Sex Lethal Gene Manipulates Gonadal Development of Medaka, Oryzias latipes, through Estrogenic Interventions
Abstract
:1. Introduction
2. Results
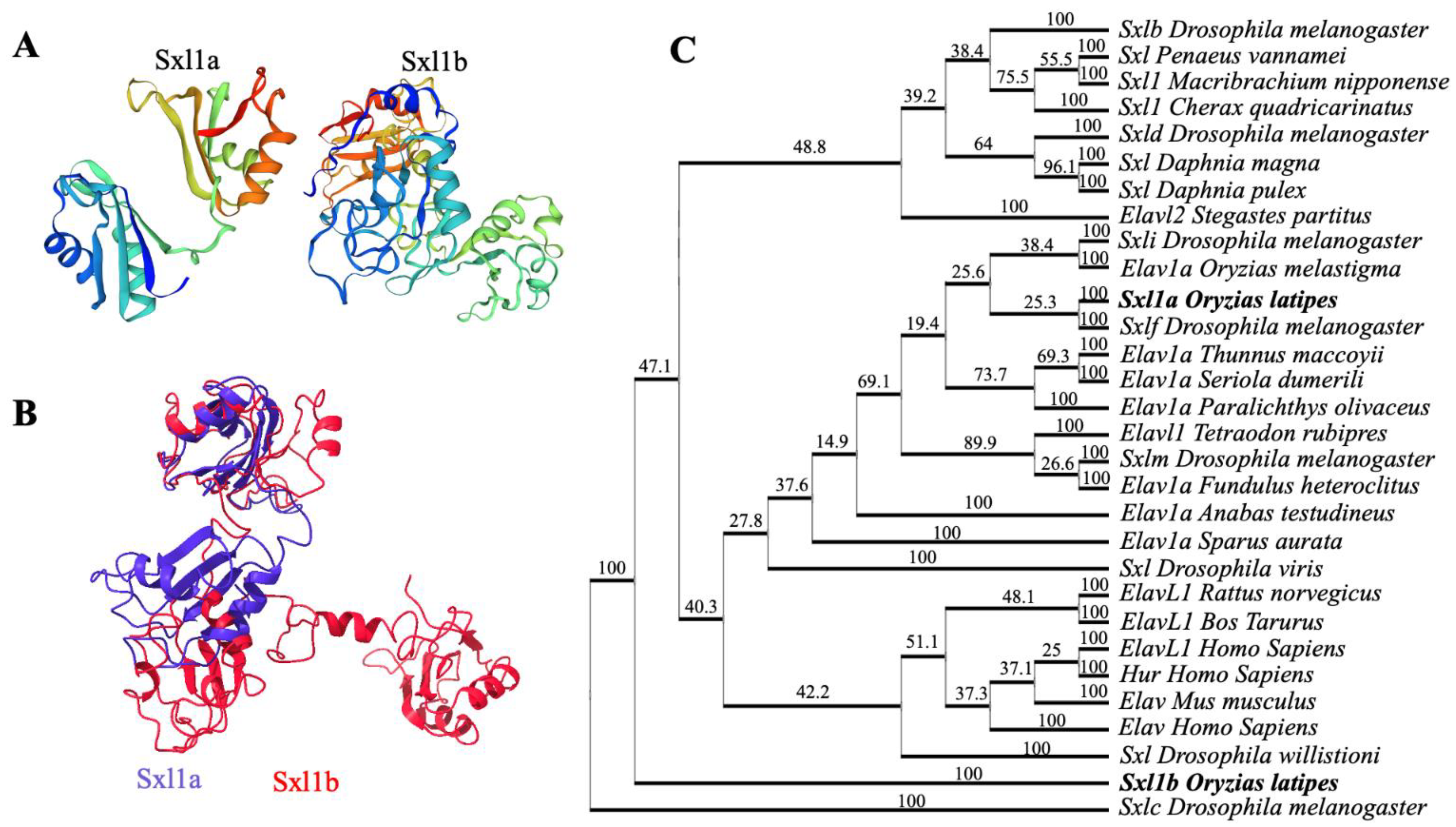
2.1. Cloning and Phylogenetic Analysis
2.2. Tissue Distribution, Ontogeny and Cellular Localization
2.3. Functional Validation of sxl Action in Gonad Development
2.4. Effect of Sex Steroids
2.5. Effect of Steroid Receptors
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Experimental Animals
4.3. Sample Collection
4.4. Phylogenetics and Protein Analysis
4.5. Tissue Distribution Analysis
4.6. Quantification of Changes in Gene Expression by Realtime PCR
4.7. Histology In Situ Hybridization (ISH) and Immunohistochemistry
4.8. Promoter Analysis
4.9. Overexpression and Knockdown of sxl Genes
4.10. Chemical Treatment
4.11. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Fox, R.J.; Fromhage, L.; Jennions, M.D. Sexual selection, phenotypic plasticity and female reproductive output. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, M.; Chen, X.; Zhao, Y.; Itoh, Y.; McQuillen, C.N.; Han, Y.; Shou, B.L.; McClusky, R.; Reue, K.; Arnold, A.P.; et al. Relative contributions of sex hormones, sex chromosomes, and gonads to sex differences in tissue gene regulation. Genome Res. 2022, 32, 807–824. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.; Capel, B. Gonad morphogenesis in vertebrates: Divergent means to a convergent end. Annu. Rev. Cell Dev. Biol. 2009, 25, 457–482. [Google Scholar] [CrossRef]
- Kim-Ha, J.; Kim, J.; Kim, Y.-J. Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol. Cell. Biol. 1999, 19, 2505–2514. [Google Scholar] [CrossRef] [Green Version]
- Sekii, K.; Salvenmoser, W.; de Mulder, K.; Scharer, L.; Ladurner, P. Melav2, an elav-like gene, is essential for spermatid differentiation in the flatworm Macrostomum lignano. BMC Dev. Biol. 2009, 9, 62. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Yuan, Y.; Cheng, F.; Fang, J.; Zhou, F.; Ma, W.; Jiang, Y.; Huang, X.; Wang, Y.; Shan, L.; et al. Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development 2017, 144, 128–138. [Google Scholar] [CrossRef] [Green Version]
- Meise, M.; Hilfiker-Kleiner, D.; Dübendorfer, A.; Brunner, C.; Nöthiger, R.; Bopp, D. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development 1998, 125, 1487–1494. [Google Scholar] [CrossRef]
- Camara, N.; Whitworth, C.; Van Doren, M. The creation of sexual dimorphism in the Drosophila soma. Curr. Top. Dev. Biol. 2008, 83, 65–107. [Google Scholar] [CrossRef]
- Siwicki, K.K.; Kravitz, E.A. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr. Opin. Neurobiol. 2009, 19, 200–206. [Google Scholar] [CrossRef]
- Evans, D.S.; Cline, T.W. Drosophila switch gene Sex-lethal can bypass its switch-gene target transformer to regulate aspects of female behavior. Proc. Natl. Acad. Sci. USA 2013, 110, E4474–E4481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maris, C.; Dominguez, C.; Allain, F.H.T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Schüpbach, T. Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of Sex-lethal in Drosophila melanogaster. Genetics 1985, 109, 529–548. [Google Scholar] [CrossRef] [PubMed]
- Steinmann-Zwicky, M.; Schmid, H.; Nöthiger, R. Cell-autonomous and inductive signals can determine the sex of the germ line of Drosophila by regulating the gene Sxl. Cell 1989, 57, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Bopp, D.; Horabin, J.I.; Lersch, R.A.; Cline, T.W.; Schedl, P. Expression of the Sex-lethal gene is controlled at multiple levels during Drosophila oogenesis. Development 1993, 118, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, P.K.; Southard, S.; Baxter, K.; van Doren, M. Germline sex determination regulates sex specific signaling between germline stem cells and their niche. Cell Rep. 2022, 39, 110620. [Google Scholar] [CrossRef]
- Hashiyama, K.; Hayashi, Y.; Kobayashi, S. Drosophila sex lethal gene initiates female development in germline progenitors. Science 2011, 333, 885–888. [Google Scholar] [CrossRef]
- López-Cuadros, I.; García-Gasca, A.; Gomez-Anduro, G.; Escobedo-Fregoso, C.; Llera-Herrera, R.A.; Ibarra, A.M. Isolation of the sex-determining gene Sex-lethal (Sxl) in Penaeus (Litopenaeus) vannamei (Boone, 1931) and characterization of its embryogenic, gametogenic, and tissue-specific expression. Gene 2018, 668, 33–47. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Herpin, A.; Fischer, P.; Liedtke, D.; Kluever, N.; Neuner, C.; Raz, E.; Schartl, M. Sequential SDF1a and b-induced mobility guides medaka PGC migration. Dev. Biol. 2008, 320, 319–327. [Google Scholar] [CrossRef]
- Kurokawa, H.; Saito, D.; Nakamura, S.; Katoh-Fukui, Y.; Ohta, K.; Baba, T.; Morohashi, K.-I.; Tanaka, M. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl. Acad. Sci. USA 2007, 104, 16958–16963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiszniak, S.E.; Dredge, B.K.; Jensen, K.B. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS ONE 2011, 6, e20773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herpin, A.; Schmidt, C.; Kneitz, S.; Gobé, C.; Regensburger, M.; Le Cam, A.; Montfort, J.; Adolfi, M.C.; Lillesaar, C.; Wilhelm, D.; et al. A novel evolutionary conserved mechanism of RNA stability regulates synexpression of primordial germ cell-specific genes prior to the sex-determination stage in medaka. PLoS Biol. 2019, 17, e3000185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, T.; Zhou, L.Y.; Chaudhari, A.; Iguchi, T.; Nagahama, Y. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 2016, 6, srep19480. [Google Scholar] [CrossRef] [Green Version]
- Mohapatra, S.; Chakraborty, T.; Shimizu, S.; Ohta, K.; Nagahama, Y.; Ohta, K. Estrogen and estrogen receptors chauffeur the sex-biased autophagic action in liver. Cell Death Differ. 2020, 27, 3117–3130. [Google Scholar] [CrossRef]
- Chakraborty, T.; Mohapatra, S.; Zhou, L.Y.; Ohta, K.; Matsubara, T.; Iguchi, T.; Nagahama, Y. Estrogen Receptor β2 Oversees Germ Cell Maintenance and Gonadal Sex Differentiation in Medaka, Oryzias latipes. Stem Cell Rep. 2019, 13, 419–433. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970; ISBN 978-3-642-86659-3. [Google Scholar] [CrossRef] [Green Version]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef] [Green Version]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Good, P.J. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. USA 1995, 92, 4557–4561. [Google Scholar] [CrossRef] [Green Version]
- Cronmiller, C.; Salz, H.K. The Feminine Mystique: The Initiation of Sex Determination in Drosophila. In Molecular Biology of Sex Differentiation Led; Wachtel, S.S., Ed.; Academic Press: San Diego, CA, USA, 1994; pp. 171–203. [Google Scholar]
- Cline, T.W.; Meyer, B.J. Vive la difference: Males vs females in flies vs. worms. Annu. Rev. Genet. 1996, 30, 637–702. [Google Scholar] [CrossRef] [PubMed]
- Saccone, G.; Pane, A.; Polito, L.C. Sex determination in flies, fruitflies and butterflies. Genetica 2002, 116, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Zaharieva, E.; Haussmann, I.U.; Bräuer, U.; Soller, M. Concentration and Localization of Coexpressed ELAV/Hu Proteins Control Specificity of mRNA Processing. Mol. Cell. Biol. 2015, 35, 3104–3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, U.; Buschmann, J.; Wahle, E. The nuclear poly(A) binding protein of mammals, but not of fission yeast, participates in mRNA polyadenylation. RNA 2017, 23, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Yang, H.; Sturgill, D.; Oliver, B.; Rabinow, L.; Samson, M.L. Sxl-Dependent, tra/tra2-Independent Alternative Splicing of the Drosophila melanogaster X-Linked Gene found in neurons. G3 Genes Genomes Genet. 2015, 5, 2865–2874. [Google Scholar] [CrossRef] [Green Version]
- Shults, C.L. Mechanisms of Estrogen Receptor Alternative Splicing and the Consequences for Aging in the Female Brain. Ph.D. Thesis, Loyola University Chicago, Chicago, IL, USA, 2015. Available online: https://ecommons.luc.edu/luc_diss/1969 (accessed on 7 July 2022).
- Yamamoto, T.M.; Cook, J.M.; Kotter, C.V.; Khat, T.; Silva, K.D.; Ferreyros, M.; Holt, J.W.; Knight, J.D.; Charlesworth, A. Zar1 represses translation in Xenopus oocytes and binds to the TCS in maternal mRNAs with different characteristics than Zar2. Biochim. Biophys. Acta 2013, 1829, 1034–1046. [Google Scholar] [CrossRef] [Green Version]
- Pryzbylkowski, P.; Obajimi, O.; Keen, J.C. Trichostatin A and 5 Aza-2′ deoxycytidine decrease estrogen receptor mRNA stability in ER positive MCF7 cells through modulation of HuR. Breast Cancer Res. Treat. 2008, 111, 15–25. [Google Scholar] [CrossRef]
- Moschall, R.; Rass, M.; Rossbach, O.; Lehmann, G.; Kullmann, L.; Eichner, N.; Strauss, D.; Meister, G.; Schneuwly, S.; Krahn, M.P.; et al. Drosophila Sister-of-Sex-lethal reinforces a male-specific gene expression pattern by controlling Sex-lethal alternative splicing. Nucleic Acids Res. 2019, 47, 2276–2288. [Google Scholar] [CrossRef] [Green Version]
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004, 121, 605–618. [Google Scholar] [CrossRef]
- Chakraborty, T.; Shibata, Y.; Zhou, L.-Y.; Katsu, Y.; Iguchi, T.; Nagahama, Y. Differential expression of three estrogen receptor subtype mRNAs in gonads and liver from embryos to adults of the medaka, Oryzias latipes. Mol. Cell. Endocrinol. 2011, 333, 47–54. [Google Scholar] [CrossRef]
- Mohapatra, S.; Chakraborty, T.; Miyagawa, S.; Zhou, L.Y.; Ohta, K.; Iguchi, T.; Nagahama, Y. Steroid responsive regulation of IFNγ2 alternative splicing and its possible role in germ cell proliferation in medaka. Mol. Cell. Endocrinol. 2015, 400, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, D.; Wittbrodt, J. One for all--a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PLoS ONE 2011, 6, e19713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, H.S.; Bond, B.L.; Parslow, T.G. Estrogen regulates the IFN-gamma promoter. J. Immunol. 1991, 146, 4362–4367. [Google Scholar] [PubMed]






| Group (Total Injected Embryos) | 1daf | 2daf | 4daf | 6daf | Hatching | Total |
|---|---|---|---|---|---|---|
| Control (151) | 5.30 ±1.1 | 2.65 ± 1.2 | 2.65 ± 1.1 | 0.66 ± 0.4 | 1.99 ± 1.1 | 13.25 ± 0.9 |
| sxl1a-OV (164) | 4.27 ± 1.4 | 3.05 ± 1.1 | 2.44 ± 1.1 | 1.83 ± 1.0 | 1.22 ± 0.5 | 12.80 ± 0.8 |
| sxl1b-OV (173) | 5.20 ± 1.3 | 2.89 ± 1.4 | 2.31 ± 0.4 | 2.31 ± 0.8 | 1.16 ± 0.3 | 13.87 ± 0.6 |
| sxl1a-KD (171) | 4.68 ± 1.4 | 2.92 ± 1.2 | 3.51 ± 1.5 | 2.34 ± 1.0 | 0.00 ± 0.0 | 13.45 ± 1.0 |
| sxl1b-KD (175) | 75.43 ± 12.4 | 18.86 ± 6.5 | 5.71 ± 4.8 | 0.00 ± 0.0 | 0.00 ± 0.0 | 100.00 ± 0.0 |
| Gene | Correlation with sxl1a |
|---|---|
| erβ2 | 0.91 |
| foxl2 | 0.34 |
| foxl3 | 0.39 |
| cyp19a1 | 0.21 |
| olvas | 0.84 |
| gsdf1 | −0.87 |
| dmrt1 | −0.27 |
| sox9a2 | −0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, T.; Mohapatra, S.; Matsuyama, M.; Nagahama, Y.; Ohta, K. Sex Lethal Gene Manipulates Gonadal Development of Medaka, Oryzias latipes, through Estrogenic Interventions. Int. J. Mol. Sci. 2022, 23, 15496. https://doi.org/10.3390/ijms232415496
Chakraborty T, Mohapatra S, Matsuyama M, Nagahama Y, Ohta K. Sex Lethal Gene Manipulates Gonadal Development of Medaka, Oryzias latipes, through Estrogenic Interventions. International Journal of Molecular Sciences. 2022; 23(24):15496. https://doi.org/10.3390/ijms232415496
Chicago/Turabian StyleChakraborty, Tapas, Sipra Mohapatra, Michiya Matsuyama, Yoshitaka Nagahama, and Kohei Ohta. 2022. "Sex Lethal Gene Manipulates Gonadal Development of Medaka, Oryzias latipes, through Estrogenic Interventions" International Journal of Molecular Sciences 23, no. 24: 15496. https://doi.org/10.3390/ijms232415496





