Cis-Regulation by NACs: A Promising Frontier in Wheat Crop Improvement
Abstract
1. Introduction
2. Structural Attributes of NAC
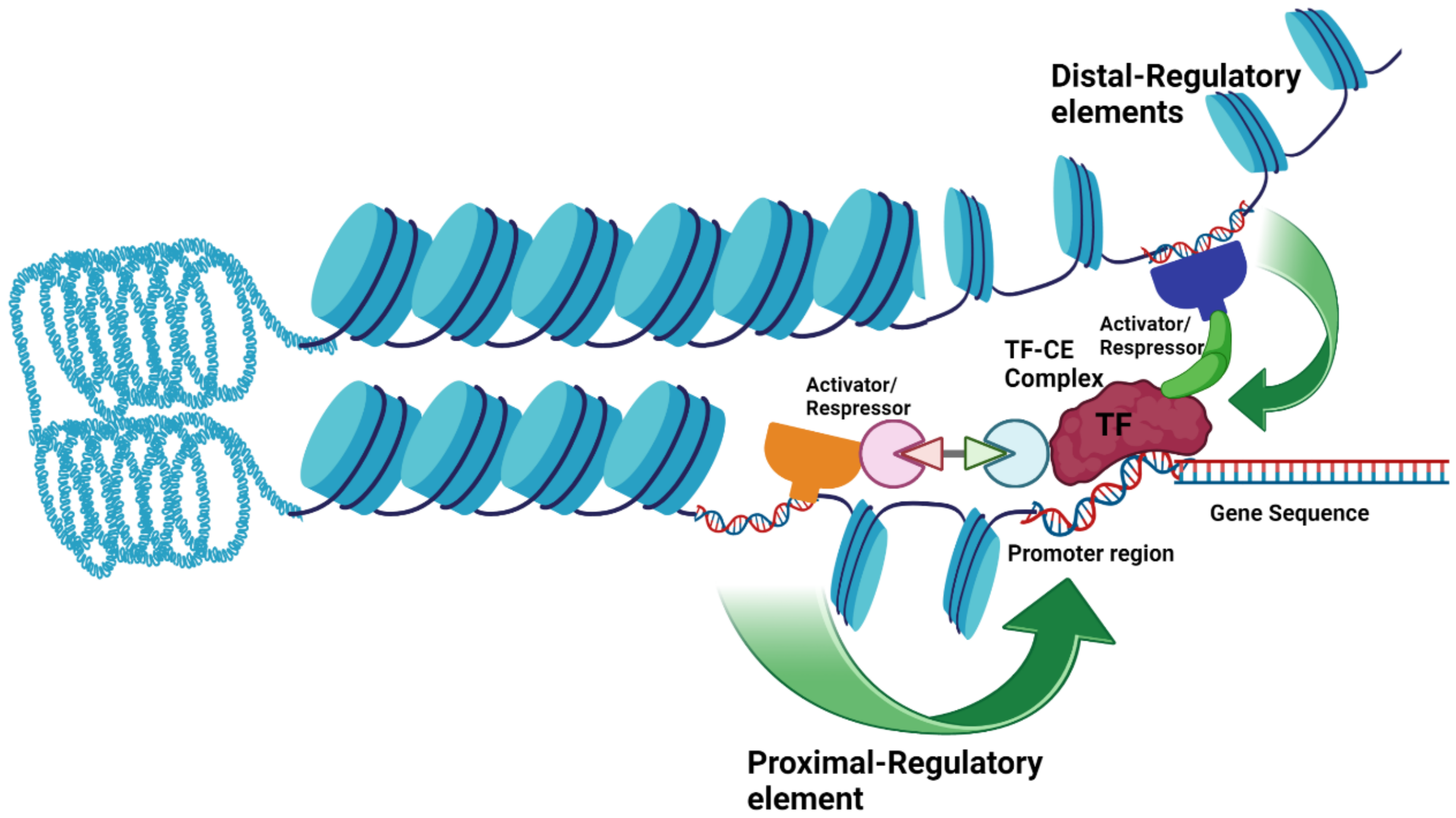
3. Cis-Regulation
4. Yield-Related Traits in Wheat and Other Cereals
5. Functional Features of TaNACs
5.1. Regulation of Phytohormonal Homeostasis by NAC TFs
5.2. Leaf Senescence
5.3. Seed-Associated Traits Regulated by Wheat NACs
5.4. NAC-Dependent Root Modulation in Wheat
5.5. Role of Wheat NACs in Abiotic Stresses
5.6. Role of Wheat NACs in Biotic Stress
6. Future Prospects and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Faris, J.D. Wheat domestication: Key to agricultural revolutions past and future. In Genomics of Plant Genetic Resources; Springer: Berlin/Heidelberg, Germany, 2014; pp. 439–464. [Google Scholar]
- Schnurbusch, T. Wheat and barley biology: Towards new frontiers. J. Integr. Plant Biol. 2019, 61, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Luo, G.; Song, Y.; Xu, J.; Ji, J.; Zhang, C.; Gregová, E.; Yang, W.; Li, X.; Sun, J. A novel NAC family transcription factor SPR suppresses seed storage protein synthesis in wheat. Plant Biotechnol. J. 2021, 19, 992–1007. [Google Scholar] [PubMed]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—Being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef] [PubMed]
- Romani, F.; Moreno, J.E. Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytol. 2021, 230, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-S.; Lai, F.-J. Functional redundancy of transcription factors explains why most binding targets of a transcription factor are not affected when the transcription factor is knocked out. BMC Syst. Biol. 2015, 9, S2. [Google Scholar]
- Johnson, A.D. The rewiring of transcription circuits in evolution. Curr. Opin. Genet. Dev. 2017, 47, 121–127. [Google Scholar]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Klíma, M.; Roy, A.; Prášil, I.T. Biological networks underlying abiotic stress tolerance in temperate crops—A proteomic perspective. Int. J. Mol. Sci. 2015, 16, 20913–20942. [Google Scholar] [CrossRef]
- Singh, S.; Grover, A.; Nasim, M. Biofuel potential of plants transformed genetically with NAC family genes. Front. Plant Sci. 2016, 7, 22. [Google Scholar] [CrossRef]
- Ernst, H.A.; Nina Olsen, A.; Skriver, K.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar]
- Liu, G.-S.; Li, H.-L.; Grierson, D.; Fu, D.-Q. NAC transcription factor family regulation of fruit ripening and quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63. [Google Scholar]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar]
- Grover, A.; Singh, S.; Pandey, P.; Patade, V.Y.; Gupta, S.M.; Nasim, M. Overexpression of NAC gene from Lepidium latifolium L. enhances biomass, shortens life cycle and induces cold stress tolerance in tobacco: Potential for engineering fourth generation biofuel crops. Mol. Biol. Rep. 2014, 41, 7479–7489. [Google Scholar] [CrossRef]
- Marand, A.P.; Schmitz, R.J. Single-cell analysis of cis-regulatory elements. Curr. Opin. Plant Biol. 2022, 65, 102094. [Google Scholar] [PubMed]
- Uygun, S.; Azodi, C.B.; Shiu, S.-H. Cis-regulatory code for predicting plant cell-type transcriptional response to high salinity. Plant Physiol. 2019, 181, 1739–1751. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Marand, A.P.; Ricci, W.A.; Ethridge, C.L.; Zhang, X.; Schmitz, R.J. The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 2019, 5, 1250–1259. [Google Scholar] [PubMed]
- Ricci, W.A.; Lu, Z.; Ji, L.; Marand, A.P.; Ethridge, C.L.; Murphy, N.G.; Noshay, J.M.; Galli, M.; Mejía-Guerra, M.K.; Colomé-Tatché, M. Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 2019, 5, 1237–1249. [Google Scholar] [PubMed]
- Sun, J.; He, N.; Niu, L.; Huang, Y.; Shen, W.; Zhang, Y.; Li, L.; Hou, C. Global quantitative mapping of enhancers in rice by STARR-seq. Genom. Proteom. Bioinform. 2019, 17, 140–153. [Google Scholar] [CrossRef]
- Jana, T.; Brodsky, S.; Barkai, N. Speed–specificity trade-offs in the transcription factors search for their genomic binding sites. Trends Genet. 2021, 37, 421–432. [Google Scholar]
- Dror, I.; Golan, T.; Levy, C.; Rohs, R.; Mandel-Gutfreund, Y. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 2015, 25, 1268–1280. [Google Scholar] [CrossRef]
- Gurdon, J.; Javed, K.; Vodnala, M.; Garrett, N. Long-term association of a transcription factor with its chromatin binding site can stabilize gene expression and cell fate commitment. Proc. Natl. Acad. Sci. USA 2020, 117, 15075–15084. [Google Scholar]
- Huang, S.s.C.; Ecker, J.R. Piecing together cis-regulatory networks: Insights from epigenomics studies in plants. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1411. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, L.; Zhang, Y.; Lin, D.; Xu, W.; Ke, C.; Han, L.; Deng, L.; Li, G.; Jackson, D. 3D genome architecture coordinates trans and cis regulation of differentially expressed ear and tassel genes in maize. Genome Biol. 2020, 21, 143. [Google Scholar]
- Worsley Hunt, R.; Wasserman, W.W. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biol. 2014, 15, 412. [Google Scholar] [CrossRef]
- Biswas, A.; Narlikar, L. Resolving diverse protein–DNA footprints from exonuclease-based ChIP experiments. Bioinformatics 2021, 37, i367–i375. [Google Scholar] [CrossRef]
- Li, J.; Xie, L.; Tian, X.; Liu, S.; Xu, D.; Jin, H.; Song, J.; Dong, Y.; Zhao, D.; Li, G. TaNAC100 acts as an integrator of seed protein and starch synthesis exerting pleiotropic effects on agronomic traits in wheat. Plant J. 2021, 108, 829–840. [Google Scholar] [CrossRef]
- Hajheidari, M.; Huang, S.-s.C. Elucidating the biology of transcription factor–DNA interaction for accurate identification of cis-regulatory elements. Curr. Opin. Plant Biol. 2022, 68, 102232. [Google Scholar]
- Lu, Z.; Hofmeister, B.T.; Vollmers, C.; DuBois, R.M.; Schmitz, R.J. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 2017, 45, e41. [Google Scholar] [CrossRef]
- Stalder, J.; Larsen, A.; Engel, J.D.; Dolan, M.; Groudine, M.; Weintraub, H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 1980, 20, 451–460. [Google Scholar]
- Minnoye, L.; Marinov, G.K.; Krausgruber, T.; Pan, L.; Marand, A.P.; Secchia, S.; Greenleaf, W.J.; Furlong, E.E.; Zhao, K.; Schmitz, R.J. Chromatin accessibility profiling methods. Nat. Rev. Methods Prim. 2021, 1, 10. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, J.; Wang, X.; Li, T.; Majeed, U.; Hao, C.; Zhang, X. The NAC transcription factor NAC019-A1 is a negative regulator of starch synthesis in wheat developing endosperm. J. Exp. Bot. 2020, 71, 5794–5807. [Google Scholar]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 2020, 18, 614–630. [Google Scholar] [CrossRef]
- Feng, N.; Song, G.; Guan, J.; Chen, K.; Jia, M.; Huang, D.; Wu, J.; Zhang, L.; Kong, X.; Geng, S. Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol. 2017, 174, 1779–1794. [Google Scholar]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [PubMed]
- Gauley, A.; Boden, S.A. Genetic pathways controlling inflorescence architecture and development in wheat and barley. J. Integr. Plant Biol. 2019, 61, 296–309. [Google Scholar] [PubMed]
- Brocklehurst, P. Factors controlling grain weight in wheat. Nature 1977, 266, 348–349. [Google Scholar] [CrossRef]
- Millet, E.; Pinthus, M. The association between grain volume and grain weight in wheat. J. Cereal Sci. 1984, 2, 31–35. [Google Scholar] [CrossRef]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef]
- Landrein, B.; Formosa-Jordan, P.; Malivert, A.; Schuster, C.; Melnyk, C.W.; Yang, W.; Turnbull, C.; Meyerowitz, E.M.; Locke, J.C.; Jönsson, H. Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. USA 2018, 115, 1382–1387. [Google Scholar]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar]
- Sakakibara, H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 2021, 105, 421–430. [Google Scholar] [CrossRef]
- Przetakiewicz, A.; Orczyk, W.; Nadolska-Orczyk, A. The effect of auxin on plant regeneration of wheat, barley and triticale. Plant Cell Tissue Organ Cult. 2003, 73, 245–256. [Google Scholar]
- Nguyen, H.N.; Perry, L.; Kisiala, A.; Olechowski, H.; Emery, R. Cytokinin activity during early kernel development corresponds positively with yield potential and later stage ABA accumulation in field-grown wheat (Triticum aestivum L.). Planta 2020, 252, 76. [Google Scholar]
- Youssef, H.M.; Hansson, M. Crosstalk among hormones in barley spike contributes to the yield. Plant Cell Rep. 2019, 38, 1013–1016. [Google Scholar] [CrossRef]
- Shanks, C.M.; Hecker, A.; Cheng, C.Y.; Brand, L.; Collani, S.; Schmid, M.; Schaller, G.E.; Wanke, D.; Harter, K.; Kieber, J.J. Role of BASIC PENTACYSTEINE transcription factors in a subset of cytokinin signaling responses. Plant J. 2018, 95, 458–473. [Google Scholar] [CrossRef]
- Christiansen, M.W.; Holm, P.B.; Gregersen, P.L. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 2011, 4, 302. [Google Scholar] [CrossRef]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Mao, C.; He, J.; Liu, L.; Deng, Q.; Yao, X.; Liu, C.; Qiao, Y.; Li, P.; Ming, F. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 2020, 18, 429–442. [Google Scholar]
- Jablonski, B.; Bajguz, A.; Bocian, J.; Orczyk, W.; Nadolska-Orczyk, A. Genotype-Dependent Effect of Silencing of TaCKX1 and TaCKX2 on Phytohormone Crosstalk and Yield-Related Traits in Wheat. Int. J. Mol. Sci. 2021, 22, 11494. [Google Scholar]
- Sultana, N.; Islam, S.; Juhasz, A.; Ma, W. Wheat leaf senescence and its regulatory gene network. Crop J. 2021, 9, 703–717. [Google Scholar] [CrossRef]
- Chapman, E.A.; Orford, S.; Lage, J.; Griffiths, S. Delaying or delivering: Identification of novel NAM-1 alleles that delay senescence to extend wheat grain fill duration. J. Exp. Bot. 2021, 72, 7710–7728. [Google Scholar] [CrossRef]
- Gregersen, P.; Holm, P.; Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008, 10, 37–49. [Google Scholar] [CrossRef]
- See, D.; Kanazin, V.; Kephart, K.; Blake, T. Mapping genes controlling variation in barley grain protein concentration. Crop Sci. 2002, 42, 680–685. [Google Scholar]
- Balibrea Lara, M.E.; Gonzalez Garcia, M.-C.; Fatima, T.; Ehneß, R.; Lee, T.K.; Proels, R.; Tanner, W.; Roitsch, T. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.W.; Matthewman, C.; Podzimska-Sroka, D.; O’Shea, C.; Lindemose, S.; Møllegaard, N.E.; Holme, I.B.; Hebelstrup, K.; Skriver, K.; Gregersen, P.L. Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J. Exp. Bot. 2016, 67, 5259–5273. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Derkx, A.; Liu, D.C.; Buchner, P.; Hawkesford, M. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar]
- Sultana, N. Characterization of TaNAC-S Gene in Australian Wheat Cultivars in Relation to Senescence and Nitrogen Stress Response. Ph.D. Dissertation, Murdoch University, Perth, Australia, 2020. [Google Scholar]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar]
- Li, W.; He, X.; Chen, Y.; Jing, Y.; Shen, C.; Yang, J.; Teng, W.; Zhao, X.; Hu, W.; Hu, M. A wheat transcription factor positively sets seed vigour by regulating the grain nitrate signal. New Phytol. 2020, 225, 1667–1680. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Guo, Y. Wheat transcription factor TaSNAC11-4B positively regulates leaf senescence through promoting ROS production in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7672. [Google Scholar] [CrossRef]
- Harrington, S.A.; Overend, L.E.; Cobo, N.; Borrill, P.; Uauy, C. Conserved residues in the wheat (Triticum aestivum) NAM-A1 NAC domain are required for protein binding and when mutated lead to delayed peduncle and flag leaf senescence. BMC Plant Biol. 2019, 19, 407. [Google Scholar] [CrossRef]
- Asplund, L.; Hagenblad, J.; Leino, M.W. Re-evaluating the history of the wheat domestication gene NAM-B1 using historical plant material. J. Archaeol. Sci. 2010, 37, 2303–2307. [Google Scholar] [CrossRef]
- Brevis, J.C.; Morris, C.F.; Manthey, F.; Dubcovsky, J. Effect of the grain protein content locus Gpc-B1 on bread and pasta quality. J. Cereal Sci. 2010, 51, 357–365. [Google Scholar] [CrossRef]
- Distelfeld, A.; Pearce, S.P.; Avni, R.; Scherer, B.; Uauy, C.; Piston, F.; Slade, A.; Zhao, R.; Dubcovsky, J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 2012, 78, 515–524. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef]
- Luo, G.; Shen, L.; Zhao, S.; Li, R.; Song, Y.; Song, S.; Yu, K.; Yang, W.; Li, X.; Sun, J. Genome-wide identification of seed storage protein gene regulators in wheat through coexpression analysis. Plant J. 2021, 108, 1704–1720. [Google Scholar]
- Shewry, P.R.; Underwood, C.; Wan, Y.; Lovegrove, A.; Bhandari, D.; Toole, G.; Mills, E.C.; Denyer, K.; Mitchell, R.A. Storage product synthesis and accumulation in developing grains of wheat. J. Cereal Sci. 2009, 50, 106–112. [Google Scholar] [CrossRef]
- Sonnewald, U.; Kossmann, J. Starches—From current models to genetic engineering. Plant Biotechnol. J. 2013, 11, 223–232. [Google Scholar]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef]
- Kumar, R.; Mukherjee, S.; Ayele, B.T. Molecular aspects of sucrose transport and its metabolism to starch during seed development in wheat: A comprehensive review. Biotechnol. Adv. 2018, 36, 954–967. [Google Scholar] [CrossRef]
- Wang, J.-C.; Xu, H.; Zhu, Y.; Liu, Q.-Q.; Cai, X.-L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef]
- Bai, A.-N.; Lu, X.-D.; Li, D.-Q.; Liu, J.-X.; Liu, C.-M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016, 26, 384–388. [Google Scholar] [CrossRef]
- Xiong, Y.; Ren, Y.; Li, W.; Wu, F.; Yang, W.; Huang, X.; Yao, J. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [PubMed]
- Zhang, J.; Chen, J.; Yi, Q.; Hu, Y.; Liu, H.; Liu, Y.; Huang, Y. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol. 2014, 84, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [PubMed]
- Wang, J.; Chen, Z.; Zhang, Q.; Meng, S.; Wei, C. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020, 184, 1775–1791. [Google Scholar] [CrossRef] [PubMed]
- Wasson, A.; Richards, R.; Chatrath, R.; Misra, S.; Prasad, S.S.; Rebetzke, G.; Kirkegaard, J.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar]
- Chen, D.; Chai, S.; McIntyre, C.L.; Xue, G.-P. Overexpression of a predominantly root-expressed NAC transcription factor in wheat roots enhances root length, biomass and drought tolerance. Plant Cell Rep. 2018, 37, 225–237. [Google Scholar] [CrossRef]
- Mao, H.; Li, S.; Wang, Z.; Cheng, X.; Li, F.; Mei, F.; Chen, N.; Kang, Z. Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 2020, 18, 1078–1092. [Google Scholar] [CrossRef]
- Qing, C.; Du, L.-y.; Wen, M.; Niu, R.-y.; Wu, B.-w.; Guo, L.-j.; Meng, M.; Liu, X.-l.; Zhao, H.-x. MiR164-TaNAC14 module regulates root development and abiotic-stress tolerance of wheat seedlings. J. Integr. Agric. 2022, in press. [Google Scholar]
- Long, S.P.; Ort, D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010, 13, 240–247. [Google Scholar]
- Rogers, E.D.; Benfey, P.N. Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Jensen, M.K.; Skriver, K. NAC transcription factor gene regulatory and protein–protein interaction networks in plant stress responses and senescence. Iubmb Life 2014, 66, 156–166. [Google Scholar]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, J.; Zhang, N.; Xin, M.; Peng, H.; Hu, Z.; Ni, Z.; Du, J. The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS ONE 2015, 10, e0135667. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Pang, H.; Fu, J. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J. Plant Physiol. 2012, 169, 146–156. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, C.; Xie, B.; Liu, Y.; Cui, J.; Chen, G.; Zhang, Y. Model analysing the antioxidant responses of leaves and roots of switchgrass to NaCl-salinity stress. Plant Physiol. Biochem. 2012, 58, 288–296. [Google Scholar] [CrossRef]
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015, 168, 1122–1139. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T. JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 2016, 6, 1174. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Saad, A.S.I.; Li, X.; Li, H.-P.; Huang, T.; Gao, C.-S.; Guo, M.-W.; Cheng, W.; Zhao, G.-Y.; Liao, Y.-C. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013, 203, 33–40. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [PubMed]
- Xue, G.-P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [PubMed]
- Xue, G.-P.; Bower, N.I.; McIntyre, C.L.; Riding, G.A.; Kazan, K.; Shorter, R. TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct. Plant Biol. 2006, 33, 43–57. [Google Scholar] [PubMed]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 2014, 9, e84359. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Newman, I.; Zhou, M.; Mendham, N.; Zhang, G.; Shabala, S. Screening plants for salt tolerance by measuring K+ flux: A case study for barley. Plant Cell Environ. 2005, 28, 1230–1246. [Google Scholar]
- Chen, Z.; Pottosin, I.I.; Cuin, T.A.; Fuglsang, A.T.; Tester, M.; Jha, D.; Zepeda-Jazo, I.; Zhou, M.; Palmgren, M.G.; Newman, I.A. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 2007, 145, 1714–1725. [Google Scholar] [CrossRef]
- Cuin, T.; Zhou, M.; Parsons, D.; Shabala, S. Genetic behaviour of physiological traits conferring cytosolic K+/Na+ homeostasis in wheat. Plant Biol. 2012, 14, 438–446. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, M.; Lv, W.; Tang, X.; Zhao, D.; Wang, L.; Li, C.; Jiang, L. Overexpression of TaSNAC4-3D in Common Wheat (Triticum aestivum L.) Negatively Regulates Drought Tolerance. Front. Plant Sci. 2022, 13, 945272. [Google Scholar] [CrossRef]
- Mei, F.; Chen, B.; Li, F.; Zhang, Y.; Kang, Z.; Wang, X.; Mao, H. Overexpression of the wheat NAC transcription factor TaSNAC4-3A gene confers drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 160, 37–50. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 10.1146. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar]
- Kaneda, T.; Taga, Y.; Takai, R.; Iwano, M.; Matsui, H.; Takayama, S.; Isogai, A.; Che, F.S. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009, 28, 926–936. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeon, H.S.; Kim, H.G.; Park, O.K. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017, 214, 343–360. [Google Scholar] [CrossRef]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar]
- Lv, S.; Guo, H.; Zhang, M.; Wang, Q.; Zhang, H.; Ji, W. Large-scale cloning and comparative analysis of TaNAC genes in response to stripe rust and powdery mildew in wheat (Triticum aestivum L.). Genes 2020, 11, 1073. [Google Scholar] [CrossRef]
- Zhang, N.; Yuan, S.; Zhao, C.; Park, R.F.; Wen, X.; Yang, W.; Liu, D. TaNAC35 acts as a negative regulator for leaf rust resistance in a compatible interaction between common wheat and Puccinia triticina. Mol. Genet. Genom. 2021, 296, 279–287. [Google Scholar]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef]
- Zhang, X.-m.; Zhang, Q.; Pei, C.-l.; Li, X.; Huang, X.-l.; Chang, C.-y.; Wang, X.-j.; Huang, L.-l.; Kang, Z.-s. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiol. Mol. Plant Pathol. 2018, 102, 144–153. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Sun, Y.-F.; Zhu, L.; Xu, L.-S.; Chen, X.-M.; Liu, B.; Yu, Y.-T.; Wang, X.-J.; Huang, L.-L. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 2010, 74, 394–402. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Liu, X.-Y.; Deng, L.; Cai, G.-L.; Zhang, Y.; Wang, X.-J.; Zhao, J.; Huang, L.-L.; Kang, Z.-S. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 2010, 37, 3703–3712. [Google Scholar] [PubMed]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [PubMed]
- Zhou, W.; Qian, C.; Li, R.; Zhou, S.; Zhang, R.; Xiao, J.; Wang, X.; Zhang, S.; Xing, L.; Cao, A. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018, 277, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Kahla, A.; Vranić, M.; Jia, J.; Malla, K.B.; Craze, M.; Wallington, E.; Doohan, F.M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol. J. 2019, 17, 1892–1904. [Google Scholar] [PubMed]




| TaNACss | Associated Function | Positive/ Negative (+/−) Regulation | Method of Functional Characterization | Co-Regulation /Interaction /Activation of Other Genes | Cis-Regulatory Sites | References |
|---|---|---|---|---|---|---|
| TaNAC74 | Seed storage proteins (SSPs), seed germination rate | − regulator of SSPs + regulator of seed germination rate | Overexpression and knockdown | TaSPR | 5′-CANNTG-3′ | [3] |
| TaNAC100 | Seed storage proteins (SSPs), starch, grain size, and weight | − regulator of SSPs + regulator of seed starch, seed size, and thousand seed weight | Overexpression | TaGBSS1 and TaSUS2 | - | [34] |
| TaNAC-S-A1 | Grain yield, chlorophyll contents | + regulator of grain yield and chlorophyll contents | Transcriptome analysis | - | - | [59,67] |
| TaNAC-S-7B2 | Grain protein contents | + regulator of grain protein contents | Transcriptome analysis | - | - | [59,67] |
| TaNAC19-A1 | Starch synthesis | − regulator of starch biosynthesis in endosperms | Overexpression | TaAGPS1-A1 and TaAGPS1-B1 | 5′-ACGCAG-3′ | [39] |
| TaNAC2-5A | Seeds vigor | + regulator of seeds vigor | Overexpression | TaNRT2.5-3B | - | [69] |
| NAM-B1 | Grain protein, leaf senescence | + regulator of grain protein, leaf senescence | Knockdown | - | - | [47] |
| TaNAC-S | Leaf senescence, grain yield, and grain protein contents | − regulator of leaf senescence + regulator of grain yield and grain protein contents | Overexpression | - | - | [66] |
| TaSNAC11-4B | Leaf senescence | + regulator of leaf senescence | ABA-induced expression | ABA-pathway responsive | - | [70] |
| NAM-1 | Leaf senescence | + regulator of leaf senescence | Bulk segregant analysis, Missense mutations | - | - | [60,71] |
| TaRNAC1 | Roots length, above-ground biomass, drought tolerance | + regulator of roots length, above-ground biomass, drought tolerance | Overexpression | GA3-ox2 | - | [88] |
| TaSNAC8-6A | Lateral roots development | + regulator of | Overexpression | activate various drought-responsive and auxin-signaling genes | - | [89] |
| TaNAC14 | Root development | − regulator of root development | Overexpression | tae-miR-164 | - | [90] |
| TaNAC29 | Leaf senescence, Drought, and salt stresses | + regulator of leaf senescence, drought, and salt stress tolerance | Overexpression | CAT and SOD enzyme and ABA-pathway responsive | - | [75] |
| TaNAC2 | Drought, salt, and freezing stress | + regulator of drought, salt, and freezing stress tolerance | Overexpression | - | - | [94] |
| TaNAC2L | Heat | + regulator of thermotolerance | Overexpression | - | - | [95] |
| TaNAC47 | Cold, salt, polyethylene glycol (PEG), and ABA | + regulator of cold, salt, polyethylene glycol (PEG), and ABA stress tolerance | Overexpression | - | - | [100] |
| TaSNAC8-6A | Drought stress | + regulator of drought tolerance | Overexpression | ABA-pathway responsive | - | [89] |
| TaRNAC1 | Drought stress, grain weight, and biomass | + regulator of drought tolerance, grain weight, and biomass | Overexpression | PEG pathway responsive | [88] | |
| TaNAC69 | Drought stress | + regulator of drought tolerance, | Overexpression | PEG pathway responsive | [104] | |
| TaNAC67 | Drought, salinity, and freezing stress | + regulator of Drought, salinity and freezing tolerance, cell membrane stability, cell membrane stability | Overexpression | DREB2A, COR15, ABI1 and ABI2. DREB2A | [106] | |
| TaSNAC4-3D | Drought stress | − regulator of drought tolerance | Overexpression | ABA-pathway responsive | - | [111] |
| TaNAC35 | Leaf rust stress | − regulator of wheat resistance to leaf rust | Knockdown | - | - | [119] |
| TaNAC30 | Strip rust stress | − regulator of wheat resistance to strip rust | Knockdown | ABA-pathway responsive | [120] | |
| TaNAC2 | Strip rust stress | − regulator of wheat resistance to strip rust | Knockdown | ABA-pathway responsive | [121] | |
| TaNAC8 | Strip rust stress | − regulator of wheat resistance to strip rust | Ethylene and methyl-jasmonate-induced expression | Ethylene and methyl-jasmonate pathway responsive | - | [122] |
| TaNAC4 | Strip rust stress | − regulator of wheat resistance to strip rust | Methyl-jasmonate, ethylene, and ABA-induced expression | Methyl-jasmonate, ethylene, and ABA pathway responsive | - | [123] |
| TaNAC21/22 | Strip rust stress | − regulator of wheat resistance to strip rust | Knockdown | tae-miR164 | - | [124] |
| TaNAC6A | Powdery mildew stress | + regulator of powdery mildew resistance | Overexpression | jasmonic acid pathway responsive | - | [125] |
| TaNACL-D1 | Fusarium head blight stress | + regulator of Fusarium head blight resistance | Overexpression | TaFROG | - | [126] |
| TaNAC2-5A | Possibly Phytohormonal homeostasis | - | Coregulation | TaCKX2 | - | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, A.; Bocian, J.; Hameed, A.; Orczyk, W.; Nadolska-Orczyk, A. Cis-Regulation by NACs: A Promising Frontier in Wheat Crop Improvement. Int. J. Mol. Sci. 2022, 23, 15431. https://doi.org/10.3390/ijms232315431
Iqbal A, Bocian J, Hameed A, Orczyk W, Nadolska-Orczyk A. Cis-Regulation by NACs: A Promising Frontier in Wheat Crop Improvement. International Journal of Molecular Sciences. 2022; 23(23):15431. https://doi.org/10.3390/ijms232315431
Chicago/Turabian StyleIqbal, Adnan, Joanna Bocian, Amir Hameed, Waclaw Orczyk, and Anna Nadolska-Orczyk. 2022. "Cis-Regulation by NACs: A Promising Frontier in Wheat Crop Improvement" International Journal of Molecular Sciences 23, no. 23: 15431. https://doi.org/10.3390/ijms232315431
APA StyleIqbal, A., Bocian, J., Hameed, A., Orczyk, W., & Nadolska-Orczyk, A. (2022). Cis-Regulation by NACs: A Promising Frontier in Wheat Crop Improvement. International Journal of Molecular Sciences, 23(23), 15431. https://doi.org/10.3390/ijms232315431







