Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome
Abstract
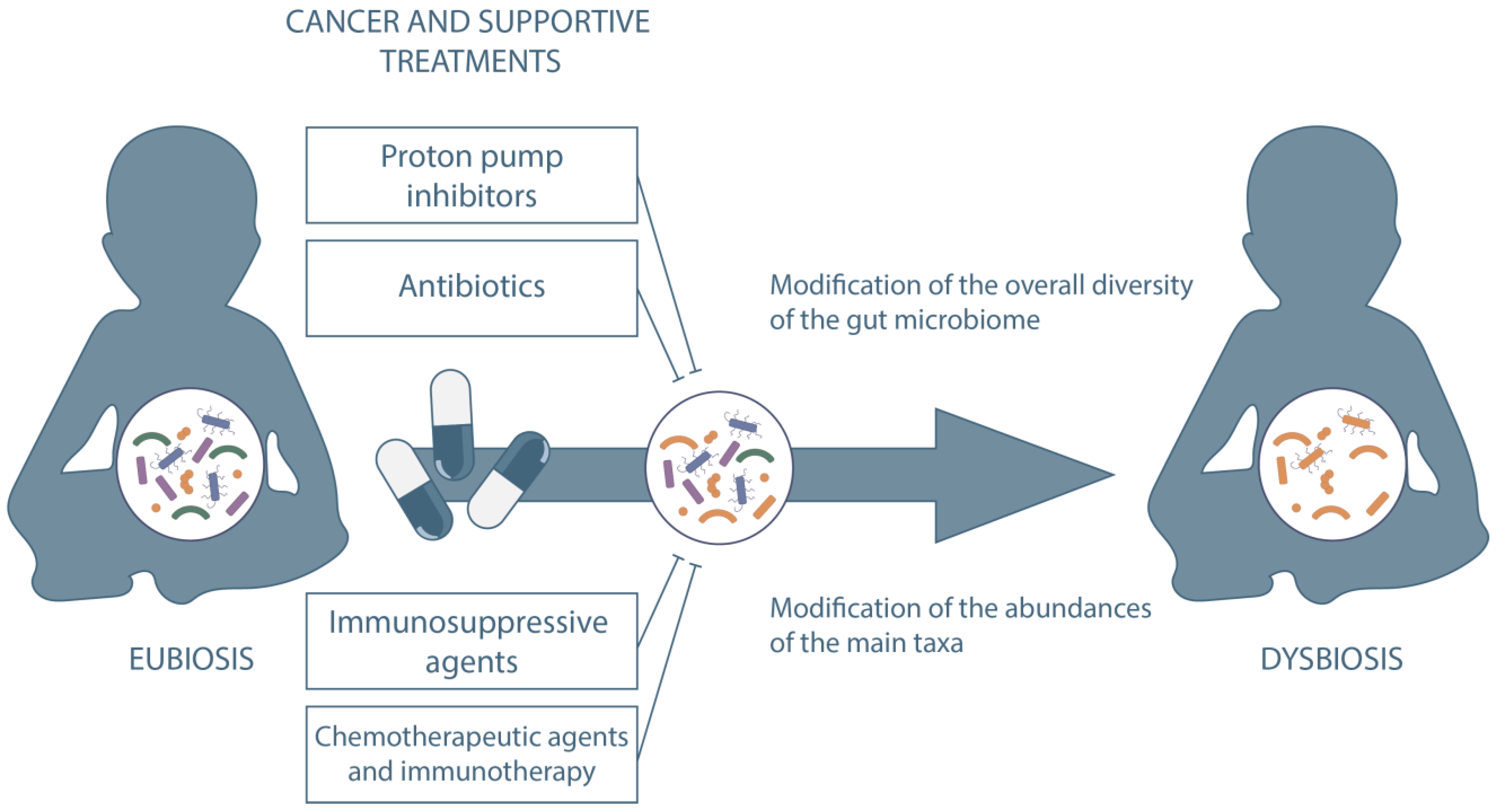
1. Introduction
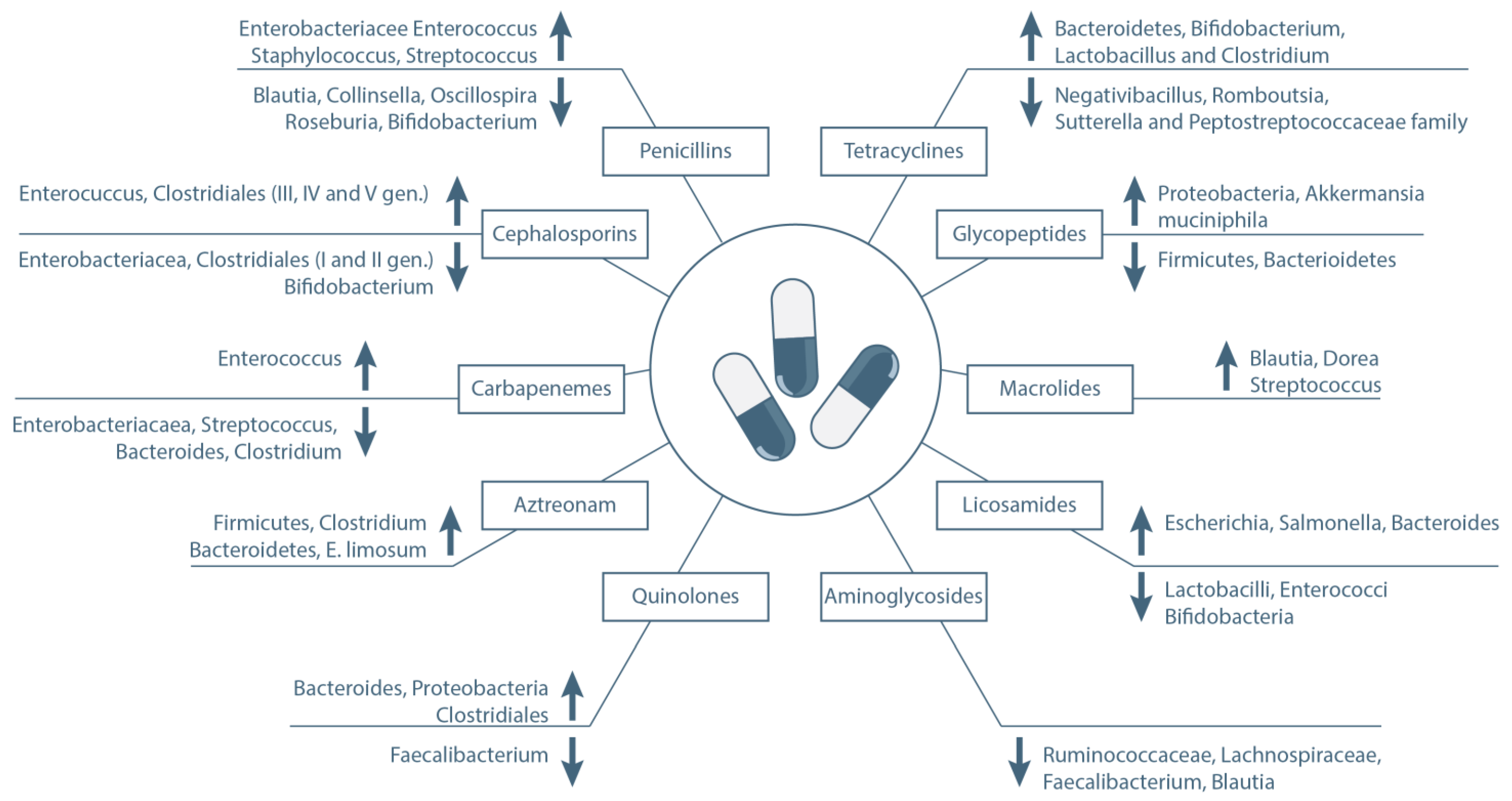
2. Antibiotics
2.1. β-Lactams
2.2. Quinolones
2.3. Tetracyclines
2.4. Glycopeptides
2.5. Macrolides
2.6. Other Antibiotics
3. Chemotherapeutic Agents
3.1. Irinotecan
3.2. Cyclophosphamide
3.3. L-Asparaginase
3.4. Other Chemotherapeutic Drugs
4. Anti-Programmed Cell Death Proteins
5. Immunosuppressive Agents
5.1. Cyclosporine
5.2. Tacrolimus
5.3. Other Immunosuppressive Drugs
6. Steroids
7. Protonic Pump Inhibitors
8. Ursodeoxycholic Acid
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schluter, J.; Peled, J.U.; Taylor, B.P.; Markey, K.A.; Smith, M.; Taur, Y.; Niehus, R.; Staffas, A.; Dai, A.; Fontana, E.; et al. The Gut Microbiota Is Associated with Immune Cell Dynamics in Humans. Nature 2020, 588, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and Cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Masetti, R.; Muratore, E.; Leardini, D.; Zama, D.; Turroni, S.; Brigidi, P.; Esposito, S.; Pession, A. Gut Microbiome in Pediatric Acute Leukemia: From Predisposition to Cure. Blood Adv. 2021, 5, 4619–4629. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; Van Den Brink, M.R.M. Gut Microbiota Injury in Allogeneic Haematopoietic Stem Cell Transplantation. Nat. Rev. Cancer 2018, 18, 283–295. [Google Scholar] [CrossRef]
- Messina, J.A.; Tan, C.Y.; Ren, Y.; Hill, L.; Bush, A.; Lew, M.; Andermann, T.; Peled, J.U.; Gomes, A.; van den Brink, M.R.M.; et al. Enterococcus Intestinal Domination Is Associated with Increased Mortality in the Acute Leukemia Chemotherapy Population. Clin. Infect. Dis. 2021, ciab1043. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut Microbiome Correlates of Response and Toxicity Following Anti-CD19 CAR T Cell Therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal Microbiota Transplant Overcomes Resistance to Anti-PD-1 Therapy in Melanoma Patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
- Masetti, R.; Zama, D.; Leardini, D.; Muratore, E.; Turroni, S.; Prete, A.; Brigidi, P.; Pession, A. The Gut Microbiome in Pediatric Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Pediatr. Blood Cancer 2020, 67, e28711. [Google Scholar] [CrossRef]
- Henig, I.; Yehudai-Ofir, D.; Zuckerman, T. The Clinical Role of the Gut Microbiome and Fecal Microbiota Transplantation in Allogeneic Stem Cell Transplantation. Haematologica 2021, 106, 933–946. [Google Scholar] [CrossRef]
- Zama, D.; Gori, D.; Muratore, E.; Leardini, D.; Rallo, F.; Turroni, S.; Prete, A.; Brigidi, P.; Pession, A.; Masetti, R. Enteral versus Parenteral Nutrition as Nutritional Support after Allogeneic Hematopoietic Stem Cell Transplantation: A Systematic Review and Meta-Analysis. Transplant. Cell. Ther. 2021, 27, e1–e180. [Google Scholar] [CrossRef]
- Weber, D.; Jenq, R.R.; Peled, J.U.; Taur, Y.; Hiergeist, A.; Koestler, J.; Dettmer, K.; Weber, M.; Wolff, D.; Hahn, J.; et al. Microbiota Disruption Induced by Early Use of Broad-Spectrum Antibiotics Is an Independent Risk Factor of Outcome after Allogeneic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2017, 23, 845–852. [Google Scholar] [CrossRef]
- Ingham, A.C.; Kielsen, K.; Cilieborg, M.S.; Lund, O.; Holmes, S.; Aarestrup, F.M.; Müller, K.G.; Pamp, S.J. Specific Gut Microbiome Members Are Associated with Distinct Immune Markers in Pediatric Allogeneic Hematopoietic Stem Cell Transplantation. Microbiome 2019, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Masetti, R.; Zama, D.; Leardini, D.; Muratore, E.; Turroni, S.; Brigidi, P.; Pession, A. Microbiome-Derived Metabolites in Allogeneic Hematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2021, 22, 1197. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, F.; Soverini, M.; Zama, D.; Consolandi, C.; Severgnini, M.; Prete, A.; Pession, A.; Barone, M.; Turroni, S.; Biagi, E.; et al. Gut Resistome Plasticity in Pediatric Patients Undergoing Hematopoietic Stem Cell Transplantation. Sci. Rep. 2019, 9, 5649. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of Therapeutic Drugs by Human Gut Bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]
- Lynn, M.A.; Eden, G.; Ryan, F.J.; Bensalem, J.; Wang, X.; Blake, S.J.; Choo, J.M.; Chern, Y.T.; Sribnaia, A.; James, J.; et al. The Composition of the Gut Microbiota Following Early-Life Antibiotic Exposure Affects Host Health and Longevity in Later Life. Cell Rep. 2021, 36, 109564. [Google Scholar] [CrossRef]
- Zhang, W.; An, Y.; Qin, X.; Wu, X.; Wang, X.; Hou, H.; Song, X.; Liu, T.; Wang, B.; Huang, X.; et al. Gut Microbiota-Derived Metabolites in Colorectal Cancer: The Bad and the Challenges. Front. Oncol. 2021, 11, 739648. [Google Scholar] [CrossRef]
- Pearson, T.; Caporaso, J.G.; Yellowhair, M.; Bokulich, N.A.; Padi, M.; Roe, D.J.; Wertheim, B.C.; Linhart, M.; Martinez, J.A.; Bilagody, C.; et al. Effects of Ursodeoxycholic Acid on the Gut Microbiome and Colorectal Adenoma Development. Cancer Med. 2019, 8, 617–628. [Google Scholar] [CrossRef]
- Rotz, S.J.; Dandoy, C.E. The Microbiome in Pediatric Oncology. Cancer 2020, 126, 3629–3637. [Google Scholar] [CrossRef] [PubMed]
- Muratore, E.; Leardini, D.; Baccelli, F.; Venturelli, F.; Prete, A.; Masetti, R. Nutritional Modulation of the Gut Microbiome in Allogeneic Hematopoietic Stem Cell Transplantation Recipients. Front. Nutr. 2022, 9, 993668. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Roberts, A.B.; Pollet, R.M.; Ingle, J.D.; Biernat, K.A.; Pellock, S.J.; Venkatesh, M.K.; Guthrie, L.; O’Neal, S.K.; Robinson, S.J.; et al. Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol. 2015, 22, 1238–1249. [Google Scholar] [CrossRef]
- Stringer, A.M.; Gibson, R.J.; Logan, R.M.; Bowen, J.M.; Yeoh, A.S.J.; Keefe, D.M.K. Faecal Microflora and Beta-Glucuronidase Expression Are Altered in an Irinotecan-Induced Diarrhea Model in Rats. Cancer Biol. Ther. 2008, 7, 1919–1925. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, X. Effects of Cyclophosphamide on Immune System and Gut Microbiota in Mice. Microbiol. Res. 2015, 171, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.A.; Connors, J.; Bielawski, J.P.; Nearing, J.T.; Langille, M.G.I.; Van Limbergen, J.; Fernandez, C.V.; MacDonald, T.; Kulkarni, K. Investigating the Gut Microbial Community and Genes in Children with Differing Levels of Change in Serum Asparaginase Activity during Pegaspargase Treatment for Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2021, 62, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Jia, J.; Tian, X.; Jiang, J.; Ren, Z.; Lu, H.; He, N.; Xie, H.; Zhou, L.; Zheng, S. Structural Shifts in the Intestinal Microbiota of Rats Treated with Cyclosporine A after Orthotropic Liver Transplantation. Front. Med. 2019, 13, 451–460. [Google Scholar] [CrossRef]
- O’Reilly, C.; O’Sullivan, Ó.; Cotter, P.D.; O’Connor, P.M.; Shanahan, F.; Cullen, A.; Rea, M.C.; Hill, C.; Coulter, I.; Paul Ross, R. Encapsulated Cyclosporine Does Not Change the Composition of the Human Microbiota When Assessed Ex Vivo and In Vivo. J. Med. Microbiol. 2020, 69, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, L.; Tang, H.; Jiao, W.; Zeng, S.; Xu, Y.; Zhang, Q.; Sun, Z.; Mukherjee, A.; Zhang, X.; et al. Immunosuppressive Effect of the Gut Microbiome Altered by High-Dose Tacrolimus in Mice. Am. J. Transplant. 2018, 18, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.; Pasini, E.; Copeland, J.; Angeli, M.; Husain, S.; Kumar, D.; Renner, E.; Teterina, A.; Allard, J.; Guttman, D.S.; et al. Impact of Immunosuppression on the Metagenomic Composition of the Intestinal Microbiome: A Systems Biology Approach to Post-Transplant Diabetes. Sci. Rep. 2017, 7, 10277. [Google Scholar] [CrossRef]
- Toral, M.; Romero, M.; Rodríguez-Nogales, A.; Jiménez, R.; Robles-Vera, I.; Algieri, F.; Chueca-Porcuna, N.; Sánchez, M.; de la Visitación, N.; Olivares, M.; et al. Lactobacillus Fermentum Improves Tacrolimus-Induced Hypertension by Restoring Vascular Redox State and Improving ENOS Coupling. Mol. Nutr. Food Res. 2018, 62, 1–13. [Google Scholar] [CrossRef]
- Jiang, J.W.; Ren, Z.G.; Lu, H.F.; Zhang, H.; Li, A.; Cui, G.Y.; Jia, J.J.; Xie, H.Y.; Chen, X.H.; He, Y.; et al. Optimal Immunosuppressor Induces Stable Gut Microbiota after Liver Transplantation. World J. Gastroenterol. 2018, 24, 3871–3883. [Google Scholar] [CrossRef]
- Flannigan, K.L.; Taylor, M.R.; Pereira, S.K.; Rodriguez-Arguello, J.; Moffat, A.W.; Alston, L.; Wang, X.; Poon, K.K.; Beck, P.L.; Rioux, K.P.; et al. An Intact Microbiota Is Required for the Gastrointestinal Toxicity of the Immunosuppressant Mycophenolate Mofetil. J. Heart Lung Transplant. 2018, 37, 1047–1059. [Google Scholar] [CrossRef]
- Jung, M.J.; Lee, J.; Shin, N.R.; Kim, M.S.; Hyun, D.W.; Yun, J.H.; Kim, P.S.; Whon, T.W.; Bae, J.W. Chronic Repression of MTOR Complex 2 Induces Changes in the Gut Microbiota of Diet-Induced Obese Mice. Sci. Rep. 2016, 6, 30887. [Google Scholar] [CrossRef]
- Li, Q.R.; Wang, C.Y.; Tang, C.; He, Q.; Li, N.; Li, J.S. Reciprocal Interaction between Intestinal Microbiota and Mucosal Lymphocyte in Cynomolgus Monkeys after Alemtuzumab Treatment. Am. J. Transplant. 2013, 13, 899–910. [Google Scholar] [CrossRef]
- Lee, J.R.; Muthukumar, T.; Dadhania, D.; Toussaint, N.C.; Ling, L.; Pamer, E.; Suthantiran, M. Gut Microbial Community Structure and Complications Following Kidney Transplantation: A Pilot Study. Transplantation 2014, 98, 697–705. [Google Scholar] [CrossRef]
- Tourret, J.; Willing, B.P.; Dion, S.; MacPherson, J.; Denamur, E.; Finlay, B.B. Immunosuppressive Treatment Alters Secretion of Ileal Antimicrobial Peptides and Gut Microbiota, and Favors Subsequent Colonization by Uropathogenic Escherichia coli. Transplantation 2017, 101, 74–82. [Google Scholar] [CrossRef]
- Wu, T.; Yang, L.; Jiang, J.; Ni, Y.; Zhu, J.; Zheng, X.; Wang, Q.; Lu, X.; Fu, Z. Chronic Glucocorticoid Treatment Induced Circadian Clock Disorder Leads to Lipid Metabolism and Gut Microbiota Alterations in Rats. Life Sci. 2018, 192, 173–182. [Google Scholar] [CrossRef]
- He, Z.; Kong, X.; Shao, T.; Zhang, Y.; Wen, C. Alterations of the Gut Microbiota Associated with Promoting Efficacy of Prednisone by Bromofuranone in MRL/Lpr Mice. Front. Microbiol. 2019, 10, 978. [Google Scholar] [CrossRef]
- Vich Vila, A.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.M.A.E.; Masclee, A.A.M.; Fu, J.; et al. Impact of Commonly Used Drugs on the Composition and Metabolic Function of the Gut Microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Jackson, M.A.; Goodrich, J.K.; Maxan, M.E.; Freedberg, D.E.; Abrams, J.A.; Poole, A.C.; Sutter, J.L.; Welter, D.; Ley, R.E.; Bell, J.T.; et al. Proton Pump Inhibitors Alter the Composition of the Gut Microbiota. Gut 2016, 65, 749–756. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vila, A.V.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.M.; et al. Proton Pump Inhibitors Affect the Gut Microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Toussaint, N.C.; Chen, S.P.; Ratner, A.J.; Whittier, S.; Wang, T.C.; Wang, H.H.; Abrams, J.A. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology 2015, 149, 883–885.e9. [Google Scholar] [CrossRef]
- Tsuda, A.; Suda, W.; Morita, H.; Takanashi, K.; Takagi, A.; Koga, Y.; Hattori, M. Influence of Proton-Pump Inhibitors on the Luminal Microbiota in the Gastrointestinal Tract. Clin. Transl. Gastroenterol. 2015, 6, e89. [Google Scholar] [CrossRef]
- Simakachorn, L.; Tanpowpong, P.; Chanprasertyothin, S.; Thongpradit, S.; Treepongkaruna, S. Gut Microbiota Characteristics in Children after the Use of Proton Pump Inhibitors. Turkish J. Gastroenterol. 2021, 32, 70–75. [Google Scholar] [CrossRef]
- Tang, R.; Wei, Y.; Li, Y.; Chen, W.; Chen, H.; Wang, Q.; Yang, F.; Miao, Q.; Xiao, X.; Zhang, H.; et al. Gut Microbial Profile Is Altered in Primary Biliary Cholangitis and Partially Restored after UDCA Therapy. Gut 2018, 67, 534–571. [Google Scholar] [CrossRef]
- Ianiro, G.; Tilg, H.; Gasbarrini, A. Antibiotics as Deep Modulators of Gut Microbiota: Between Good and Evil. Gut 2016, 65, 1906–1915. [Google Scholar] [CrossRef]
- Theriot, C.M.; Young, V.B. Interactions Between the Gastrointestinal Microbiome and Clostridium Difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lieberman, S.R.; Bhatt, A.S.; Jenq, R.R.; Xu, K.; Gomes, C.; Gyurkocza, B.; Moss, E.L.; Jay, H.V.; Calarfiore, M.; et al. Increased GVHD-Related Mortality with Broad-Spectrum Antibiotic Use after Allogeneic Hematopoietic Stem Cell Transplantation in Human Patients and Mice. Sci. Transl. Med. 2016, 8, 339ra71. [Google Scholar] [CrossRef]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-Term Impacts of Antibiotic Exposure on the Human Intestinal Microbiota. Microbiology 2010, 156, 3216–3223. [Google Scholar] [CrossRef] [PubMed]
- Muratore, E.; Baccelli, F.; Leardini, D.; Campoli, C.; Belotti, T.; Viale, P.; Prete, A.; Pession, A.; Masetti, R.; Zama, D. Antimicrobial Stewardship Interventions in Pediatric Oncology: A Systematic Review. J. Clin. Med. 2022, 11, 4545. [Google Scholar] [CrossRef] [PubMed]
- Nel Van Zyl, K.; Matukane, S.R.; Hamman, B.L.; Whitelaw, A.C.; Newton-Foot, M. Effect of Antibiotics on the Human Microbiome: A Systematic Review. Int. J. Antimicrob. Agents 2022, 59, 106502. [Google Scholar] [CrossRef]
- Livadiotti, S.; Milano, G.M.; Serra, A.; Folgori, L.; Jenkner, A.; Castagnola, E.; Cesaro, S.; Rossi, M.R.; Barone, A.; Zanazzo, G.; et al. A Survey on Hematology-Oncology Pediatric AIEOP Centers: Prophylaxis, Empirical Therapy and Nursing Prevention Procedures of Infectious Complications. Haematologica 2012, 97, 147–150. [Google Scholar] [CrossRef][Green Version]
- Zama, D.; Masetti, R.; Baccelli, F.; Leardini, D.; Muratore, E.; Abram, N.; Vendemini, F.; Biffi, A.; Perruccio, K.; D’Amico, M.R.; et al. Antibiotic Prophylaxis and Management of Infections in Pediatric Hematopoietic Stem Cell Transplantation: A Survey from the Stem Cell Transplant and the Infectious Disease Working Groups of the AIEOP Network. Bone Marrow Transplant. 2022, 57, 1851–1853. [Google Scholar] [CrossRef]
- Lehrnbecher, T.; Averbuch, D.; Castagnola, E.; Cesaro, S.; Ammann, R.A.; Garcia-Vidal, C.; Kanerva, J.; Lanternier, F.; Mesini, A.; Mikulska, M.; et al. 8th European Conference on Infections in Leukaemia: 2020 Guidelines for the Use of Antibiotics in Paediatric Patients with Cancer or Post-Haematopoietic Cell Transplantation. Lancet Oncol. 2021, 22, e270–e280. [Google Scholar] [CrossRef]
- Raymond, F.; Ouameur, A.A.; Déraspe, M.; Iqbal, N.; Gingras, H.; Dridi, B.; Leprohon, P.; Plante, P.L.; Giroux, R.; Bérubé, È.; et al. The Initial State of the Human Gut Microbiome Determines Its Reshaping by Antibiotics. ISME J. 2016, 10, 707–720. [Google Scholar] [CrossRef]
- MacPherson, C.W.; Mathieu, O.; Tremblay, J.; Champagne, J.; Nantel, A.; Girard, S.A.; Tompkins, T.A. Gut Bacterial Microbiota and Its Resistome Rapidly Recover to Basal State Levels after Short-Term Amoxicillin-Clavulanic Acid Treatment in Healthy Adults. Sci. Rep. 2018, 8, 11192. [Google Scholar] [CrossRef]
- Pallav, K.; Dowd, S.E.; Villafuerte, J.; Yang, X.; Kabbani, T.; Hansen, J.; Dennis, M.; Leffler, D.A.; Newburg, D.S.; Kelly, C.P. Effects of Polysaccharopeptide from Trametes Versicolor and Amoxicillin on the Gut Microbiome of Healthy Volunteers: A Randomized Clinical Trial. Gut Microbes 2014, 5, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Brismar, B.; Edlund, C.; Nord, C.E. Impact of Cefpodoxime Proxetil and Amoxicillin on the Normal Oral and Intestinal Microflora. Eur. J. Clin. Microbiol. Infect. Dis. 1993, 12, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Brandt, B.W.; de Mattos, M.J.T.; Buijs, M.J.; Caspers, M.P.M.; Rashid, M.U.; Weintraub, A.; Nord, C.E.; Savell, A.; Hu, Y.; et al. Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces. MBio 2015, 6, e01693-15. [Google Scholar] [CrossRef] [PubMed]
- De La Cochetière, M.F.; Durand, T.; Lepage, P.; Bourreille, A.; Galmiche, J.P.; Doré, J. Resilience of the Dominant Human Fecal Microbiota upon Short-Course Antibiotic Challenge. J. Clin. Microbiol. 2005, 43, 5588–5592. [Google Scholar] [CrossRef]
- Romick-Rosendale, L.E.; Haslam, D.B.; Lane, A.; Denson, L.; Lake, K.; Wilkey, A.; Watanabe, M.; Bauer, S.; Litts, B.; Luebbering, N.; et al. Antibiotic Exposure and Reduced Short Chain Fatty Acid Production after Hematopoietic Stem Cell Transplant. Biol. Blood Marrow Transplant. 2018, 24, 2418–2424. [Google Scholar] [CrossRef]
- Heimdahl, A.; Nord, C.E. Effect of Phenoxymethylpenicillin and Clindamycin on the Oral, Throat and Faecal Microflora of Man. Scand. J. Infect. Dis. 1979, 11, 233–242. [Google Scholar] [CrossRef]
- Calitri, C.; Ruberto, E.; Castagnola, E. Antibiotic Prophylaxis in Neutropenic Children with Acute Leukemia: Do the Presently Available Data Really Support This Practice? Eur. J. Haematol. 2018, 101, 721–727. [Google Scholar] [CrossRef]
- Rashid, M.-U.; Rosenborg, S.; Panagiotidis, G.; Löfdal, K.S.; Weintraub, A.; Nord, C.E. Ecological Effect of Ceftazidime/Avibactam on the Normal Human Intestinal Microbiota. Int. J. Antimicrob. Agents 2015, 46, 60–65. [Google Scholar] [CrossRef]
- Stein-Thoeringer, C.K.; Nichols, K.B.; Lazrak, A.; Docampo, M.D.; Slingerland, A.E.; Slingerland, J.B.; Clurman, A.G.; Armijo, G.; Gomes, A.L.C.; Shono, Y.; et al. Lactose Drives Enterococcus Expansion to Promote Graft-versus-Host Disease. Science 2019, 366, 1143–1149. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. The Effect of Antibiotics on the Composition of the Intestinal Microbiota—A Systematic Review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef]
- Meijer-Severs, G.J.; Van Santen, E.; Meijer, B.C. Short-Chain Fatty Acid and Organic Acid Concentrations in Feces of Healthy Human Volunteers and Their Correlations with Anaerobe Cultural Counts during Systemic Ceftriaxone Administration. Scand. J. Gastroenterol. 1990, 25, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Artacho, A.; Knecht, H.; Ferrús, M.L.; Friedrichs, A.; Ott, S.J.; Moya, A.; Latorre, A.; Gosalbes, M.J. Differential Effects of Antibiotic Therapy on the Structure and Function of Human Gut Microbiota. PLoS ONE 2013, 8, e80201. [Google Scholar] [CrossRef] [PubMed]
- Zwittink, R.D.; Renes, I.B.; van Lingen, R.A.; van Zoeren-Grobben, D.; Konstanti, P.; Norbruis, O.F.; Martin, R.; Groot Jebbink, L.J.M.; Knol, J.; Belzer, C. Association between Duration of Intravenous Antibiotic Administration and Early-Life Microbiota Development in Late-Preterm Infants. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 475–483. [Google Scholar] [CrossRef]
- Payne, L.E.; Gagnon, D.J.; Riker, R.R.; Seder, D.B.; Glisic, E.K.; Morris, J.G.; Fraser, G.L. Cefepime-Induced Neurotoxicity: A Systematic Review. Crit. Care 2017, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Lim, J.Y.; Bin Ryu, D.; Kim, T.W.; Park, S.S.; Jeon, Y.W.; Yoon, J.H.; Cho, B.S.; Eom, K.S.; Kim, Y.J.; et al. Alteration of the Intestinal Microbiota by Broad-Spectrum Antibiotic Use Correlates with the Occurrence of Intestinal Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2019, 25, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Bächer, K.; Schaeffer, M.; Lode, H.; Nord, C.E.; Borner, K.; Koeppe, P. Multiple Dose Pharmacokinetics, Safety, and Effects on Faecal Microflora, of Cefepime in Healthy Volunteers. J. Antimicrob. Chemother. 1992, 30, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Pletz, M.W.R.; Rau, M.; Bulitta, J.; De Roux, A.; Burkhardt, O.; Kruse, G.; Kurowski, M.; Nord, C.E.; Lode, H. Ertapenem Pharmacokinetics and Impact on Intestinal Microflora, in Comparison to Those of Ceftriaxone, after Multiple Dosing in Male and Female Volunteers. Antimicrob. Agents Chemother. 2004, 48, 3765–3772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Limaye, P.B.; Renaud, H.J.; Klaassen, C.D. Effect of Various Antibiotics on Modulation of Intestinal Microbiota and Bile Acid Profile in Mice. Toxicol. Appl. Pharmacol. 2014, 277, 138–145. [Google Scholar] [CrossRef]
- Gu, S.-L.; Gong, Y.; Zhang, J.; Chen, Y.; Wu, Z.; Xu, Q.; Fang, Y.; Wang, J.; Tang, L.-L. Effect of the Short-Term Use of Fluoroquinolone and β-Lactam Antibiotics on Mouse Gut Microbiota. Infect. Drug Resist. 2020, 13, 4547–4558. [Google Scholar] [CrossRef]
- Peled, J.U.; Devlin, S.M.; Staffas, A.; Lumish, M.; Khanin, R.; Littmann, E.R.; Ling, L.; Kosuri, S.; Maloy, M.; Slingerland, J.B.; et al. Intestinal Microbiota and Relapse after Hematopoietic-Cell Transplantation. J. Clin. Oncol. 2017, 35, 1650–1659. [Google Scholar] [CrossRef]
- Alexander, S.; Fisher, B.B.T.B.T.; Gaur, A.H.H.; Dvorak, C.C.C.; Villa Luna, D.; Dang, H.; Chen, L.; Green, M.; Nieder, M.L.L.; Fisher, B.B.T.B.T.; et al. Effect of Levofloxacin Prophylaxis on Bacteremia in Children With Acute Leukemia or Undergoing Hematopoietic Stem Cell Transplantation. JAMA 2018, 320, 995. [Google Scholar] [CrossRef] [PubMed]
- Leardini, D.; Muratore, E.; Abram, N.; Baccelli, F.; Belotti, T.; Prete, A.; Gori, D.; Masetti, R.; Oncology, P.; Unit, H.; et al. Effectiveness of Quinolone Prophylaxis in Pediatric Acute Leukemia and Hematopoietic Stem Cell Transplantation: A Systematic Review and Meta-Analysis. Open Forum Infect. Dis. 2022, ofac594. [Google Scholar] [CrossRef]
- Olson, J.; Mehra, S.; Hersh, A.L.; Thorell, E.A.; Stoddard, G.J.; Maese, L.; Barnette, P.E.; Lemons, R.S.; Pavia, A.T.; Knackstedt, E.D. Oral Step-Down Therapy With Levofloxacin for Febrile Neutropenia in Children With Cancer. J. Pediatr. Infect. Dis. Soc. 2021, 10, 27–33. [Google Scholar] [CrossRef]
- Stewardson, A.J.; Gaïa, N.; François, P.; Malhotra-Kumar, S.; Delémont, C.; Martinez de Tejada, B.; Schrenzel, J.; Harbarth, S.; Lazarevic, V.; Vervoort, J.; et al. Collateral Damage from Oral Ciprofloxacin versus Nitrofurantoin in Outpatients with Urinary Tract Infections: A Culture-Free Analysis of Gut Microbiota. Clin. Microbiol. Infect. 2015, 21, e1–e344. [Google Scholar] [CrossRef]
- Rashid, M.U.; Zaura, E.; Buijs, M.J.; Keijser, B.J.F.; Crielaard, W.; Nord, C.E.; Weintraub, A. Determining the Long-Term Effect of Antibiotic Administration on the Human Normal Intestinal Microbiota Using Culture and Pyrosequencing Methods. Clin. Infect. Dis. 2015, 60, S77–S84. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; Von Bergen, M.; et al. Gut Microbiota Disturbance during Antibiotic Therapy: A Multi-Omic Approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef]
- Jothishankar, B.; Di Raimondo, C.; Mueller, L.; Zain, J.; Parekh, V.; Abdulla, F. Primary Cutaneous Marginal Zone Lymphoma Treated with Doxycycline in a Pediatric Patient. Pediatr. Dermatol. 2020, 37, 759–761. [Google Scholar] [CrossRef]
- Thompson, K.G.; Rainer, B.M.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Minocycline and Its Impact on Microbial Dysbiosis in the Skin and Gastrointestinal Tract of Acne Patients. Ann. Dermatol. 2020, 32, 21–30. [Google Scholar] [CrossRef]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of Oral Vancomycin on Gut Microbiota, Bile Acid Metabolism, and Insulin Sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef]
- Hwang, I.; Park, Y.J.; Kim, Y.-R.; Kim, Y.N.; Ka, S.; Lee, H.Y.; Seong, J.K.; Seok, Y.-J.; Kim, J.B. Alteration of Gut Microbiota by Vancomycin and Bacitracin Improves Insulin Resistance via Glucagon-like Peptide 1 in Diet-Induced Obesity. FASEB J. 2015, 29, 2397–2411. [Google Scholar] [CrossRef]
- Hansen, C.H.F.; Krych, L.; Nielsen, D.S.; Vogensen, F.K.; Hansen, L.H.; Sørensen, S.J.; Buschard, K.; Hansen, A.K. Early Life Treatment with Vancomycin Propagates Akkermansia Muciniphila and Reduces Diabetes Incidence in the NOD Mouse. Diabetologia 2012, 55, 2285–2294. [Google Scholar] [CrossRef]
- Masetti, R.; D’Amico, F.; Zama, D.; Leardini, D.; Muratore, E.; Ussowicz, M.; Fraczkiewicz, J.; Cesaro, S.; Caddeo, G.; Pezzella, V.; et al. Febrile Neutropenia Duration Is Associated with the Severity of Gut Microbiota Dysbiosis in Pediatric Allogeneic Hematopoietic Stem Cell Transplantation Recipients. Cancers 2022, 14, 1932. [Google Scholar] [CrossRef]
- Doan, T.; Arzika, A.M.; Ray, K.J.; Cotter, S.Y.; Kim, J.; Maliki, R.; Zhong, L.; Zhou, Z.; Porco, T.C.; Vanderschelden, B.; et al. Gut Microbial Diversity in Antibiotic-Naive Children after Systemic Antibiotic Exposure: A Randomized Controlled Trial. Clin. Infect. Dis. 2017, 64, 1147–1153. [Google Scholar] [CrossRef]
- Abeles, S.R.; Jones, M.B.; Santiago-Rodriguez, T.M.; Ly, M.; Klitgord, N.; Yooseph, S.; Nelson, K.E.; Pride, D.T. Microbial Diversity in Individuals and Their Household Contacts Following Typical Antibiotic Courses. Microbiome 2016, 4, 39. [Google Scholar] [CrossRef]
- Nikolaou, E.; Kamilari, E.; Savkov, D.; Sergeev, A.; Zakharova, I.; Vogazianos, P.; Tomazou, M.; Antoniades, A.; Shammas, C. Intestinal Microbiome Analysis Demonstrates Azithromycin Post-Treatment Effects Improve When Combined with Lactulose. World J. Pediatr. 2020, 16, 168–176. [Google Scholar] [CrossRef]
- Hakim, H.; Dallas, R.; Wolf, J.; Tang, L.; Schultz-Cherry, S.; Darling, V.; Johnson, C.; Karlsson, E.A.; Chang, T.C.; Jeha, S.; et al. Gut Microbiome Composition Predicts Infection Risk during Chemotherapy in Children with Acute Lymphoblastic Leukemia. Clin. Infect. Dis. 2018, 67, 541–548. [Google Scholar] [CrossRef]
- Heinsen, F.A.; Knecht, H.; Neulinger, S.C.; Schmitz, R.A.; Knecht, C.; Kühbacher, T.; Rosenstiel, P.C.; Schreiber, S.; Friedrichs, A.K.; Ott, S.J. Dynamic Changes of the Luminal and Mucosaassociated Gut Microbiota during and after Antibiotic Therapy with Paromomycin. Gut Microbes 2015, 6, 243–254. [Google Scholar] [CrossRef]
- Park, E.M.; Chelvanambi, M.; Bhutiani, N.; Kroemer, G.; Zitvogel, L.; Wargo, J.A. Targeting the Gut and Tumor Microbiota in Cancer. Nat. Med. 2022, 28, 690–703. [Google Scholar] [CrossRef]
- Yohe, M.E.; Heske, C.M.; Stewart, E.; Adamson, P.C.; Ahmed, N.; Antonescu, C.R.; Chen, E.; Collins, N.; Ehrlich, A.; Galindo, R.L.; et al. Insights into Pediatric Rhabdomyosarcoma Research: Challenges and Goals. Pediatr. Blood Cancer 2019, 66, e27869. [Google Scholar] [CrossRef]
- Bagatell, R.; London, W.B.; Wagner, L.M.; Voss, S.D.; Stewart, C.F.; Maris, J.M.; Kretschmar, C.; Cohn, S.L. Phase II Study of Irinotecan and Temozolomide in Children with Relapsed or Refractory Neuroblastoma: A Children’s Oncology Group Study. J. Clin. Oncol. 2011, 29, 208–213. [Google Scholar] [CrossRef]
- Kawato, Y.; Aonuma, M.; Hirota, Y.; Kuga, H.; Sato, K. Intracellular Roles of SN-38, a Metabolite of the Camptothecin Derivative CPT-11, in the Antitumor Effect of CPT-11. Cancer Res. 1991, 51, 4187–4191. [Google Scholar]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.A.; Mani, S.; et al. Alleviating Cancer Drug Toxicity by Inhibiting a Bacterial Enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Gibson, R.J.; Stringer, A.M. Chemotherapy-Induced Diarrhoea. Curr. Opin. Support. Palliat. Care 2009, 3, 31–35. [Google Scholar] [CrossRef] [PubMed]
- McQuade, R.M.; Stojanovska, V.; Donald, E.L.; Rahman, A.A.; Campelj, D.G.; Abalo, R.; Rybalka, E.; Bornstein, J.C.; Nurgali, K. Irinotecan-Induced Gastrointestinal Dysfunction Is Associated with Enteric Neuropathy, but Increased Numbers of Cholinergic Myenteric Neurons. Front. Physiol. 2017, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Takasuna, K.; Hagiwara, T.; Hirohashi, M.; Kato, M.; Nomura, M.; Nagai, E.; Yokoi, T.; Kamataki, T. Inhibition of Intestinal Microflora β-Glucuronidase Modifies the Distribution of the Active Metabolite of the Antitumor Agent, Irinotecan Hydrochloride (CPT-11) in Rats. Cancer Chemother. Pharmacol. 1998, 42, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Modulation of Irinotecan-Induced Diarrhea by Cotreatment with Neomycin in Cancer Patients—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/11350876/ (accessed on 23 April 2022).
- De Jong, F.A.; Kehrer, D.F.S.; Mathijssen, R.H.J.; Creemers, G.-J.; de Bruijn, P.; van Schaik, R.H.N.; Planting, A.S.T.; van der Gaast, A.; Eskens, F.A.L.M.; Janssen, J.T.P.; et al. Prophylaxis of Irinotecan-Induced Diarrhea with Neomycin and Potential Role for UGT1A1*28 Genotype Screening: A Double-Blind, Randomized, Placebo-Controlled Study. Oncologist 2006, 11, 944–954. [Google Scholar] [CrossRef]
- Kong, R.; Liu, T.; Zhu, X.; Ahmad, S.; Williams, A.L.; Phan, A.T.; Zhao, H.; Scott, J.E.; Yeh, L.A.; Wong, S.T.C. Old Drug New Use--Amoxapine and Its Metabolites as Potent Bacterial β-Glucuronidase Inhibitors for Alleviating Cancer Drug Toxicity. Clin. Cancer Res. 2014, 20, 3521–3530. [Google Scholar] [CrossRef]
- Yue, B.; Gao, R.; Lv, C.; Yu, Z.; Wang, H.; Geng, X.; Wang, Z.; Dou, W. Berberine Improves Irinotecan-Induced Intestinal Mucositis Without Impairing the Anti-Colorectal Cancer Efficacy of Irinotecan by Inhibiting Bacterial β-Glucuronidase. Front. Pharmacol. 2021, 12, 2880. [Google Scholar] [CrossRef]
- Emadi, A.; Jones, R.J.; Brodsky, R.A. Cyclophosphamide and Cancer: Golden Anniversary. Nat. Rev. Clin. Oncol. 2009, 6, 638–647. [Google Scholar] [CrossRef]
- Viaud, S.; Flament, C.; Zoubir, M.; Pautier, P.; LeCesne, A.; Ribrag, V.; Soria, J.C.; Marty, V.; Vielh, P.; Robert, C.; et al. Cyclophosphamide Induces Differentiation of Th17 Cells in Cancer Patients. Cancer Res. 2011, 71, 661–665. [Google Scholar] [CrossRef]
- Sistigu, A.; Viaud, S.; Chaput, N.; Bracci, L.; Proietti, E.; Zitvogel, L. Immunomodulatory Effects of Cyclophosphamide and Implementations for Vaccine Design. Semin. Immunopathol. 2011, 33, 369–383. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus Hirae and Barnesiella Intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Wang, J.; Li, M.; Gao, Y.; Li, H.; Fang, L.; Liu, C.; Liu, X.; Min, W. Effects of Exopolysaccharides from Lactiplantibacillus Plantarum JLAU103 on Intestinal Immune Response, Oxidative Stress, and Microbial Communities in Cyclophosphamide-Induced Immunosuppressed Mice. J. Agric. Food Chem. 2022, 70, 2197–2210. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zeng, Z.; Xie, Z.; Chen, G.; Chen, D.; Sun, Y.; Zeng, X.; Liu, Z. Effects of Polysaccharides from Fuzhuan Brick Tea on Immune Function and Gut Microbiota of Cyclophosphamide-Treated Mice. J. Nutr. Biochem. 2022, 101, 108947. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wang, Y.; Liu, R.; Ran, B.; Li, W. Structural Characterization and Protective Effect of Lonicerae Flos Polysaccharide on Cyclophosphamide-Induced Immunosuppression in Mice. Ecotoxicol. Environ. Saf. 2022, 230, 113174. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; Yu, Q.; Zheng, B.; Wang, H.; Wang, J.; Chen, S.; Nie, S.; Xie, M. Cultured Cordyceps Sinensis Polysaccharides Modulate Intestinal Mucosal Immunity and Gut Microbiota in Cyclophosphamide-Treated Mice. Carbohydr. Polym. 2020, 235, 115957. [Google Scholar] [CrossRef]
- Masetti, R.; Pession, A. First-Line Treatment of Acute Lymphoblastic Leukemia with Pegasparaginase. Biol. Targets Ther. 2009, 3, 359–368. [Google Scholar] [CrossRef][Green Version]
- Pession, A.; Valsecchi, M.G.; Masera, G.; Kamps, W.A.; Magyarosy, E.; Rizzari, C.; Van Wering, E.R.; Lo Nigro, L.; Van Der Does, A.; Locatelli, F.; et al. Long-Term Results of a Randomized Trial on Extended Use of High Dose L-Asparaginase for Standard Risk Childhood Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2005, 23, 7161–7167. [Google Scholar] [CrossRef]
- Silverman, L.B.; Gelber, R.D.; Dalton, V.K.; Asselin, B.L.; Barr, R.D.; Clavell, L.A.; Hurwitz, C.A.; Moghrabi, A.; Samson, Y.; Schorin, M.A.; et al. Improved Outcome for Children with Acute Lymphoblastic Leukemia: Results of Dana-Farber Consortium Protocol 91-01. Blood 2001, 97, 1211–1218. [Google Scholar] [CrossRef]
- Asselin, B.L.; Whitin, J.C.; Coppola, D.J.; Rupp, I.P.; Sallan, S.E.; Cohen, H.J. Comparative Pharmacokinetic Studies of Three Asparaginase Preparations. J. Clin. Oncol. 1993, 11, 1780–1786. [Google Scholar] [CrossRef]
- George, D.T.; Mathesius, U.; Behm, C.A.; Verma, N.K. The Periplasmic Enzyme, AnsB, of Shigella Flexneri Modulates Bacterial Adherence to Host Epithelial Cells. PLoS ONE 2014, 9, e94954. [Google Scholar] [CrossRef]
- Westman, E.L.; Canova, M.J.; Radhi, I.J.; Koteva, K.; Kireeva, I.; Waglechner, N.; Wright, G.D. Bacterial Inactivation of the Anticancer Drug Doxorubicin. Chem. Biol. 2012, 19, 1255–1264. [Google Scholar] [CrossRef]
- Blaustein, R.A.; Seed, P.C.; Hartmann, E.M. Biotransformation of Doxorubicin Promotes Resilience in Simplified Intestinal Microbial Communities. mSphere 2021, 6, e0006821. [Google Scholar] [CrossRef]
- Yan, A.; Culp, E.; Perry, J.; Lau, J.T.; Macneil, L.T.; Surette, M.G.; Wright, G.D. Transformation of the Anticancer Drug Doxorubicin in the Human Gut Microbiome. ACS Infect. Dis. 2018, 4, 68–76. [Google Scholar] [CrossRef]
- Sunshine, J.; Taube, J.M. PD-1/PD-L1 Inhibitors. Curr. Opin. Pharmacol. 2015, 23, 32–38. [Google Scholar] [CrossRef]
- Bie, F.; Tian, H.; Sun, N.; Zang, R.; Zhang, M.; Song, P.; Liu, L.; Peng, Y.; Bai, G.; Zhou, B.; et al. Research Progress of Anti-PD-1/PD-L1 Immunotherapy Related Mechanisms and Predictive Biomarkers in NSCLC. Front. Oncol. 2022, 12, 331. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The Commensal Microbiome Is Associated with Anti-PD-1 Efficacy in Metastatic Melanoma Patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A Defined Commensal Consortium Elicits CD8 T Cells and Anti-Cancer Immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Wadud Khan, M.A.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary Fiber and Probiotics Influence the Gut Microbiome and Melanoma Immunotherapy Response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef]
- Messaoudene, M.; Pidgeon, R.; Richard, C.; Ponce, M.; Diop, K.; Benlaifaoui, M.; Nolin-Lapalme, A.; Cauchois, F.; Malo, J.; Belkaid, W.; et al. A Natural Polyphenol Exerts Antitumor Activity and Circumvents Anti-PD-1 Resistance through Effects on the Gut Microbiota. Cancer Discov. 2022, 12, 1070–1087. [Google Scholar] [CrossRef]
- Ferrere, G.; Alou, M.T.; Liu, P.; Goubet, A.G.; Fidelle, M.; Kepp, O.; Durand, S.; Iebba, V.; Fluckiger, A.; Daillère, R.; et al. Ketogenic Diet and Ketone Bodies Enhance the Anticancer Effects of PD-1 Blockade. JCI Insight 2021, 6, e145207. [Google Scholar] [CrossRef]
- Miller, P.L.; Carson, T.L. Mechanisms and Microbial Influences on CTLA-4 and PD-1-Based Immunotherapy in the Treatment of Cancer: A Narrative Review. Gut Pathog. 2020, 12, 43. [Google Scholar] [CrossRef]
- Dees, K.J.; Koo, H.; Humphreys, J.F.; Hakim, J.A.; Crossman, D.K.; Crowley, M.R.; Nabors, L.B.; Benveniste, E.N.; Morrow, C.D.; McFarland, B.C. Human Gut Microbial Communities Dictate Efficacy of Anti-PD-1 Therapy in a Humanized Microbiome Mouse Model of Glioma. Neuro-Oncology Adv. 2021, 3, vdab023. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.F.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, T.; Tu, X.; Huang, Y.; Zhang, H.; Tan, D.; Jiang, W.; Cai, S.; Zhao, P.; Song, R.; et al. Gut Microbiome Affects the Response to Anti-PD-1 Immunotherapy in Patients with Hepatocellular Carcinoma. J. Immunother. Cancer 2019, 7, 193. [Google Scholar] [CrossRef]
- Gabarre, P.; Loens, C.; Tamzali, Y.; Barrou, B.; Jaisser, F.; Tourret, J. Immunosuppressive Therapy after Solid Organ Transplantation and the Gut Microbiota: Bidirectional Interactions with Clinical Consequences. Am. J. Transplant. 2021, 22, 1014–1030. [Google Scholar] [CrossRef]
- Kapturczak, M.H.; Meier-Kriesche, H.U.; Kaplan, B. Pharmacology of Calcineurin Antagonists. Transplant. Proc. 2004, 36, S25–S32. [Google Scholar] [CrossRef]
- Lee, J.R.; Muthukumar, T.; Dadhania, D.; Taur, Y.; Jenq, R.R.; Toussaint, N.C.; Ling, L.; Pamer, E.; Suthanthiran, M. Gut Microbiota and Tacrolimus Dosing in Kidney Transplantation. PLoS ONE 2015, 10, e0122399. [Google Scholar] [CrossRef]
- Cain, D.W.; Cidlowski, J.A. Immune Regulation by Glucocorticoids. Nat. Rev. Immunol. 2017, 17, 233–247. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Ikegawa, S.; Alves, J.M.P.; Zhou, B.; Kobayashi, A.; Iida, T.; Mitamura, K.; Tanabe, G.; Serrano, M.; De Guzman, A.; et al. Clostridium Scindens: A Human Gut Microbe with a High Potential to Convert Glucocorticoids into Androgens. J. Lipid Res. 2013, 54, 2437–2449. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.M.; Kearns, G.L. Proton Pump Inhibitors in Pediatrics: Mechanism of Action, Pharmacokinetics, Pharmacogenetics, and Pharmacodynamics. Pediatr. Drugs 2013, 15, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.A.; Verdi, S.; Maxan, M.E.; Shin, C.M.; Zierer, J.; Bowyer, R.C.E.; Martin, T.; Williams, F.M.K.; Menni, C.; Bell, J.T.; et al. Gut Microbiota Associations with Common Diseases and Prescription Medications in a Population-Based Cohort. Nat. Commun. 2018, 9, 2655. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive Impact of Non-Antibiotic Drugs on Human Gut Bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Masetti, R.; Biagi, E.; Zama, D.; Muratore, E.; D’Amico, F.; Leardini, D.; Turroni, S.; Prete, A.; Brigidi, P.; Pession, A. Early Modifications of the Gut Microbiome in Children with Hepatic Sinusoidal Obstruction Syndrome after Hematopoietic Stem Cell Transplantation. Sci. Rep. 2021, 11, 14307. [Google Scholar] [CrossRef]
- Winston, J.A.; Rivera, A.; Cai, J.; Patterson, A.D.; Theriot, C.M. Secondary Bile Acid Ursodeoxycholic Acid Alters Weight, the Gut Microbiota, and the Bile Acid Pool in Conventional Mice. PLoS ONE 2021, 16, e0246161. [Google Scholar] [CrossRef]
- Pession, A.; Zama, D.; Muratore, E.; Leardini, D.; Gori, D.; Guaraldi, F.; Prete, A.; Turroni, S.; Brigidi, P.; Masetti, R. Fecal Microbiota Transplantation in Allogeneic Hematopoietic Stem Cell Transplantation Recipients: A Systematic Review. J. Pers. Med. 2021, 11, 100. [Google Scholar] [CrossRef]
| Drug Name | Year | First Author | Setting | Sample Size | Interaction with Gut Microbiome | Ref |
|---|---|---|---|---|---|---|
| Irinotecan (CPT-11) | 2015 | Wallace, BD | Preclinical | / | Intestine bacteria producing β-glucuronidase can convert non-toxic CPT-11 metabolite (SN-38-G) to toxic metabolite (SN-38), causing diarrhea. | [23] |
| Irinotecan (CPT-11) | 2008 | Stringer, AM | Preclinical, rats | / | ↑ number of β-glucuronidases-expressing species. | [24] |
| Cyclophosphamide | 2013 | Viaud, S | Preclinical | / | Translocation of specific Gram-positive bacteria from the intestine to secondary lymphoid organs was critical for the differentiation of CD4+ T cells into Th1 and Th17 cells. | [25] |
| Cyclophosphamide | 2015 | Xu, X | Preclinical | / | ↑ Firmicutes, ↓ Bacteroidetes. | [26] |
| L-asparaginase | 2021 | Dunn, KA | Pediatric ALL | 12 patients | ↑ Escherichia in the community if decreased-activity, ↑ Bacteroides and Streptococcus in the community if increased-activity. | [27] |
| Anti-PD1 | 2018 | Gopalakrishnan, V | Adults, melanoma | 112 patients | ↑ α-diversity of responders to anti-PD1 therapy. Higher proportion of Ruminococcaceae, Faecalibacterium, and Bifidobacterium spp. reported in responders. | [28] |
| Anti-PD1 | 2018 | Routy, B | Mice, Adults | Mice, 249 treated | ↑ Akkermansia, Ruminococcus spp., Alistipes spp., and Eubacterium spp in responders. ↓ Bifidobacterium adolescentis, B. longum, and Parabacteroids distasonis in responders. | [29] |
| Cyclosporine | 2019 | Jia et al. | Preclinical | 8 treated | ↑ gut microbial richness, Enterobacteriaceae. ↓ F. prausnitzii, Clostridium clusters I and XIV. | [30] |
| Cyclosporine | 2020 | O Reilly et al. | Adults | 6 ex vivo, 8 in vivo | No significant α and β diversity before and after treatment. | [31] |
| Tacrolimus | 2017 | Zhang et al. | Mice | 8 treated | No change in bacterial richness and diversity. ↑ genera Allobaculum, Bacteroides and Lactobacillus. ↓ Clostridiales, Ruminococcaceae, Rikenella, Ruminococcaceae and Oscillospira. | [32] |
| Tacrolimus | 2017 | Bhat et al. | Mice | 5 treated | ↓ Mollicutes, Micrococcaceae, Actinomycetales, Roseburia, Oscillospira, Rothia and Staphylococcus. ↑ A. muciniphila. | [33] |
| Tacrolimus | 2018 | Toral et al. | Mice | 8 treated | ↓ microbial diversity. ↑ Firmicutes/Bacteroidetes ratio. | [34] |
| Tacrolimus | 2018 | Jiang et al. | Mice | 8 high dosage, 8 medium dosage, 8 low dosage | Intermediate dose: ↑ Bifidobacterium, Faecalibacterium prausnitzii ↓ less Enterobacteriaceae, Bacteroides-Prevotella Low and high doses: ↑ Enterobacteriaceae ↓ Bifidobacterium, Faecalibacterium prausnitzii. | [35] |
| MMF | 2018 | Flannigan et al. | Mice | 9 treated | ↓ overall diversity ↑ Proteobacteria (Escherichia/Shigella), Deferribacteres, Firmicutes ↓ Bacteroidetes and Verrucomicrobia phyla, Akkermansia, Parabacteroides and Clostridium genera. | [36] |
| Rapamycin | 2017 | Bhat et al., | Mice | 5 treated | ↓ bacterial diversity. ↓ Roseburia, Oscillospira, Mollicutes, Rothia, Micrococcaceae, Acninomycetales and Staphylococcus. | [33] |
| Rapamycin | 2016 | Jung et al. | Mice | 5 treated | ↓ Turicibacter, unclassified Marinilabiliaceae, Alloprevotella. ↑ Ruminococcus. | [37] |
| Alemtuzumab | 2013 | Li et al. | Monkeys | 15 treated | ↑ Enterobacteriales order and Prevotella genus. ↓ Lactobacillales order. | [38] |
| Steroids | 2014 | Lee et al. | Humans | 4 treated | ↓ Clostridiales ↑ Erysipelotrichales. | [39] |
| Steroids | 2016 | Tourret et al. | Mice | 8–10 treated | ↑ Firmicutes/Bacteroidetes ratio ↓ Clostridium sensu stricto. | [40] |
| Steroids | 2017 | Wu et al. | Mice | 30 lower dose, 30 higher dose | ↓ bacterial richness and diversity. ↓ Firmicutes, Bacteroides, Actinobacteria, α and γ Proteobacteria, Clostridiales and Lactobacillus. ↑ Proteobacteria. | [41] |
| Steroids | 2019 | He et al. | Mice | 10 treated | ↓ Proteobacteria, Deferribacteres, Rikenella, Mucispirillum, Oscillospira and Bilophila. ↑ Prevotella and Anaerostipes. | [42] |
| Steroids | 2020 | Vich Vila et al. | Adults | 17 treated | ↑ Methanobrevibacter smithii and Streptococcus salivarius. | [43] |
| PPI | 2016 | Jackson et al. | Adults | 1827 | ↓ diversity in PPI users. ↑ Lactobacillales order, families Micrococcaceae and Streptococcaceae, genera Rothia and Streptococcus, species Rothia mucilaginosa and Streptococcus anginosus. ↓ families Erysipelotrichaceae, Lachnospiraceae, Ruminococcaceae, genera Firmicutes, species Erysipelotrichales and Clostridiales. | [44] |
| PPI | 2015 | Imhann et al. | Adults | 99 treated | ↓ species richness and ↓ Shannon diversity, although not significant. ↑ Gammaproteobacteria class, Actinomycetales order, families Streptococcaceae and Micrococcaceae, genera Rothia, Streptococcus and Veilonella, species Lactobacillus salivarius. | [45] |
| PPI | 2015 | Freedberg et al. | Adults | 12 treated | No changes in diversity. ↑ families Enterococcaceae, Streptococcaceae, Micrococcaceae and Staphylococcaceae. ↓ Clostridiales. | [46] |
| PPI | 2015 | Tsuda et al. | Adults | 18 treated | No changes in α diversity, increased β diversity. ↓ genus Faecalibacterium. | [47] |
| PPI | 2020 | Vich Vila et al. | Adults | 108 treated | ↑ species Veillonella parvula, Streptococcus salivarius, Streptococcus parasanguinis, Streptococcus vestibularis, Bifidobacterium dentium, Haemophilus parainfluenzae. | [43] |
| PPI | 2021 | Simakachorn et al. | Pediatrics | 20 treated | No significant change in α and β diversity. No change in total number of species-level taxonomy categories. | [48] |
| UDCA | 2018 | Pearson et al. | Adults | 661 treated | No change in microbial richness. ↑ Streptocuccus, Escherichia and Bilophila spp., Faecalibacterium prausnitzii; ↓ Fusobacterium spp., Ruminococcus gnavus. | [20] |
| UDCA | 2018 | Tang et al. | Adults | 60 treated | ↑ Enterobacteriaceae. | [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leardini, D.; Venturelli, F.; Baccelli, F.; Cerasi, S.; Muratore, E.; Brigidi, P.; Pession, A.; Prete, A.; Masetti, R. Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome. Int. J. Mol. Sci. 2022, 23, 15387. https://doi.org/10.3390/ijms232315387
Leardini D, Venturelli F, Baccelli F, Cerasi S, Muratore E, Brigidi P, Pession A, Prete A, Masetti R. Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome. International Journal of Molecular Sciences. 2022; 23(23):15387. https://doi.org/10.3390/ijms232315387
Chicago/Turabian StyleLeardini, Davide, Francesco Venturelli, Francesco Baccelli, Sara Cerasi, Edoardo Muratore, Patrizia Brigidi, Andrea Pession, Arcangelo Prete, and Riccardo Masetti. 2022. "Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome" International Journal of Molecular Sciences 23, no. 23: 15387. https://doi.org/10.3390/ijms232315387
APA StyleLeardini, D., Venturelli, F., Baccelli, F., Cerasi, S., Muratore, E., Brigidi, P., Pession, A., Prete, A., & Masetti, R. (2022). Pharmacomicrobiomics in Pediatric Oncology: The Complex Interplay between Commonly Used Drugs and Gut Microbiome. International Journal of Molecular Sciences, 23(23), 15387. https://doi.org/10.3390/ijms232315387









