]
- Modlich, U.; Navarro, S.; Zychlinski, D.; Maetzig, T.; Knoess, S.; Brugman, M.H.; Schambach, A.; Charrier, S.; Galy, A.; Thrasher, A.J.; et al. Insertional Transformation of Hematopoietic Cells by Self-Inactivating Lentiviral and Gammaretroviral Vectors. Mol. Ther. 2009, 17, 1919–1928. [Google Scholar] [CrossRef]
- Kaufmann, K.B.; Büning, H.; Galy, A.; Schambach, A.; Grez, M. Gene Therapy on the Move. EMBO Mol. Med. 2013, 5, 1642–1661. [Google Scholar] [CrossRef]
- Gardlík, R.; Pálffy, R.; Hodosy, J.; Lukács, J.; Turna, J.; Celec, P. Vectors and Delivery Systems in Gene Therapy. Med. Sci. Monit. 2005, 11, 110–121. [Google Scholar]
- Cavazzana-Calvo, M.; Thrasher, A.; Mavilio, F. The Future of Gene Therapy. Nature 2004, 427, 779–781. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. Viruses, MicroRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Leung, A.K.L.; Sharp, P.A. MicroRNA Functions in Stress Responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Gottwein, E.; Cullen, B.R. Viral and Cellular MicroRNAs as Determinants of Viral Pathogenesis and Immunity. Cell Host Microbe 2008, 3, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schüttler, C.G.; Fehr, C.; Jünemann, C.; Niepmann, M. MicroRNA-122 Stimulates Translation of Hepatitis C Virus RNA. EMBO J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.P.E.; Lewis, A.P.; Jopling, C.L. MiR-122 Activates Hepatitis C Virus Translation by a Specialized Mechanism Requiring Particular RNA Components. Nucleic Acids Res. 2011, 39, 7716–7729. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-G.; Bharaj, P.; Abraham, S.; Ma, H.; Yi, G.; Ye, C.; Dang, Y.; Manjunath, N.; Wu, H.; Shankar, P. Multiplexing Seven MiRNA-Based ShRNAs to Suppress HIV Replication. Mol. Ther. 2015, 23, 310–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zhou, P.; Wang, X.; Chen, F.; Christensen, E.; Thompson, J.; Ren, X.; Kells, A.; Stanek, L.; Carter, T.; et al. Efficient and Precise Processing of the Optimized Primary Artificial MicroRNA in a Huntingtin-Lowering Adeno-Associated Viral Gene Therapy In Vitro and in Mice and Nonhuman Primates. Hum. Gene Ther. 2022, 33, 37–60. [Google Scholar] [CrossRef]
- Khan, A.A.; Betel, D.; Miller, M.L.; Sander, C.; Leslie, C.S.; Marks, D.S. Transfection of Small RNAs Globally Perturbs Gene Regulation by Endogenous MicroRNAs. Nat. Biotechnol. 2009, 27, 549–555. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, C.; Wang, B.; Wang, F.; Pei, B.; Cheng, C.; Yang, W.; Zhao, Z. Transduction with Lentiviral Vectors Altered the Expression Profile of Host MicroRNAs. J. Virol. 2018, 92, e00503-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Kong, X.; Wu, X. RNAi-Based Immunity in Insects against Baculoviruses and the Strategies of Baculoviruses Involved in SiRNA and MiRNA Pathways to Weaken the Defense. Dev. Comp. Immunol. 2021, 122, 104116. [Google Scholar] [CrossRef]
- Arora, S.; Rana, R.; Chhabra, A.; Jaiswal, A.; Rani, V. MiRNA–Transcription Factor Interactions: A Combinatorial Regulation of Gene Expression. Mol. Genet. Genom. 2013, 288, 77–87. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Sullivan, C.S. Virus-Encoded MicroRNAs: An Overview and a Look to the Future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef] [Green Version]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral Noncoding RNAs: More Surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef]
- Stutika, C.; Mietzsch, M.; Gogol-Döring, A.; Weger, S.; Sohn, M.; Chen, W.; Heilbronn, R. Comprehensive Small RNA-Seq of Adeno-Associated Virus (AAV)-Infected Human Cells Detects Patterns of Novel, Non-Coding AAV RNAs in the Absence of Cellular MiRNA Regulation. PLoS ONE 2016, 11, e0161454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arumugam, S.; Mary, B.; Kumar, M.; Jayandharan, G.R. Analysis of Hepatic and Retinal Cell MicroRNAome during AAV Infection Reveals Their Diverse Impact on Viral Transduction and Cellular Physiology. Gene 2020, 724, 144157. [Google Scholar] [CrossRef] [PubMed]
- Spassieva, S.D.; Mullen, T.D.; Townsend, D.M.; Obeid, L.M. Disruption of Ceramide Synthesis by CerS2 Down-Regulation Leads to Autophagy and the Unfolded Protein Response. Biochem. J. 2009, 424, 273–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathews, M.B.; Shenk, T. Adenovirus Virus-Associated RNA and Translation Control. J. Virol. 1991, 65, 5657–5662. [Google Scholar] [CrossRef] [Green Version]
- Kitajewski, J.; Schneider, R.J.; Safer, B.; Munemitsu, S.M.; Samuel, C.E.; Thimmappaya, B.; Shenk, T. Adenovirus VAI RNA Antagonizes the Antiviral Action of Interferon by Preventing Activation of the Interferon-Induced EIF-2α Kinase. Cell 1986, 45, 195–200. [Google Scholar] [CrossRef]
- Minamitani, T.; Iwakiri, D.; Takada, K. Adenovirus Virus-Associated RNAs Induce Type I Interferon Expression through a RIG-I-Mediated Pathway. J. Virol. 2011, 85, 4035–4040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, S.Y.; Patel, R.C.; Sen, G.C.; Malhotra, P.; Ghadge, G.D.; Thimmapaya, B. Activation of Interferon-Inducible 2′−5′ Oligoadenylate Synthetase by Adenoviral VAI RNA. J. Biol. Chem. 1995, 270, 3454–3461. [Google Scholar] [CrossRef] [Green Version]
- Piedade, D.; Azevedo-Pereira, J.M. MicroRNAs as Important Players in Host–Adenovirus Interactions. Front. Microbiol. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Lu, S.; Cullen, B.R. Adenovirus VA1 Noncoding RNA Can Inhibit Small Interfering RNA and MicroRNA Biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef] [Green Version]
- Bennasser, Y.; Chable-Bessia, C.; Triboulet, R.; Gibbings, D.; Gwizdek, C.; Dargemont, C.; Kremer, E.J.; Voinnet, O.; Benkirane, M. Competition for XPO5 Binding between Dicer MRNA, Pre-MiRNA and Viral RNA Regulates Human Dicer Levels. Nat. Struct. Mol. Biol. 2011, 18, 323–327. [Google Scholar] [CrossRef] [PubMed]
- McIndoo, E.R.; Burgoyne, H.M.; Shin, H.-S.; Radke, J.R. Characterization of Viral MiRNAs during Adenovirus 14 Infection and Their Differential Expression in the Emergent Strain Adenovirus 14p1. Viruses 2022, 14, 898. [Google Scholar] [CrossRef] [PubMed]
- Machitani, M.; Katayama, K.; Sakurai, F.; Matsui, H.; Yamaguchi, T.; Suzuki, T.; Miyoshi, H.; Kawabata, K.; Mizuguchi, H. Development of an Adenovirus Vector Lacking the Expression of Virus-Associated RNAs. J. Control. Release 2011, 154, 285–289. [Google Scholar] [CrossRef]
- Xu, N.; Segerman, B.; Zhou, X.; Akusjärvi, G. Adenovirus Virus-Associated RNAII-Derived Small RNAs Are Efficiently Incorporated into the RNA-Induced Silencing Complex and Associate with Polyribosomes. J. Virol. 2007, 81, 10540–10549. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, O.; Carnero, E.; Abad, X.; Razquin, N.; Guruceaga, E.; Segura, V.; Fortes, P. Adenovirus VA RNA-Derived MiRNAs Target Cellular Genes Involved in Cell Growth, Gene Expression and DNA Repair. Nucleic Acids Res. 2010, 38, 750–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Chen, M.; Tellgren-Roth, C.; Pettersson, U. Fluctuating Expression of MicroRNAs in Adenovirus Infected Cells. Virology 2015, 478, 99–111. [Google Scholar] [CrossRef]
- Qi, Y.; Tu, J.; Cui, L.; Guo, X.; Shi, Z.; Li, S.; Shi, W.; Shan, Y.; Ge, Y.; Shan, J.; et al. High-Throughput Sequencing of MicroRNAs in Adenovirus Type 3 Infected Human Laryngeal Epithelial Cells. J. Biomed. Biotechnol. 2010, 2010, 915980. [Google Scholar] [CrossRef] [Green Version]
- Manríquez, V.; Gutierrez, A.; Morales, A.; Brito, R.; Pavez, M.; Sapunar, J.; Fonseca, L.; Molina, V.; Ortiz, E.; Barra, M.I.; et al. Influence of Adenovirus 36 Seropositivity on the Expression of Adipogenic MicroRNAs in Obese Subjects. Int. J. Obes. 2020, 44, 2303–2312. [Google Scholar] [CrossRef]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Nakatani, K.; Tachibana, M.; Mizuguchi, H. MicroRNA MiR-27 Inhibits Adenovirus Infection by Suppressing the Expression of SNAP25 and TXN2. J. Virol. 2017, 91, e00159-17. [Google Scholar] [CrossRef] [Green Version]
- Hodzic, J.; Sie, D.; Vermeulen, A.; van Beusechem, V.W. Functional Screening Identifies Human MiRNAs That Modulate Adenovirus Propagation in Prostate Cancer Cells. Hum. Gene Ther. 2017, 28, 766–780. [Google Scholar] [CrossRef]
- Houzet, L.; Yeung, M.L.; de Lame, V.; Desai, D.; Smith, S.M.; Jeang, K.-T. MicroRNA Profile Changes in Human Immunodeficiency Virus Type 1 (HIV-1) Seropositive Individuals. Retrovirology 2008, 5, 118. [Google Scholar] [CrossRef]
- Sardo, L.; Vakil, P.R.; Elbezanti, W.; El-Sayed, A.; Klase, Z. The Inhibition of MicroRNAs by HIV-1 Tat Suppresses Beta Catenin Activity in Astrocytes. Retrovirology 2016, 13, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Del Cojo, M.; López-Huertas, M.R.; Díez-Fuertes, F.; Rodríguez-Mora, S.; Bermejo, M.; López-Campos, G.; Mateos, E.; Jiménez-Tormo, L.; Gómez-Esquer, F.; Díaz-Gil, G.; et al. Changes in the Cellular MicroRNA Profile by the Intracellular Expression of HIV-1 Tat Regulator: A Potential Mechanism for Resistance to Apoptosis and Impaired Proliferation in HIV-1 Infected CD4+ T Cells. PLoS ONE 2017, 12, e0185677. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and Validation of Plasma MiRNA Biomarker Signature Panel for the Detection of Early HIV-1 Infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consuegra, I.; Gasco, S.; Serramía, M.J.; Jiménez, J.L.; Mellado, M.J.; Muñoz-Fernández, M.Á. Establishment of a MiRNA Profile in Paediatric HIV-1 Patients and Its Potential as a Biomarker for Effectiveness of the Combined Antiretroviral Therapy. Sci. Rep. 2021, 11, 23477. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qiu, C.; Dai, L.; Zhang, L.; Feng, M.; Yang, Y.; Qiu, C.; Zhang, A.; Huang, J.; Wang, Y.; et al. Hsa-MiR-31 Governs T-Cell Homeostasis in HIV Protection via IFN-γ-Stat1-T-Bet Axis. Front. Immunol. 2021, 12, 771279. [Google Scholar] [CrossRef]
- Hariharan, M.; Scaria, V.; Pillai, B.; Brahmachari, S.K. Targets for Human Encoded MicroRNAs in HIV Genes. Biochem. Biophys. Res. Commun. 2005, 337, 1214–1218. [Google Scholar] [CrossRef]
- Wang, P.; Qu, X.; Zhou, X.; Shen, Y.; Ji, H.; Fu, Z.; Deng, J.; Lu, P.; Yu, W.; Lu, H.; et al. Two Cellular MicroRNAs, MiR-196b and MiR-1290, Contribute to HIV-1 Latency. Virology 2015, 486, 228–238. [Google Scholar] [CrossRef]
- Chiang, K.; Sung, T.-L.; Rice, A.P. Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4 + T Lymphocytes. J. Virol. 2012, 86, 3244–3252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular MicroRNAs Contribute to HIV-1 Latency in Resting Primary CD4+ T Lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef]
- Wang, X.; Ye, L.; Hou, W.; Zhou, Y.; Wang, Y.-J.; Metzger, D.S.; Ho, W.-Z. Cellular MicroRNA Expression Correlates with Susceptibility of Monocytes/Macrophages to HIV-1 Infection. Blood 2009, 113, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, J.K.; Khan, S.Z.; Soni, K.; Rawat, P.; Gupta, A.; Hariharan, M.; Scaria, V.; Lalwani, M.; Pillai, B.; Mitra, D.; et al. Human Cellular MicroRNA Hsa-MiR-29a Interferes with Viral Nef Protein Expression and HIV-1 Replication. Retrovirology 2008, 5, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, G.; Li, H.; Wu, X.; Covarrubias, M.; Scherer, L.; Meinking, K.; Luk, B.; Chomchan, P.; Alluin, J.; Gombart, A.F.; et al. Interplay between HIV-1 Infection and Host MicroRNAs. Nucleic Acids Res. 2012, 40, 2181–2196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ye, L.; Zhou, Y.; Liu, M.-Q.; Zhou, D.-J.; Ho, W.-Z. Inhibition of Anti-HIV MicroRNA Expression. Am. J. Pathol. 2011, 178, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Modai, S.; Farberov, L.; Herzig, E.; Isakov, O.; Hizi, A.; Shomron, N. HIV-1 Infection Increases MicroRNAs That Inhibit Dicer1, HRB and HIV-EP2, Thereby Reducing Viral Replication. PLoS ONE 2019, 14, e0211111. [Google Scholar] [CrossRef] [Green Version]
- Qiao, J.; Peng, Q.; Qian, F.; You, Q.; Feng, L.; Hu, S.; Liu, W.; Huang, L.; Shu, X.; Sun, B. HIV-1 Vpr Protein Upregulates MicroRNA-210-5p Expression to Induce G2 Arrest by Targeting TGIF2. PLoS ONE 2021, 16, e0261971. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.L.; Plante, I.; Landry, P.; Barat, C.; Janelle, M.-E.; Flamand, L.; Tremblay, M.J.; Provost, P. Identification of Functional MicroRNAs Released through Asymmetrical Processing of HIV-1 TAR Element. Nucleic Acids Res. 2008, 36, 2353–2365. [Google Scholar] [CrossRef] [Green Version]
- Harwig, A.; Berkhout, B.; Das, A. Characterization of MicroRNAs Derived from the HIV-1 TAR RNA Hairpin. Retrovirology 2013, 10, P24. [Google Scholar] [CrossRef] [Green Version]
- Ouellet, D.L.; Vigneault-Edwards, J.; Létourneau, K.; Gobeil, L.-A.; Plante, I.; Burnett, J.C.; Rossi, J.J.; Provost, P. Regulation of Host Gene Expression by HIV-1 TAR MicroRNAs. Retrovirology 2013, 10, 86. [Google Scholar] [CrossRef] [Green Version]
- Kaul, D.; Ahlawat, A.; Gupta, S.D. HIV-1 Genome-Encoded Hiv1-Mir-H1 Impairs Cellular Responses to Infection. Mol. Cell Biochem. 2009, 323, 143–148. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, M.; Geng, G.; Liu, B.; Huang, Z.; Luo, H.; Zhou, J.; Guo, X.; Cai, W.; Zhang, H. A Novel HIV-1-Encoded MicroRNA Enhances Its Viral Replication by Targeting the TATA Box Region. Retrovirology 2014, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.A.; Zhao, H.; Yue, S.C.; Anandaiah, A.; Koziel, H.; Tachado, S.D. Novel HIV-1 MiRNAs Stimulate TNFα Release in Human Macrophages via TLR8 Signaling Pathway. PLoS ONE 2014, 9, e106006. [Google Scholar] [CrossRef] [PubMed]
- Omoto, S.; Ito, M.; Tsutsumi, Y.; Ichikawa, Y.; Okuyama, H.; Brisibe, E.; Saksena, N.K.; Fujii, Y.R. HIV-1 Nef Suppression by Virally Encoded MicroRNA. Retrovirology 2004, 1, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, D.D.; Vorhies, J.S.; Senzer, N.; Nemunaitis, J. SiRNA vs. ShRNA: Similarities and Differences. Adv. Drug Deliv. Rev. 2009, 61, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhang, J.; Wu, H. Designing Ago2-Specific SiRNA/ShRNA to Avoid Competition with Endogenous MiRNAs. Mol. Ther. Nucleic Acids 2014, 3, e176. [Google Scholar] [CrossRef]
- Guda, S.; Brendel, C.; Renella, R.; Du, P.; Bauer, D.E.; Canver, M.C.; Grenier, J.K.; Grimson, A.W.; Kamran, S.C.; Thornton, J.; et al. MiRNA-Embedded ShRNAs for Lineage-Specific BCL11A Knockdown and Hemoglobin F Induction. Mol. Ther. 2015, 23, 1465–1474. [Google Scholar] [CrossRef] [Green Version]
- Valdmanis, P.N.; Gu, S.; Chu, K.; Jin, L.; Zhang, F.; Munding, E.M.; Zhang, Y.; Huang, Y.; Kutay, H.; Ghoshal, K.; et al. RNA Interference–Induced Hepatotoxicity Results from Loss of the First Synthesized Isoform of MicroRNA-122 in Mice. Nat. Med. 2016, 22, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Course, M.M.; Gudsnuk, K.; Desai, N.; Chamberlain, J.R.; Valdmanis, P.N. Endogenous MicroRNA Competition as a Mechanism of ShRNA-Induced Cardiotoxicity. Mol. Ther. Nucleic Acids 2020, 19, 572–580. [Google Scholar] [CrossRef]
- Alsing, S.; Doktor, T.K.; Askou, A.L.; Jensen, E.G.; Ahmadov, U.; Kristensen, L.S.; Andresen, B.S.; Aagaard, L.; Corydon, T.J. VEGFA-Targeting MiR-AgshRNAs Combine Efficacy with Specificity and Safety for Retinal Gene Therapy. Mol. Ther. Nucleic Acids 2022, 28, 58–76. [Google Scholar] [CrossRef]
- Harwig, A.; Kruize, Z.; Yang, Z.; Restle, T.; Berkhout, B. Analysis of AgoshRNA Maturation and Loading into Ago2. PLoS ONE 2017, 12, e0183269. [Google Scholar] [CrossRef] [Green Version]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. MicroRNAs Associated with the Different Human Argonaute Proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [PubMed]
- Mowa, M.B.; Crowther, C.; Ely, A.; Arbuthnot, P. Inhibition of Hepatitis B Virus Replication by Helper Dependent Adenoviral Vectors Expressing Artificial Anti-HBV Pri-MiRs from a Liver-Specific Promoter. Biomed. Res. Int. 2014, 2014, 718743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drobna-Śledzińska, M.; Maćkowska-Maślak, N.; Jaksik, R.; Dąbek, P.; Witt, M.; Dawidowska, M. CRISPRi for Specific Inhibition of MiRNA Clusters and MiRNAs with High Sequence Homology. Sci. Rep. 2022, 12, 6297. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, G.; Gea-Sorlí, S.; Otero-Mateo, M.; Fillat, C. Inhibition of MiR-222 by Oncolytic Adenovirus-Encoded MiRNA Sponges Promotes Viral Oncolysis and Elicits Antitumor Effects in Pancreatic Cancer Models. Cancers 2021, 13, 3233. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Sakurai, F.; Tomita, K.; Nagamoto, Y.; Nakamura, S.; Katayama, K.; Tachibana, M.; Kawabata, K.; Mizuguchi, H. Suppression of Leaky Expression of Adenovirus Genes by Insertion of MicroRNA-Targeted Sequences in the Replication-Incompetent Adenovirus Vector Genome. Mol. Ther. Methods Clin. Dev. 2014, 1, 14035. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Gironella, M.; Fillat, C. MiR-148a- and MiR-216a-Regulated Oncolytic Adenoviruses Targeting Pancreatic Tumors Attenuate Tissue Damage Without Perturbation of MiRNA Activity. Mol. Ther. 2014, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callegari, E.; Elamin, B.K.; D’Abundo, L.; Falzoni, S.; Donvito, G.; Moshiri, F.; Milazzo, M.; Altavilla, G.; Giacomelli, L.; Fornari, F.; et al. Anti-Tumor Activity of a MiR-199-Dependent Oncolytic Adenovirus. PLoS ONE 2013, 8, e73964. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Guo, G.; Zhang, Q.; Fan, L.; Wu, N.; Bo, Y. The Application of Multiple MiRNA Response Elements Enables Oncolytic Adenoviruses to Possess Specificity to Glioma Cells. Virology 2014, 458–459, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Cawood, R.; Wong, S.-L.; Di, Y.; Baban, D.F.; Seymour, L.W. MicroRNA Controlled Adenovirus Mediates Anti-Cancer Efficacy without Affecting Endogenous MicroRNA Activity. PLoS ONE 2011, 6, e16152. [Google Scholar] [CrossRef]
- Brachtlova, T.; van Ginkel, J.-W.; Luinenburg, M.J.; de Menezes, R.X.; Koppers-Lalic, D.; Pegtel, D.M.; Dong, W.; de Gruijl, T.D.; van Beusechem, V.W. Expression of Oncolytic Adenovirus-Encoded RNAi Molecules Is Most Effective in a Pri-MiRNA Precursor Format. Mol. Ther. Oncolytics 2020, 19, 332–343. [Google Scholar] [CrossRef]
- Boisgerault, F.; Gross, D.-A.; Ferrand, M.; Poupiot, J.; Darocha, S.; Richard, I.; Galy, A. Prolonged Gene Expression in Muscle Is Achieved Without Active Immune Tolerance Using MicrorRNA 142.3p-Regulated RAAV Gene Transfer. Hum. Gene Ther. 2013, 24, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Yuan, Z.; Li, J.; He, B.; Zheng, H.; Mayer, C.; Li, J.; Xiao, X. Liver-Specific MicroRNA-122 Target Sequences Incorporated in AAV Vectors Efficiently Inhibits Transgene Expression in the Liver. Gene Ther. 2011, 18, 403–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- della Peruta, M.; Badar, A.; Rosales, C.; Chokshi, S.; Kia, A.; Nathwani, D.; Galante, E.; Yan, R.; Arstad, E.; Davidoff, A.M.; et al. Preferential Targeting of Disseminated Liver Tumors Using a Recombinant Adeno-Associated Viral Vector. Hum. Gene Ther. 2015, 26, 94–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, A.; Jungmann, A.; Kurreck, J.; Poller, W.; Katus, H.A.; Vetter, R.; Fechner, H.; Müller, O.J. MicroRNA122-Regulated Transgene Expression Increases Specificity of Cardiac Gene Transfer upon Intravenous Delivery of AAV9 Vectors. Gene Ther. 2011, 18, 199–209. [Google Scholar] [CrossRef]
- Geisler, A.; Schön, C.; Größl, T.; Pinkert, S.; Stein, E.A.; Kurreck, J.; Vetter, R.; Fechner, H. Application of Mutated MiR-206 Target Sites Enables Skeletal Muscle-Specific Silencing of Transgene Expression of Cardiotropic AAV9 Vectors. Mol. Ther. 2013, 21, 924–933. [Google Scholar] [CrossRef] [Green Version]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA Sponges: Competitive Inhibitors of Small RNAs in Mammalian Cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target MRNA Abundance Dilutes MicroRNA and SiRNA Activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, C.H.; Lee, S.-W. Targeting the MicroRNA Passenger Strand for Regulating Therapeutic Transgenes. Nucleic Acid Ther. 2015, 25, 209–218. [Google Scholar] [CrossRef]
- Wu, K.; Ye, C.; Lin, L.; Chu, Y.; Ji, M.; Dai, W.; Zeng, X.; Lin, Y. Inhibiting MiR-21 Attenuates Experimental Hepatic Fibrosis by Suppressing Both the ERK1 Pathway in HSC and Hepatocyte EMT. Clin. Sci. 2016, 130, 1469–1480. [Google Scholar] [CrossRef]
- He, X.; Xie, J.; Zhang, D.; Su, Q.; Sai, X.; Bai, R.; Chen, C.; Luo, X.; Gao, G.; Pan, W. Recombinant Adeno-Associated Virus-Mediated Inhibition of MicroRNA-21 Protects Mice against the Lethal Schistosome Infection by Repressing Both IL-13 and Transforming Growth Factor Beta 1 Pathways. Hepatology 2015, 61, 2008–2017. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Su, Y.; Sun, B.; Ji, W.; Peng, Z.; Xu, Y.; Wu, M.; Su, C. An Artificially Designed Interfering LncRNA Expressed by Oncolytic Adenovirus Competitively Consumes OncomiRs to Exert Antitumor Efficacy in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016, 15, 1436–1451. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savenkova, D.A.; Makarova, A.-L.A.; Shalik, I.K.; Yudkin, D.V. miRNA Pathway Alteration in Response to Non-Coding RNA Delivery in Viral Vector-Based Gene Therapy. Int. J. Mol. Sci. 2022, 23, 14954. https://doi.org/10.3390/ijms232314954
Savenkova DA, Makarova A-LA, Shalik IK, Yudkin DV. miRNA Pathway Alteration in Response to Non-Coding RNA Delivery in Viral Vector-Based Gene Therapy. International Journal of Molecular Sciences. 2022; 23(23):14954. https://doi.org/10.3390/ijms232314954
Chicago/Turabian StyleSavenkova, Darya A., Aelita-Luiza A. Makarova, Igor K. Shalik, and Dmitry V. Yudkin. 2022. "miRNA Pathway Alteration in Response to Non-Coding RNA Delivery in Viral Vector-Based Gene Therapy" International Journal of Molecular Sciences 23, no. 23: 14954. https://doi.org/10.3390/ijms232314954





