Understanding Life at High Temperatures: Relationships of Molecular Channels in Enzymes of Methanogenic Archaea and Their Growth Temperatures
Abstract
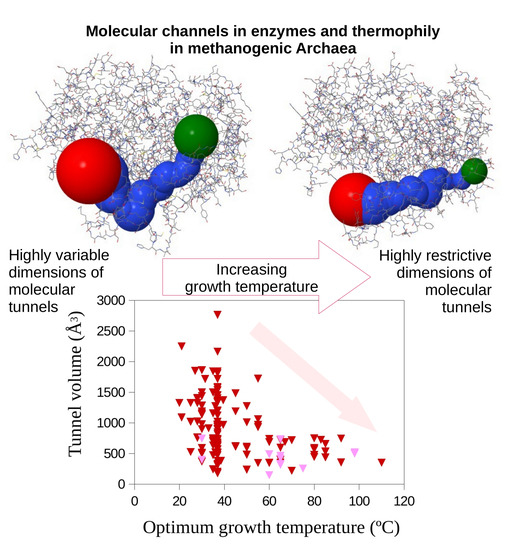
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miles, E.W.; Rhee, S.; Davies, D.R. The molecular basis of substrate channeling. J. Biol. Chem. 1999, 274, 12193–12196. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; McCammon, J.A. The gates of ion channels and enzymes. Trends Biochem. Sci. 2010, 35, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Hubner, C.A.; Jentsch, T.J. Ion channel diseases. Hum. Mol. Genet. 2002, 11, 2435–2445. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Holden, H.M.; Raushel, F.M. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 2001, 70, 149–180. [Google Scholar] [CrossRef]
- Kingsley, L.; Lill, M.A. Substrate tunnels in enzymes: Structure-function relationships and computational methodology. Proteins 2015, 83, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Pravda, L.; Sehnal, D.; Vařeková, R.S.; Navrátilova, V.; Toušek, D.; Berka, K.; Otyepka, M.; Koca, J. ChannelsDB: Database of biomacromolecular tunnels and pores. Nucleic Acids Res. 2018, 46, D399–D405. [Google Scholar] [CrossRef]
- Pravda, L.; Berka, K.; Svobodová Vařeková, R.; Sehnal, D.; Banáš, P.; Laskowski, R.A.; Koča, J.; Otyepka, M. Anatomy of enzyme channels. BMC Bioinform. 2014, 15, 379. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Pesce, A.; Bolognesi, M.; Bocedi, A.; Ascenzi, P. Substrate channeling. Biochem. Mol. Biol. Educ. 2003, 31, 228–233. [Google Scholar] [CrossRef]
- Raushel, F.M.; Thoden, J.B.; Holden, H.M. Enzymes with molecular tunnels. Acc. Chem. Res. 2003, 36, 539–548. [Google Scholar] [CrossRef]
- Furmanová, K.; Vavra, O.; Kozlíková, B.; Damborský, J.; Vonásek, V.; Bednái, D.; Byska, J. DockVis: Visual analysis of molecular docking trajectories. Comput. Graph. Forum 2020, 39, 452–464. [Google Scholar] [CrossRef]
- Vavra, O.; Damborsky, J.; Bednar, D. Fast approximative methods for study of ligand transport and rational design of improved enzymes for biotechnologies. Biotech. Adv. 2022, 60, 108009. [Google Scholar] [CrossRef] [PubMed]
- Cuecas, A.; Cruces, J.; Galisteo-López, J.F.; Peng, X.; Gonzalez, J.M. Cellular viscosity in Prokaryotes and thermal stability of low molecular weight biomolecules. Biophys. J. 2016, 111, 875–882. [Google Scholar] [CrossRef]
- Grogan, D.W. Hyperthermophiles and the problem of DNA instability. Mol. Microbiol. 1998, 28, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2000, 65, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, I.N.; Shakhnovich, E.I. Physics and evolution of thermophilic adaptation. Proc. Natl. Acad. Sci. USA 2005, 102, 12742–12747. [Google Scholar] [CrossRef]
- Daniel, R.M.; Cowan, D.A. Biomolecular stability and life at high temperatures. Cell. Mol. Life Sci. 2000, 57, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A. The upper temperature of life—Where do we draw the line? Trends Microbiol. 2004, 12, 58–60. [Google Scholar] [CrossRef]
- Brock, T.D. Life at high temperatures. Science 1967, 158, 1012–1019. [Google Scholar] [CrossRef]
- Stetter, K.O.; Fiala, G.; Huber, G.; Huber, R.; Segerer, A. Hyperthermophilic microorganisms. FEMS Microbiol. Rev. 1990, 75, 117–124. [Google Scholar] [CrossRef]
- Gonzalez, J.M. Molecular tunnels in enzymes and thermophily: A case study on the relationship to growth temperature. Microorganisms 2018, 6, 109. [Google Scholar] [CrossRef]
- Ferry, J.G. Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr. Opin. Biotechnol. 2011, 22, 351–357. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Stres, B. Trace element enzymes in reactions essential for anaerobic digestion. In Trace Elements in Anaerobic Biotechnologies; Chapter 3; Fermoso, F.G., van Hullebusch, E., Collins, G., Roussel, J., Mucha, A.P., Esposito, G., Eds.; IWA Publishing: London, UK, 2019; pp. 51–72. ISBN 9781789060225. [Google Scholar] [CrossRef]
- Bapteste, E.; Brochier, C.; Boucher, Y. Higher-level classification of the Archaea: Evolution of methanogenesis and methanogens. Archaea 2005, 1, 353–363. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Buongiorno, J.; Bird, J.; Krivushin, K.; Spirina, E.V.; Oshurkova, V.; Shcherbakova, V.A.; Wilson, G.; Lloyd, K.G.; Rivkina, E.M. Methanogens in the Antartic dry Valley permafrost. FEMS Microbiol. Ecol. 2018, 94, fiy109. [Google Scholar] [CrossRef]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell proliferation at 122 °C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Robb, F.T.; Adams, M.W. Purification and characterization of NADP-specific alcohol dehydrogenase and glutamate dehydrogenase from the hyperthermophilic archaeon Thermococcus litoralis. Appl. Environ. Microbiol. 1994, 60, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Britton, K.L.; Yip, K.S.; Sedelnikova, S.E.; Stillman, T.J.; Adams, M.W.; Ma, K.; Maeder, D.L.; Robb, F.T.; Tolliday, N.; Vetriani, C.; et al. Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus. J. Mol. Biol. 1999, 12, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Enzymology of one carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 1999, 23, 13–38. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Petřek, M.; Košinová, P.; Koča, J.; Otyepka, M. MOLE: A voronoi diagrama-based explorer of molecular channels, pores, and tunnels. Structure 2007, 15, 1357–1363. [Google Scholar] [CrossRef]
- Masood, T.B.; Sandhya, S.; Chandra, N.; Natarajan, V. CHEXVIS: A tool for molecular channel extraction and visualization. BMC Bioinform. 2015, 16, 119. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. The Principles and Practice of Statistics in Biological Research, 7th ed.; W.H. Freeman and Co.: New York, NY, USA, 2001. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ginsbach, L.F.; Gonzalez, J.M. Understanding Life at High Temperatures: Relationships of Molecular Channels in Enzymes of Methanogenic Archaea and Their Growth Temperatures. Int. J. Mol. Sci. 2022, 23, 15149. https://doi.org/10.3390/ijms232315149
Ginsbach LF, Gonzalez JM. Understanding Life at High Temperatures: Relationships of Molecular Channels in Enzymes of Methanogenic Archaea and Their Growth Temperatures. International Journal of Molecular Sciences. 2022; 23(23):15149. https://doi.org/10.3390/ijms232315149
Chicago/Turabian StyleGinsbach, Laura F., and Juan M. Gonzalez. 2022. "Understanding Life at High Temperatures: Relationships of Molecular Channels in Enzymes of Methanogenic Archaea and Their Growth Temperatures" International Journal of Molecular Sciences 23, no. 23: 15149. https://doi.org/10.3390/ijms232315149
APA StyleGinsbach, L. F., & Gonzalez, J. M. (2022). Understanding Life at High Temperatures: Relationships of Molecular Channels in Enzymes of Methanogenic Archaea and Their Growth Temperatures. International Journal of Molecular Sciences, 23(23), 15149. https://doi.org/10.3390/ijms232315149