Genome-Wide Analysis and Functional Characterization of Pyruvate Kinase (PK) Gene Family Modulating Rice Yield and Quality
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of 10 PK Genes in Rice Genome
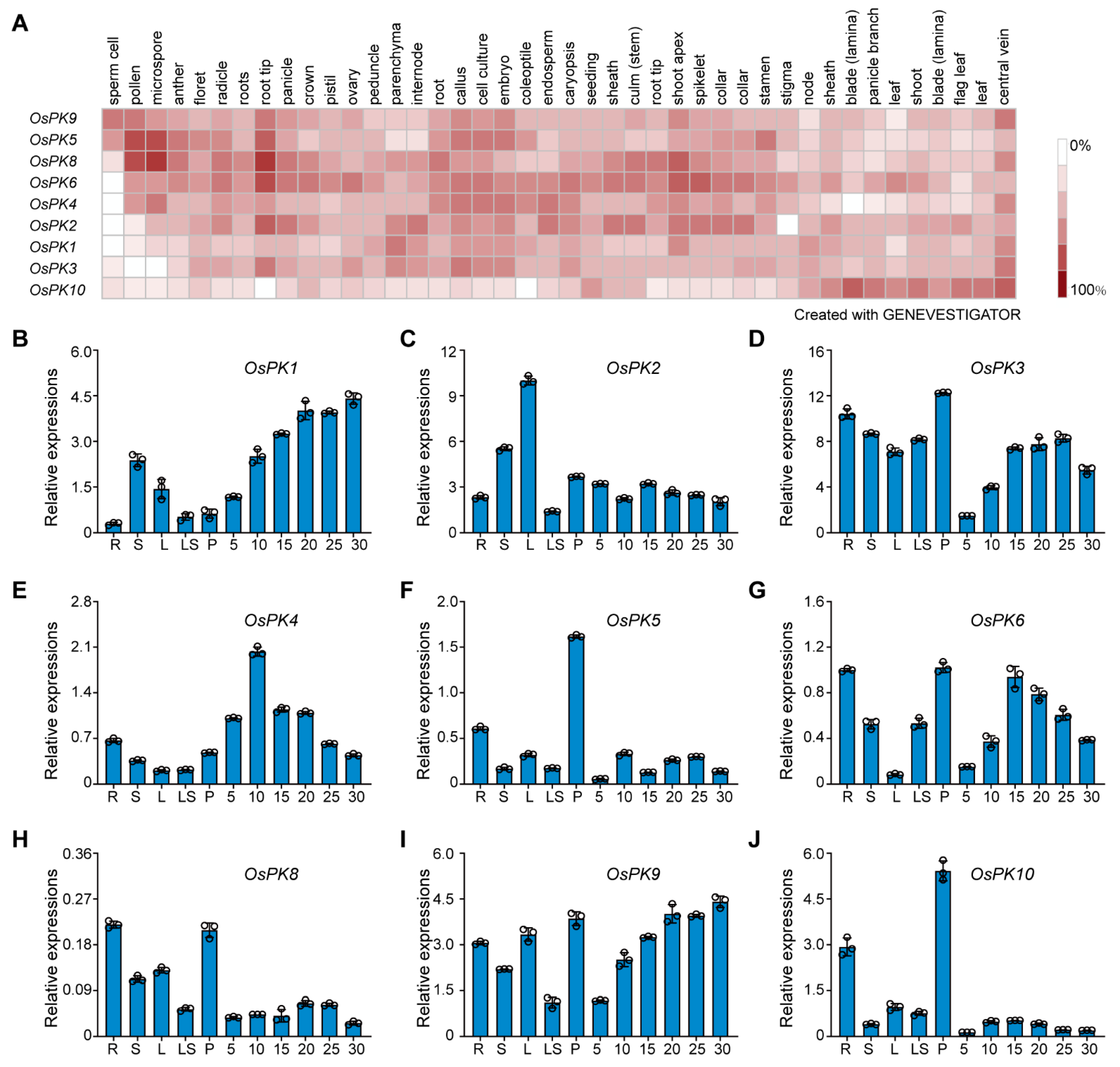
2.2. Temporal and Spatial Expression Pattern and Subcellular Localization of Rice Pks
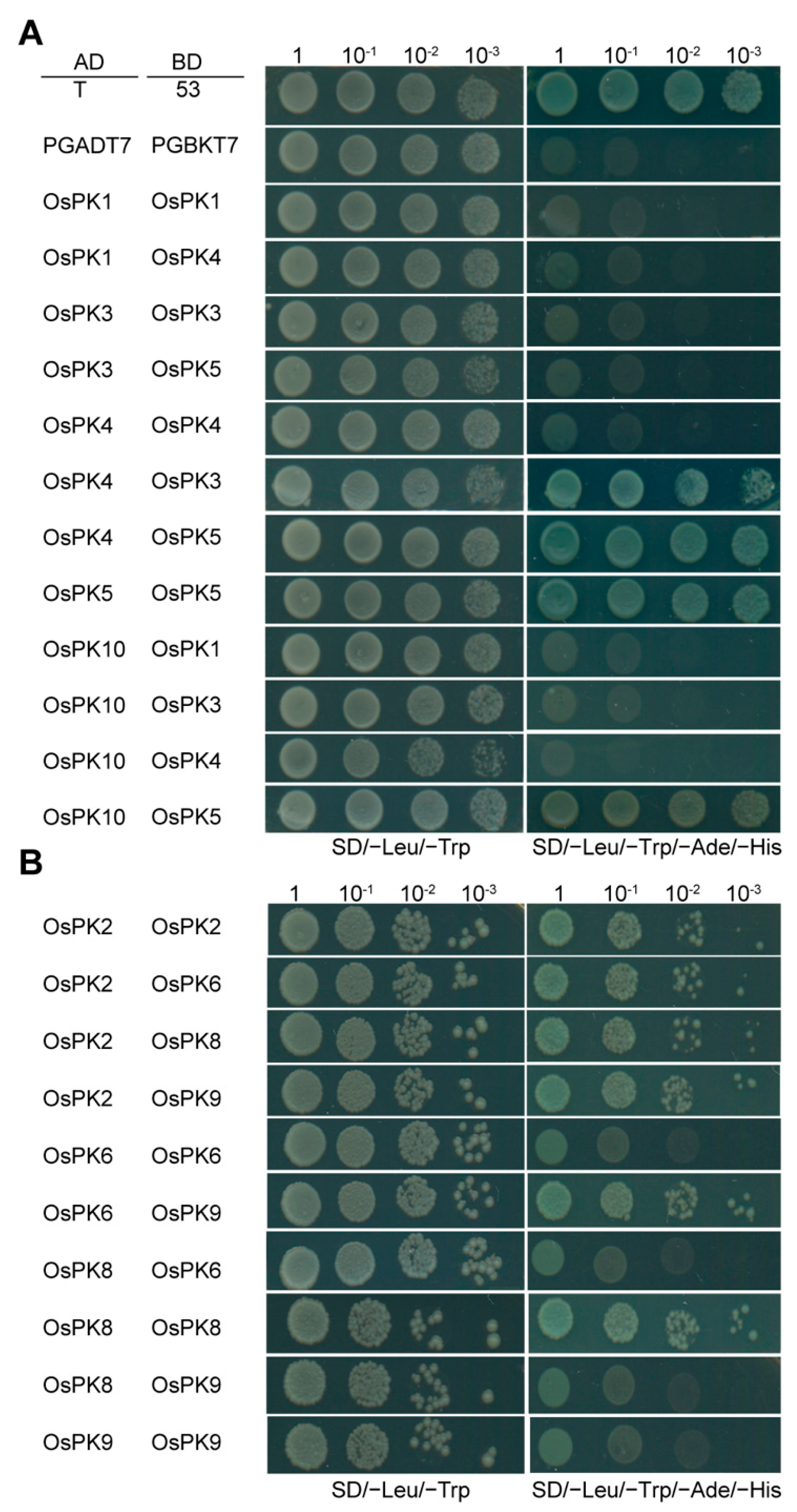
2.3. PK Proteins of Rice Form Heteropolymers
2.4. Promoter Cis-Regulatory Element (CRE) Analysis and the Response to Abscisic Acid of PK Genes in Rice
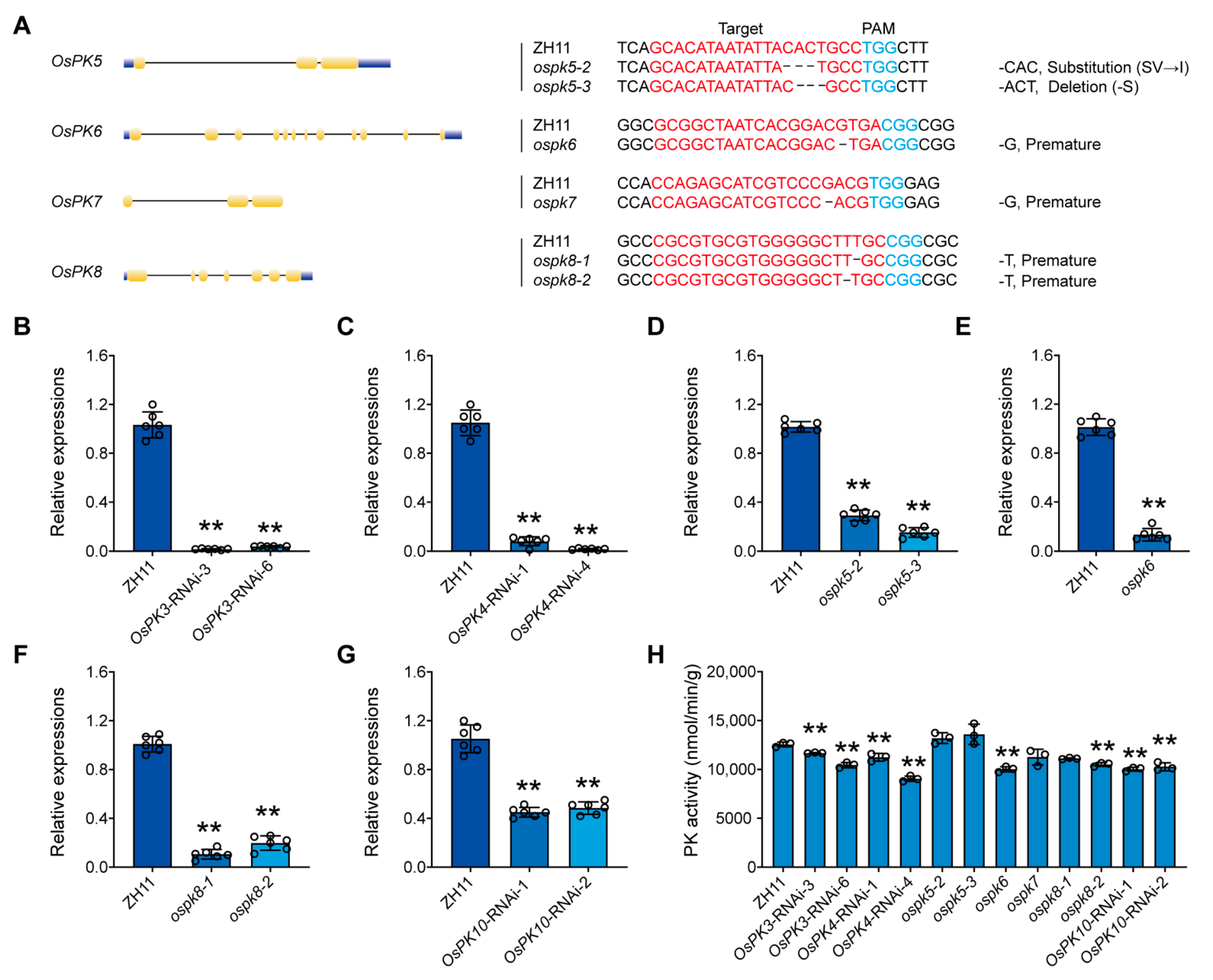
2.5. Creation of CRISPR Mutants and Rnai Lines of PK Genes in Rice
2.6. PK Modulates the Formation of Rice Yield and Quality
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of PKs
4.3. Multiple Sequence Alignment, Phylogenetic Analysis, Motifs, and Gene Structure Analysis
4.4. Expression Pattern of PK Family Genes in Rice
4.5. Subcellular Localization
4.6. Yeast Two-Hybrid Assays
4.7. Identification of Cis-Regulatory Elements in the Promoter Region
4.8. Identification of the Positions of the Genes on the Chromosome
4.9. Measurement of PK Enzyme Activity
4.10. Determination of Total Starch and Protein Contents in Endosperm
4.11. Seed Germination Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Munoz, M.E.; Ponce, E. Pyruvate kinase: Current status of regulatory and functional properties. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 135, 197–218. [Google Scholar] [CrossRef]
- Ambasht, P.K.; Kayastha, A.M. Plant pyruvate kinase. Biol. Plantarum. 2002, 45, 1–10. [Google Scholar] [CrossRef]
- Yang, B.; Chen, M.; Zhan, C.; Liu, K.; Cheng, Y.; Xie, T.; Zhu, P.; He, Y.; Zeng, P.; Tang, H.; et al. Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via GWAS. J. Exp. Bot. 2022, 73, 3446–3461. [Google Scholar] [CrossRef]
- Mierzejewska, J.; Chreptowicz, K. Lack of Maf1 enhances pyruvate kinase activity and fermentative metabolism while influencing lipid homeostasis in Saccharomyces cerevisiae. FEBS Lett. 2016, 590, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, S.N.; Lunn, J.E.; Urbanczyk-Wochniak, E.; Lytovchenko, A.; van Dongen, J.T.; Faix, B.; Schmalzlin, E.; Fernie, A.R.; Geigenberger, P. Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol. 2008, 148, 1640–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenthaler, H.K. The 1-Deoxy-D-Xylulose-5-Phosphate Pathway of Isoprenoid Biosynthesis in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Le, X.H.; Lee, C.P.; Millar, A.H. The mitochondrial pyruvate carrier (MPC) complex mediates one of three pyruvate-supplying pathways that sustain Arabidopsis respiratory metabolism. Plant Cell 2021, 33, 2776–2793. [Google Scholar] [CrossRef]
- Mattevi, A.; Bolognesi, M.; Valentini, G. The allosteric regulation of pyruvate kinase. FEBS Lett. 1996, 389, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Kayne, F.J. Thallium (I) activation of pyruvate kinase. Arch. Biochem. Biophys. 1971, 143, 232–239. [Google Scholar] [CrossRef]
- D’Auria, S.; Rossi, M.; Herman, P.; Lakowicz, J.R. Pyruvate kinase from the thermophilic eubacterium Bacillus acidocaldarius as probe to monitor the sodium concentrations in the blood. Biophys. Chem. 2000, 84, 167–176. [Google Scholar] [CrossRef]
- Waygood, E.B.; Sanwal, B.D. The control of pyruvate kinases of Escherichia coli. I. Physicochemical and regulatory properties of the enzyme activated by fructose 1,6-diphosphate. J. Biol. Chem. 1974, 249, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Shang, X.L.; Zhang, Y.; Liu, S.W.; Liang, Y.; Zhang, Y.; Wen, T.Y. A novel pyruvate kinase and its application in lactic acid production under oxygen deprivation in Corynebacterium glutamicum. BMC Biotechnol. 2016, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Abdelhamid, Y.; Brear, P.; Greenhalgh, J.; Chee, X.; Rahman, T.; Welch, M. Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA from Pseudomonas aeruginosa. J. Biol. Chem. 2019, 294, 15505–15516. [Google Scholar] [CrossRef] [PubMed]
- Muirhead, H.; Clayden, D.A.; Barford, D.; Lorimer, C.G.; Fothergill-Gilmore, L.A.; Schiltz, E.; Schmitt, W. The structure of cat muscle pyruvate kinase. EMBO J. 1986, 5, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Liu, J.Y. Cotton cytosolic pyruvate kinase GhPK6 participates in fast fiber elongation regulation in a ROS-mediated manner. Planta 2016, 244, 915–926. [Google Scholar] [CrossRef]
- Plaxton, W.C.; Dennis, D.T.; Knowles, V.L. Purification of leucoplast pyruvate kinase from developing castor bean endosperm. Plant Physiol. 1990, 94, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Negm, F.B.; Cornel, F.A.; Plaxton, W.C. Suborganellar Localization and Molecular Characterization of Nonproteolytic Degraded Leukoplastid Pyruvate-Kinase from Developing Castor-Oil Seeds. Plant Physiol. 1995, 109, 1461–1469. [Google Scholar] [CrossRef] [Green Version]
- Plaxton, W.C.; Smith, C.R.; Knowles, V.L. Molecular and regulatory properties of leucoplast pyruvate kinase from Brassica napus (rapeseed) suspension cells. Arch. Biochem. Biophys. 2002, 400, 54–62. [Google Scholar] [CrossRef] [Green Version]
- Andre, C.; Froehlich, J.E.; Moll, M.R.; Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [Google Scholar] [CrossRef]
- Turner, W.L.; Knowles, V.L.; Plaxton, W.C. Cytosolic pyruvate kinase: Subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 2005, 222, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Tu, B.; Yang, W.; Yuan, H.; Li, J.; Guo, L.; Zheng, L.; Chen, W.; Zhu, X.; Wang, Y.; et al. Mitochondria-Associated Pyruvate Kinase Complexes Regulate Grain Filling in Rice. Plant Physiol. 2020, 183, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.; Benning, C. Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol. 2007, 145, 1670–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baud, S.; Wuilleme, S.; Dubreucq, B.; de Almeida, A.; Vuagnat, C.; Lepiniec, L.; Miquel, M.; Rochat, C. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 2007, 52, 405–419. [Google Scholar] [CrossRef]
- Ruuska, S.A.; Girke, T.; Benning, C.; Ohlrogge, J.B. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 2002, 14, 1191–1206. [Google Scholar] [CrossRef] [Green Version]
- Knowles, V.L.; McHugh, S.G.; Hu, Z.; Dennis, D.T.; Miki, B.L.; Plaxton, W.C. Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol. 1998, 116, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Feng, F.; He, C.Z. Downregulation of OsPK1 Contributes to Oxidative Stress and the Variations in ABA/GA Balance in Rice. Plant Mol. Biol. Rep. 2012, 30, 1006–1013. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, W.; Luo, L.; Pang, J.; Rong, W.; He, C. Downregulation of OsPK1, a cytosolic pyruvate kinase, by T-DNA insertion causes dwarfism and panicle enclosure in rice. Planta 2012, 235, 25–38. [Google Scholar] [CrossRef]
- Cai, Y.; Li, S.; Jiao, G.; Sheng, Z.; Wu, Y.; Shao, G.; Xie, L.; Peng, C.; Xu, J.; Tang, S.; et al. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol. J. 2018, 16, 1878–1891. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Zhang, W.W.; Jin, J.; Yang, X.M.; You, X.M.; Yan, H.G.; Wang, L.; Chen, J.; Xu, J.H.; Chen, W.W.; et al. OsPKp alpha 1 encodes a plastidic pyruvate kinase that affects starch biosynthesis in the rice endosperm. J. Integr. Plant Biol. 2018, 60, 1097–1118. [Google Scholar] [CrossRef]
- Larsen, T.M.; Benning, M.M.; Wesenberg, G.E.; Rayment, I.; Reed, G.H. Ligand-induced domain movement in pyruvate kinase: Structure of the enzyme from rabbit muscle with Mg2+, K+, and L-phospholactate at 2.7 A resolution. Arch. Biochem. Biophys. 1997, 345, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Schramm, A.; Siebers, B.; Tjaden, B.; Brinkmann, H.; Hensel, R. Pyruvate kinase of the hyperthermophilic crenarchaeote Thermoproteus tenax: Physiological role and phylogenetic aspects. J. Bacteriol. 2000, 182, 2001–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fothergill-Gillmore, L.A.; Rigden, D.J.; Michels, P.A.; Phillips, S.E. Leishmania pyruvate kinase: The crystal structure reveals the structural basis of its unique regulatory properties. Biochem. Soc. Trans. 2000, 28, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.R.; Knowles, V.L.; Plaxton, W.C. Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures: Implications for the integration of glycolysis with nitrogen assimilation. Eur. J. Biochem. 2000, 267, 4477–4485. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wei, X.; Jiao, G.; Chen, W.; Wu, Y.; Sheng, Z.; Hu, S.; Xie, L.; Wang, J.; Tang, S.; et al. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. J. Integr. Plant Biol. 2020, 62, 948–966. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wei, X.; Ren, Y.; Qiu, J.; Jiao, G.; Guo, X.; Tang, S.; Wan, J.; Hu, P. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017, 7, 40124. [Google Scholar] [CrossRef] [Green Version]
- Teng, X.; Zhong, M.; Zhu, X.; Wang, C.; Ren, Y.; Wang, Y.; Zhang, H.; Jiang, L.; Wang, D.; Hao, Y.; et al. FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice. Plant Biotechnol. J. 2019, 17, 1914–1927. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Wu, M.; Ren, Y.; Wang, Y.; Li, J.; Lei, C.; Sun, Y.; Bao, X.; Wu, H.; Yang, H.; et al. Rice FLOURY ENDOSPERM 18 encodes a pentatricopeptide repeat protein required for 5’ processing of mitochondrial nad5 messenger RNA and endosperm development. J. Integr. Plant Biol. 2020, 63, 834–847. [Google Scholar] [CrossRef]
- Tao, Y.; Din, A.M.U.; An, L.; Chen, H.; Li, G.H.; Ding, Y.F.; Liu, Z.H. Metabolic Disturbance Induced by the Embryo Contributes to the Formation of Chalky Endosperm of a Notched-Belly Rice Mutant. Front. Plant Sci. 2022, 12, 761668. [Google Scholar] [CrossRef]
- Lin, Z.M.; Zhang, X.C.; Yang, X.Y.; Li, G.H.; Tang, S.; Wang, S.H.; Ding, Y.F.; Liu, Z.H. Proteomic analysis of proteins related to rice grain chalkiness using iTRAQ and a novel comparison system based on a notched-belly mutant with white-belly. BMC Plant Biology. 2014, 14, 163. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14 (Suppl. 1), S15–S45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Hua, D.; Chen, Z.; Zhou, Z.; Gong, Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009, 60, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanning, S.B.; Siebenmorgen, T.J.; Counce, P.A.; Ambardekara, A.A.; Mauromoustakos, A. Extreme nighttime air temperatures in 2010 impact rice chalkiness and milling quality. Field Crop. Res. 2011, 124, 132–136. [Google Scholar] [CrossRef]
- Siebenmorgen, T.J.; Grigg, B.C.; Lanning, S.B. Impacts of preharvest factors during kernel development on rice quality and functionality. Annu. Rev. Food Sci. Technol. 2013, 4, 101–115. [Google Scholar] [CrossRef]
- Morita, S.; Wada, H.; Matsue, Y. Countermeasures for heat damage in rice grain quality under climate change. Plant Prod. Sci. 2016, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Masutomi, Y.; Arakawa, M.; Minoda, T.; Yonekura, T.; Shimada, T. Critical air temperature and sensitivity of the incidence of chalky rice kernels for the rice cultivar “Sai-no-kagayaki”. Agr. Forest. Meteorol. 2015, 203, 11–16. [Google Scholar] [CrossRef]
- Nevame, A.Y.M.; Emon, R.M.; Malek, M.A.; Hasan, M.M.; Alam, M.A.; Muharam, F.M.; Aslani, F.; Rafii, M.Y.; Ismail, M.R. Relationship between High Temperature and Formation of Chalkiness and Their Effects on Quality of Rice. Biomed. Res. Int. 2018, 2018, 1653721. [Google Scholar] [CrossRef]
- Tabata, M.; Hirabayashi, H.; Takeuchi, Y.; Ando, I.; Iida, Y.; Ohsawa, R. Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.). Breed. Sci. 2007, 57, 47–52. [Google Scholar] [CrossRef]
- Wada, T.; Miyahara, K.; Sonoda, J.Y.; Tsukaguehi, T.; Miyazaki, M.; Tsubone, M.; Ando, T.; Ebana, K.; Yamamoto, T.; Iwasawa, N.; et al. Detection of QTLs for white-back and basal-white grains caused by high temperature during ripening period in japonica rice. Breed. Sci. 2015, 65, 216–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyahara, K.; Wada, T.; Sonoda, J.; Tsukaguchi, T.; Miyazaki, M.; Tsubone, M.; Yamaguchi, O.; Ishibashi, M.; Iwasawa, N.; Umemoto, T.; et al. Detection and validation of QTLs for milky-white grains caused by high temperature during the ripening period in Japonica rice. Breed. Sci. 2017, 67, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, A.; Bao, G.L.; Ye, S.H.; Tomita, K. Detection of quantitative trait loci for white-back and basal-white kernels under high temperature stress in japonica rice varieties. Breed. Sci. 2007, 57, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.B.; Duan, S.; Ao, Y.; Dai, J.R.; Liu, J.; Wang, P.; Li, Y.G.; Liu, B.; Feng, D.R.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, N.; Chen, L.; Ahmad, S.; Cai, Y.; Duan, Y.; Li, X.; Liu, Y.; Jiao, G.; Xie, L.; Hu, S.; et al. Genome-Wide Analysis and Functional Characterization of Pyruvate Kinase (PK) Gene Family Modulating Rice Yield and Quality. Int. J. Mol. Sci. 2022, 23, 15357. https://doi.org/10.3390/ijms232315357
Dong N, Chen L, Ahmad S, Cai Y, Duan Y, Li X, Liu Y, Jiao G, Xie L, Hu S, et al. Genome-Wide Analysis and Functional Characterization of Pyruvate Kinase (PK) Gene Family Modulating Rice Yield and Quality. International Journal of Molecular Sciences. 2022; 23(23):15357. https://doi.org/10.3390/ijms232315357
Chicago/Turabian StyleDong, Nannan, Luna Chen, Shakeel Ahmad, Yicong Cai, Yingqing Duan, Xinwei Li, Yongqiang Liu, Guiai Jiao, Lihong Xie, Shikai Hu, and et al. 2022. "Genome-Wide Analysis and Functional Characterization of Pyruvate Kinase (PK) Gene Family Modulating Rice Yield and Quality" International Journal of Molecular Sciences 23, no. 23: 15357. https://doi.org/10.3390/ijms232315357
APA StyleDong, N., Chen, L., Ahmad, S., Cai, Y., Duan, Y., Li, X., Liu, Y., Jiao, G., Xie, L., Hu, S., Sheng, Z., Shao, G., Wang, L., Tang, S., Wei, X., & Hu, P. (2022). Genome-Wide Analysis and Functional Characterization of Pyruvate Kinase (PK) Gene Family Modulating Rice Yield and Quality. International Journal of Molecular Sciences, 23(23), 15357. https://doi.org/10.3390/ijms232315357







