Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea
Abstract
1. Introduction
2. Results and Discussion
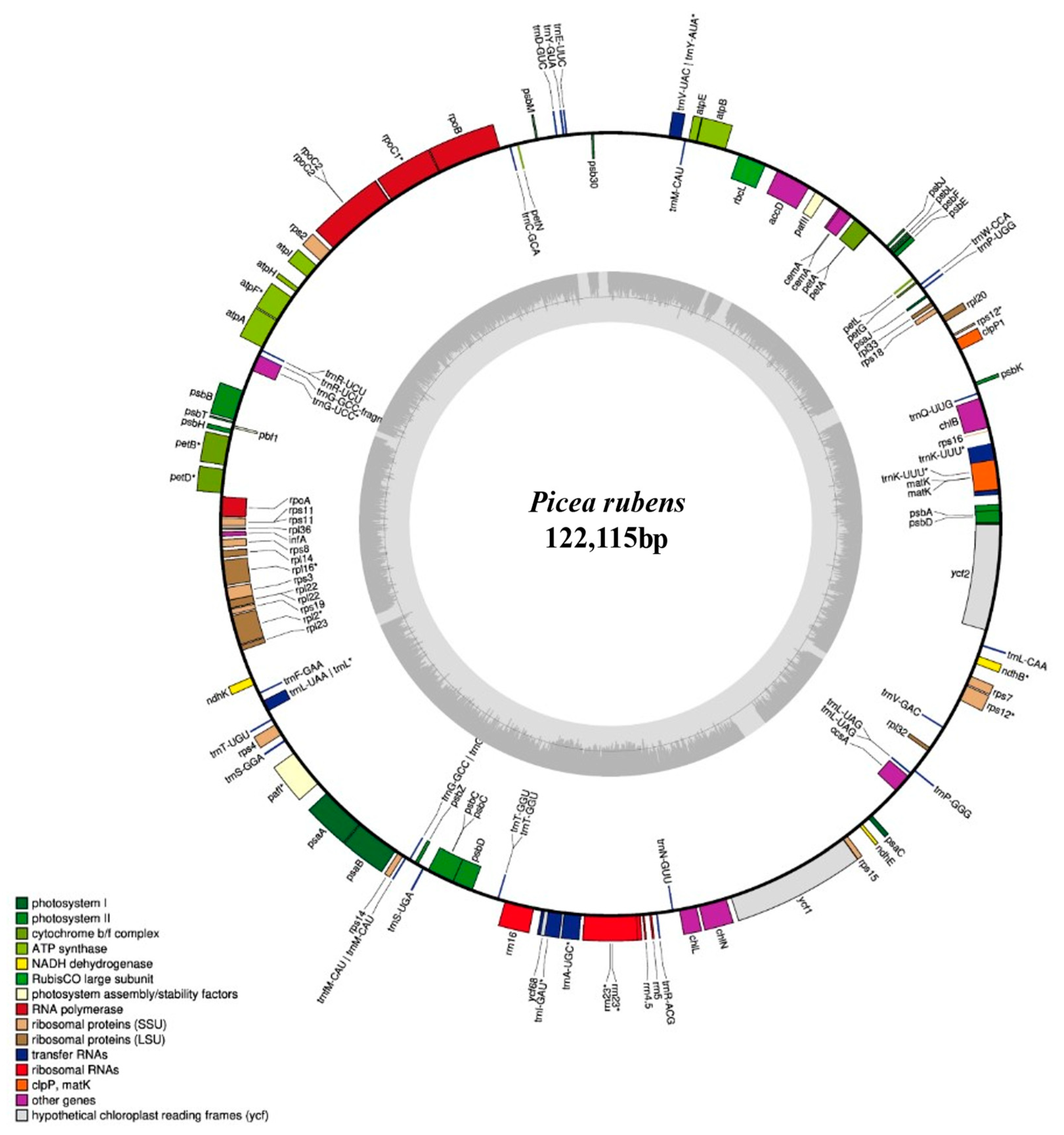
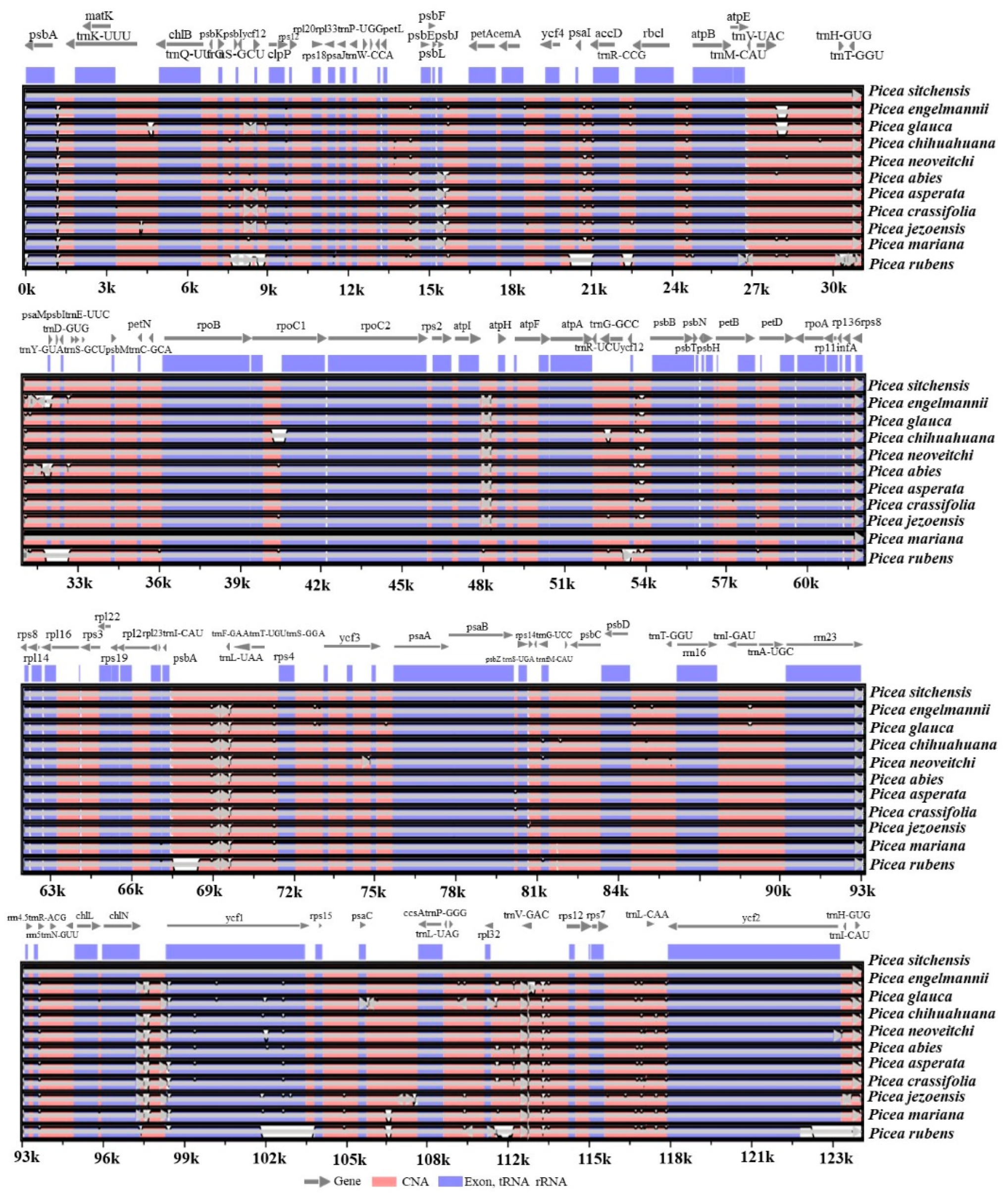
2.1. Chloroplast Genome Features and Gene Content
2.2. SSR Identification and Primer Designing
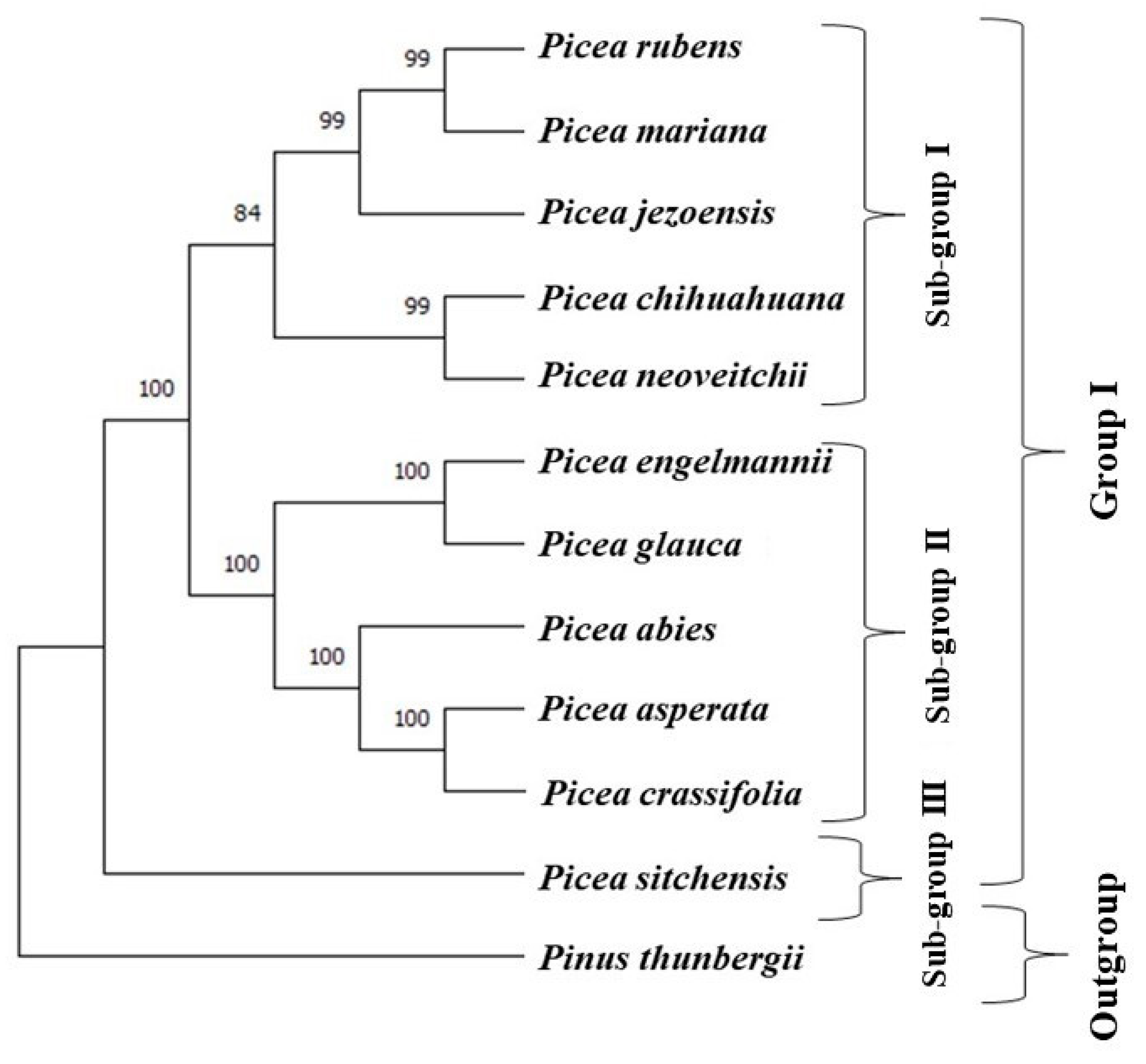
2.3. Phylogenetic Analysis
3. Materials and Methods
3.1. Chloroplast Isolation and DNA Extraction
3.2. Library Preparation and Sequencing
3.3. Chloroplast Genome Assembly, Annotation, and Sequence Architecture
3.4. Sequence Divergence and Phylogenetic Analysis
3.5. SSR Mining and Primer Designing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajora, O.P.; Dancik, B.P. Chloroplast DNA inheritance in Populus. Theor. Appl. Genet. 1992, 84, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.B.; Furnier, G.R.; Saghai-Maroof, M.A.; Williams, S.M.; Dancik, B.P.; Allard, R.W. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc. Natl. Acad. Sci. USA 1987, 84, 2097–2100. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. Chloroplast DNA and molecular phylogeny. Bioessays 1985, 2, 263–267. [Google Scholar] [CrossRef]
- Mereschkowsky, C. Uber natur und ursprung der chromatophoren im pflanzenreiche. Biol. Cent. 1905, 25, 293–604. [Google Scholar]
- Mereschkowsky, C. Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen. Biol. Cent. 1910, 30, 278–288. [Google Scholar]
- Mereschkowsky, C. La plante considérée comme un complexe symbiotique. Bull. Soc. Sci. Nat. Fr. 1920, 6, 17. [Google Scholar]
- Kutschera, U.; Niklas, K.J. Endosymbiosis, cell evolution, and speciation. Theory Biosci. 2005, 124, 1–24. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Zoschke, R.; Bock, R. Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Hosie, R.C. Native Trees of Canada; Fitzhenry and Whiteside: Toronto, ON, Canada, 1979. [Google Scholar]
- Burns, R.M.; Honkala, B.H. (Eds.) Silvics of North America. 1. Conifers. Agricultural Handbook 654; USDA Forest Service: Washington, DC, USA, 1990.
- Honkaniemi, J.; Rammer, W.; Seidl, R. Norway spruce at the trailing edge: The effect of landscape configuration and composition on climate resilience. Landsc. Ecol. 2020, 35, 591–606. [Google Scholar] [CrossRef]
- Lockwood, J.D.; Aleksić, J.M.; Zou, J.; Wang, J.; Liu, J.; Renner, S.S. A new phylogeny for the genus Picea from plastid, mitochondrial, and nuclear sequences. Mol. Phylogenetics Evol. 2013, 69, 717–727. [Google Scholar] [CrossRef]
- Sullivan, A.R.; Schiffthaler, B.; Thompson, S.L.; Street, N.R.; Wang, X.R. Interspecific plastome recombination reflects ancient reticulate evolution in Picea (Pinaceae). Mol. Biol. Evol. 2017, 34, 1689–1701. [Google Scholar] [CrossRef]
- Ran, J.H.; Wei, X.X.; Wang, X.Q. Molecular phylogeny and biogeography of Picea (Pinaceae): Implications for phylogeographical studies using cytoplasmic haplotypes. Mol. Phylogenetics Evol. 2006, 41, 405–419. [Google Scholar] [CrossRef]
- Lo, T.; Ouyang, L.; Lin, D.; Warren, R.L.; Kirk, H.; Pandoh, P.; Birol, I. Complete chloroplast genome sequence of a black spruce (Picea mariana) from Eastern Canada. Microbiol. Resour. Announc. 2020, 9, e00877-20. [Google Scholar] [CrossRef]
- Lin, D.; Coombe, L.; Jackman, S.D.; Gagalova, K.K.; Warren, R.L.; Hammond, S.A.; Birol, I. Complete chloroplast genome sequence of a white spruce (Picea glauca, genotype ws 77111) from Eastern Canada. Microbiol. Resour. Announc. 2019, 8, e00381-19. [Google Scholar] [CrossRef]
- Coombe, L.; Warren, R.L.; Jackman, S.D.; Yang, C.; Vandervalk, B.P.; Moore, R.A.; Birol, I. Assembly of the complete Sitka spruce chloroplast genome using 10X Genomics’ GemCode sequencing data. PLoS ONE 2016, 11, e0163059. [Google Scholar] [CrossRef]
- Lin, D.; Coombe, L.; Jackman, S.D.; Gagalova, K.K.; Warren, R.L.; Hammond, S.A.; Birol, I. Complete chloroplast genome sequence of an Engelmann spruce (Picea engelmannii, genotype Se404-851) from western Canada. Microbiol. Resour. Announc. 2019, 8, e00382-19. [Google Scholar] [CrossRef]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Blum, B.M. Picea rubens Sarg. Red spruce. In Silvics of North America. 1. Conifers. Agricultural Handbook 654; Burns, R.M., Honkala, B.H., Eds.; USDA Forest Service: Washington, DC, USA, 1990; pp. 250–259. [Google Scholar]
- Rajora, O.P.; Mosseler, A.; Major, J.E. Indicators of population viability in red spruce, Picea rubens. II. Genetic diversity, population structure, and mating behavior. Can. J. Bot. 2000, 78, 941–956. [Google Scholar]
- DeHayes, D.H.; Hawley, G.J. Genetic implications in the decline of red spruce. Water Air Soil Pollut. 1992, 62, 233–248. [Google Scholar] [CrossRef]
- McLaughlin, S.B.; Downing, D.J.; Blasing, T.J.; Cook, E.R.; Adams, H.S. An analysis of climate and competition as contributors to decline of red spruce in high elevation Appalachian forests of the eastern United States. Oecologia 1987, 72, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Bashalkhanov, S.; Eckert, A.J.; Rajora, O.P. Genetic signatures of selection in response to air pollution in red spruce (Picea rubens, Pinaceae). Mol. Ecol. 2013, 22, 5877–5889. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, E.K.; Farrar, J.L. Introgressive Hybridization in Red Spruce and Black Spruce; Technical Report 4; Faculty of Forestry, University of Toronto: Toronto, ON, Canada, 1964. [Google Scholar]
- Manley, S.A.M. The occurrence of hybrid swarms of red and black spruces in central New Brunswick. Can. J. For. Res. 1972, 2, 381–391. [Google Scholar] [CrossRef]
- Gordon, A.G. The taxonomy and genetics of Picea rubens and its relationship to Picea mariana. Can. J. Bot. 1976, 54, 781–813. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Ouyang, F.; Hu, J.; Wang, J.; Ling, J.; Wang, Z.; Wang, N.; Wang, J. Complete plastome sequences of Picea asperata and P crassifolia and comparative analyses with P. abies and P. morrisonicola. Genome 2019, 62, 317–328. [Google Scholar]
- Yang, J.C.; Joo, M.; So, S.; Yi, D.K.; Shin, C.H.; Lee, Y.M.; Choi, K. The complete plastid genome sequence of Picea jezoensis (Pinaceae: Piceoideae). Mitochondrial DNA Part A 2016, 27, 3761–3763. [Google Scholar] [CrossRef]
- Perron, M.; Bousquet, J. Natural hybridization between black spruce and red spruce. Mol. Ecol. 1997, 6, 725–734. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.P.; Bousquet, J. New evidence from mitochondrial DNA of a progenitor-derivative species relationship between black spruce and red spruce (Pinaceae). Am. J. Bot. 2003, 90, 1801–1806. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35 (Suppl. 2), W71–W74. [Google Scholar] [CrossRef]
- Tsudzuki, J.; Nakashima, K.; Tsudzuki, T.; Hiratsuka, J.; Shibata, M.; Wakasugi, T.; Sugiura, M. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: Nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol. Gen. Genet. 1992, 232, 206–214. [Google Scholar] [CrossRef]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef]
- Gugerli, F.; Sperisen, C.; Büchler, U.; Brunner, I.; Brodbeck, S.; Palmer, J.D.; Qiu, Y.L. The evolutionary split of Pinaceae from other conifers: Evidence from an intron loss and a multigene phylogeny. Mol. Phylogenetics Evol. 2001, 21, 167–175. [Google Scholar] [CrossRef]
- Chaw, S.M.; Parkinson, C.L.; Cheng, Y.; Vincent, T.M.; Palmer, J.D. Seed plant phylogeny inferred from all three plant genomes: Monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc. Natl. Acad. Sci. USA 2000, 97, 4086–4091. [Google Scholar] [CrossRef]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef]
- Wu, C.S.; Lin, C.P.; Hsu, C.Y.; Wang, R.J.; Chaw, S.M. Comparative chloroplast genomes of Pinaceae: Insights into the mechanism of diversified genomic organizations. Genome Biol. Evol. 2011, 3, 309–319. [Google Scholar] [CrossRef]
- Ranade, S.S.; Garcia-Gil, M.R.; Rossello, J.A. Non-functional plastid ndh gene fragments are present in the nuclear genome of Norway spruce (Picea abies L. Karsch.): Insights from in silico analysis of nuclear and organellar genomes. Mol. Genet. Genomes 2016, 291, 935–941. [Google Scholar] [CrossRef]
- Ni, Z.; Ye, Y.; Bai, T.; Xu, M.; Xu, L.A. Complete chloroplast genome of Pinus massoniana (Pinaceae): Gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, G.G.; Lelli, L.; Rossi, P.; Morgante, M. A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol. Ecol. 1996, 5, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Kim, Y.; Maunder, M.; Li, X. The conservation status and conservation strategy of Picea neoveitchii. Chin. J. Popul. Resour. Environ. 2006, 4, 58–64. [Google Scholar]
- Coombe, L.; Nikolić, V.; Chu, J.; Birol, I.; Warren, R.L. ntJoin: Fast and lightweight assembly-guided scaffolding using minimizer graphs. Bioinformatics 2020, 36, 3885–3887. [Google Scholar] [CrossRef] [PubMed]
- Bouillé, M.; Senneville, S.; Bousquet, J. Discordant mtDNA and cpDNA phylogenies indicate geographic speciation and reticulation as driving factors for the diversification of the genus Picea. Tree Genet. Genomes 2011, 7, 469–484. [Google Scholar] [CrossRef]
- Rajora, O.P.; Dancik, B.P. Population genetic variation, structure, and evolution in Engelmann spruce, white spruce, and their natural hybrid complex in Alberta. Can. J. Bot. 2000, 78, 768–780. [Google Scholar]
- Sigurgeirsson, A.; Szmidt, A.E. Phylogenetic and biogeographic implications of chloroplast DNA variation in Picea. Nord. J. Bot. 1993, 13, 233–246. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 9, 11–15. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 October 2022).
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; Wang, J. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Paulino, D.; Warren, R.L.; Vandervalk, B.P.; Raymond, A.; Jackman, S.D.; Birol, I. Sealer: A scalable gap-closing application for finishing draft genomes. BMC Bioinform. 2015, 16, 230. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32 (Suppl. 2), W273–W279. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]


| Picea sitchensis | Picea engelmannii | Picea glauca | Picea chihuahuana | Picea neoveitchii | Picea abies | Picea asperata | Picea crassifolia | Picea jezoensis | Picea mariana | |
|---|---|---|---|---|---|---|---|---|---|---|
| Misassembled contigs length | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Local misassemblies | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 |
| Mismatches | 598 | 368 | 368 | 337 | 279 | 270 | 278 | 278 | 131 | 122 |
| Indels | 105 | 66 | 72 | 58 | 56 | 72 | 64 | 64 | 36 | 31 |
| Indels (<=5 bp) | 73 | 52 | 55 | 49 | 44 | 60 | 50 | 50 | 28 | 27 |
| Indels (>5 bp) | 32 | 14 | 17 | 9 | 12 | 12 | 14 | 14 | 8 | 4 |
| Indels length (bp) | 632 | 493 | 527 | 398 | 238 | 331 | 323 | 321 | 250 | 202 |
| Functional Component | Genes |
|---|---|
| Photosystem I | psaA, psaB, psaC, and psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbM, psbT, psbZ, and ycf12 (psb30) |
| Large ribosomal subunit | rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, and rpl36 |
| Small ribosomal subunits | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps18, and rps19 |
| Subunits of cytochrome b/f complex | petA, petB, petD, petG, petL, and petN |
| ATP synthase (subunits) | atpA, atpB, atpE, atpF, atpH, and atpI |
| RNA polymerase | rpoA, rpoB, rpoC1, and rpoC2 |
| Chlorophyll biosynthesis genes | chlB, chlN, and chlL |
| Protease | clpP |
| Maturase | matK |
| Envelope membrane protein | cemA |
| Translation initiation factor | infA |
| Cytochrome c biogenesis | ccsA |
| Subunit Acetyl-CoA-Carboxylate | accD |
| Subunit of Rubisco | rbcL |
| Hypothetical open reading frames | pafI (ycf3), pafII (ycf4), ycf1, ycf2, and ycf68 |
| Ribosomal RNAs | rrn4.5, rrn5, rrn16, and rrn23 |
| Transfer RNA | trnV-UAC/trnY-AUA, trnM-CAU, trnW-CCA, trnP-UGG, trnQ-UUG, trnK-UUU, trnL-CAA, trnV-GAC, trnP-GGG, trnL-UAG, trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnT-GGU, trnS-UGA, trnG-GCC, trnF-CAU/trnM-CAU, trnS-GGA, trnT-UGU, trnL/trnL-UAA/UAG, trnF-GAA, trnG-GCC, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, and trnE-UUC |
| Repeats | Total Number Identified |
|---|---|
| A/T | 25 |
| C/G | 2 |
| AG/CT | 1 |
| AT/AT | 9 |
| AAT/ATT | 1 |
| AAAG/CTTT | 1 |
| ACCT/AGGT | 1 |
| ATCC/ATGG | 1 |
| AAAATG/ATTTTC | 1 |
| Locus | Product Size (bp) | Type of Repeat | Length | Tm | Orientation | Primer Sequence (5′-3′) |
|---|---|---|---|---|---|---|
| RPRSCP1 | 167 | (A)13 | 20 | 55.01 | Forward | ATCGGAAGATCCTCTTTTTC |
| 20 | 54.95 | Reverse | AGCTGTATTGTATGCGGAAT | |||
| RPRSCP2 | 176 | (TA)8 | 20 | 54.15 | Forward | TAAGGTGGTAACTCCCATTC |
| 20 | 54.73 | Reverse | AACAAGAGGATTGGTTCTCA | |||
| RPRSCP3 | 241 | (TA)5 | 20 | 54.90 | Forward | GTTAATGAAAGAGCCCAATG |
| 20 | 54.62 | Reverse | CCATCGATCTTGATAAGGAC | |||
| RPRSCP4 | 229 | (T)13 | 20 | 55.12 | Forward | GAAGTATCTGTCCGATCCAA |
| 20 | 54.35 | Reverse | GTTCCGAACTAGACGATGTT | |||
| RPRSCP5 | 250 | (TA)5 | 20 | 56.03 | Forward | ACAGAATCGTGGTGAATCAG |
| 20 | 54.91 | Reverse | GGATAGCGAGTATTGTCCAG | |||
| RPRSCP6 | 194 | (AT)7 | 20 | 54.94 | Forward | GTCTCTCTTCAGAGCGAAAA |
| 20 | 55.00 | Reverse | GTACCCCGTGATCTCAATAA | |||
| RPRSCP7 | 163 | (AT)5 | 20 | 55.02 | Forward | GTAAACCAAGAAGCCCCTAT |
| 20 | 55.02 | Reverse | CTTCTTCCATTTCTCGATTG | |||
| RPRSCP8 | 202 | (CT)7 | 20 | 54.97 | Forward | CAGGAAAAAGAGCTGAAGAA |
| 20 | 55.05 | Reverse | AGGGTAGATCGGGATAATGT | |||
| RPRSCP9 | 231 | (A)11 | 20 | 55.03 | Forward | CCAATCCAATGTGAGAAAGT |
| 20 | 54.95 | Reverse | CATTGGATCAAGAACAGGAT | |||
| RPRSCP10 | 207 | (T)15 | 20 | 54.44 | Forward | TTTCCTTAGTTTCCATCGAC |
| 20 | 54.40 | Reverse | CGAGAAAGGTGTTTGGTAAT | |||
| RPRSCP11 | 236 | (T)14 | 20 | 55.06 | Forward | CATTGCAGGTACAATGACAG |
| 20 | 54.89 | Reverse | TCGGAAGAGGAATAGGTACA | |||
| RPRSCP12 | 245 | (T)14 | 20 | 55.07 | Forward | CAGAGGTCAATTTCTTCTGC |
| 20 | 54.82 | Reverse | GAAAAAGGAGGAAAGAGAGG | |||
| RPRSCP13 | 213 | (T)16 | 20 | 54.79 | Forward | GATGGCTAGAGATTCATTGG |
| 20 | 55.23 | Reverse | ATTGAGCTGACATCCGTTAC | |||
| RPRSCP14 | 234 | (T)12 | 20 | 54.96 | Forward | AACAGGTATGGTTGGTATCG |
| 20 | 55.21 | Reverse | AGCCGAGCTATTCTCTTTTT | |||
| RPRSCP15 | 360 | (C)10 | 20 | 55.03 | Forward | TATCTGATCCTCGAATCACC |
| 20 | 55.11 | Reverse | ATCGGACCACGATGTAGTAG | |||
| RPRSCP16 | 214 | (T)13 | 20 | 54.57 | Forward | GTGATCCAAAAGTGAAAACC |
| 20 | 55.43 | Reverse | CGAATTACGGACAACCTAAA | |||
| RPRSCP17 | 229 | (AGGT)3 | 19 | 54.89 | Forward | TGAAGTAACCCATGCCATA |
| 20 | 55.12 | Reverse | GGAGACCTGTGTTTTTGGTA | |||
| RPRSCP18 | 234 | (TAT)4 | 20 | 55.03 | Forward | ACACCCCACCCTAGAGTTAT |
| 20 | 55.33 | Reverse | GGGCGACTGAGATATTACAA | |||
| RPRSCP19 | 249 | (AT)4 | 22 | 50.33 | Forward | CTCCTAGATAAGCTAACAGAGA |
| 20 | 56.33 | Reverse | TCGAAACTCCTTGTTGATTG | |||
| RPRSCP20 | 360 | (ATGAA)3 | 20 | 55.85 | Forward | ACATCGGTGACAAAGATGAC |
| 20 | 55.16 | Reverse | GTTCTTCTTTCGGAAGTCCT | |||
| RPRSCP21 | 221 | (T)13 | 20 | 55.78 | Forward | CGCAGTATGGGTCTAGCTTA |
| 20 | 54.92 | Reverse | GCAGATATGGGCAAACTAAC | |||
| RPRSCP22 | 229 | (AT)10 | 20 | 53.61 | Forward | TCCTTTTCCGTATACTTTCC |
| 20 | 54.93 | Reverse | CGGGTTAATGTGAGCTTATC | |||
| RPRSCP23 | 239 | (AAAG)3 | 20 | 55.12 | Forward | AGGTTCGAGTCAAATAGCAA |
| 20 | 55.37 | Reverse | AACCGTACATACGACTTTCG | |||
| RPRSCP24 | 229 | (T1)12 | 20 | 55.17 | Forward | GGACATGTGGAAAAGAGAAA |
| 20 | 55.37 | Reverse | GCGCATGTATAAGACCAAAT | |||
| RPRSCP25 | 177 | (AT)8 | 21 | 55.12 | Forward | CGATATCAATACTCGAAGACG |
| 20 | 54.75 | Reverse | TGTCTACCATTTCACCATCA | |||
| RPRSCP26 | 210 | (T)12 | 20 | 55.87 | Forward | GATCTCGGAGTGAAGAACCT |
| 20 | 54.81 | Reverse | GAAAGAGCAATGGAATATGG |
| Statistics | Picea sitchensis | Picea engelmannii | Picea glauca | Picea chihuahuana | Picea neoveitchii | Picea abies | Picea asperata | Picea crassifolia | Picea jezoensis | Picea mariana | Picea rubens |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total size of examined sequences (bp) | 124,049 | 123,542 | 123,421 | 123,488 | 124,234 | 124,084 | 124,145 | 124,126 | 124,146 | 123,961 | 122,115 |
| Total number of identified SSRs | 40 | 39 | 40 | 45 | 45 | 37 | 49 | 46 | 49 | 48 | 42 |
| Number of SSRs present in compound formation | 6 | 4 | 5 | 7 | 7 | 7 | 9 | 7 | 10 | 9 | 8 |
| Number of sequences containing more than 1 SSR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parmar, R.; Cattonaro, F.; Phillips, C.; Vassiliev, S.; Morgante, M.; Rajora, O.P. Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea. Int. J. Mol. Sci. 2022, 23, 15243. https://doi.org/10.3390/ijms232315243
Parmar R, Cattonaro F, Phillips C, Vassiliev S, Morgante M, Rajora OP. Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea. International Journal of Molecular Sciences. 2022; 23(23):15243. https://doi.org/10.3390/ijms232315243
Chicago/Turabian StyleParmar, Rajni, Federica Cattonaro, Carrie Phillips, Serguei Vassiliev, Michele Morgante, and Om P. Rajora. 2022. "Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea" International Journal of Molecular Sciences 23, no. 23: 15243. https://doi.org/10.3390/ijms232315243
APA StyleParmar, R., Cattonaro, F., Phillips, C., Vassiliev, S., Morgante, M., & Rajora, O. P. (2022). Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea. International Journal of Molecular Sciences, 23(23), 15243. https://doi.org/10.3390/ijms232315243







