Analysis of the Type 4 Effectome across the Genus Rickettsia
Abstract
1. Introduction
2. Results and Discussion
2.1. Effector Prediction Compared to Other Rickettsiales
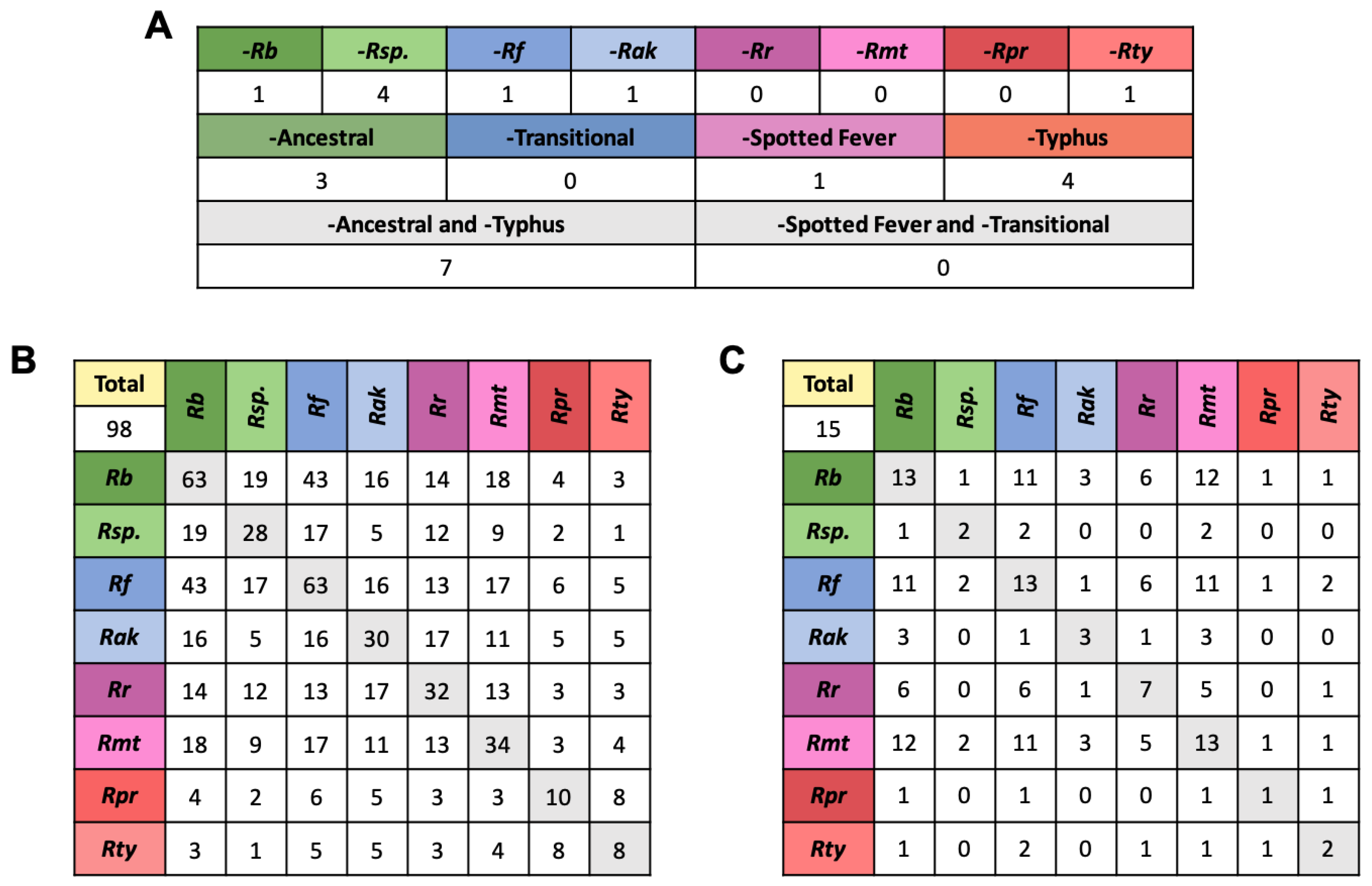
2.2. Analysis of Sets of Homologous Effectors across Rickettsia
2.3. Gene Fragment Analysis
2.4. G+C Content Analysis
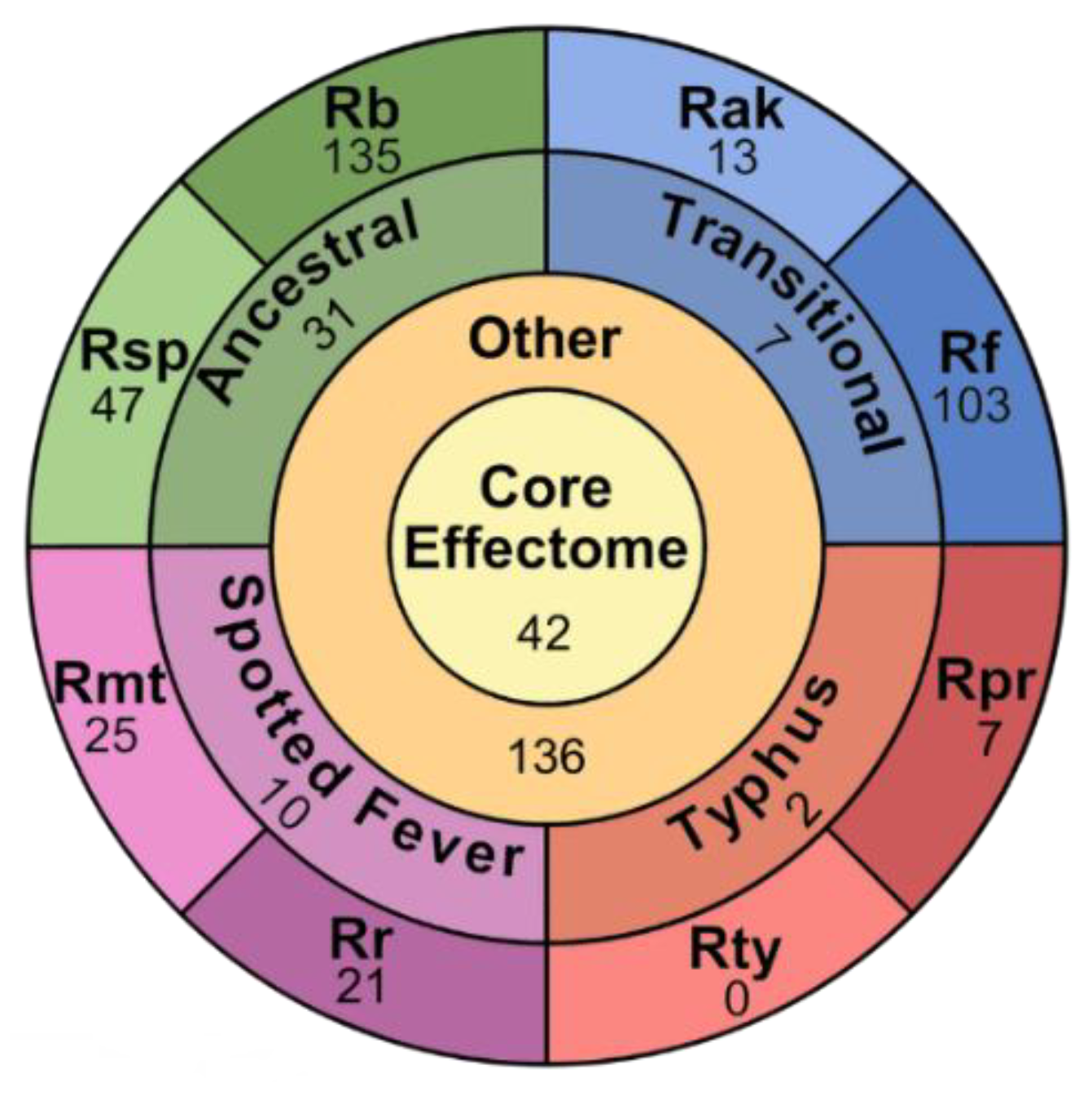
2.5. Effector Categorization
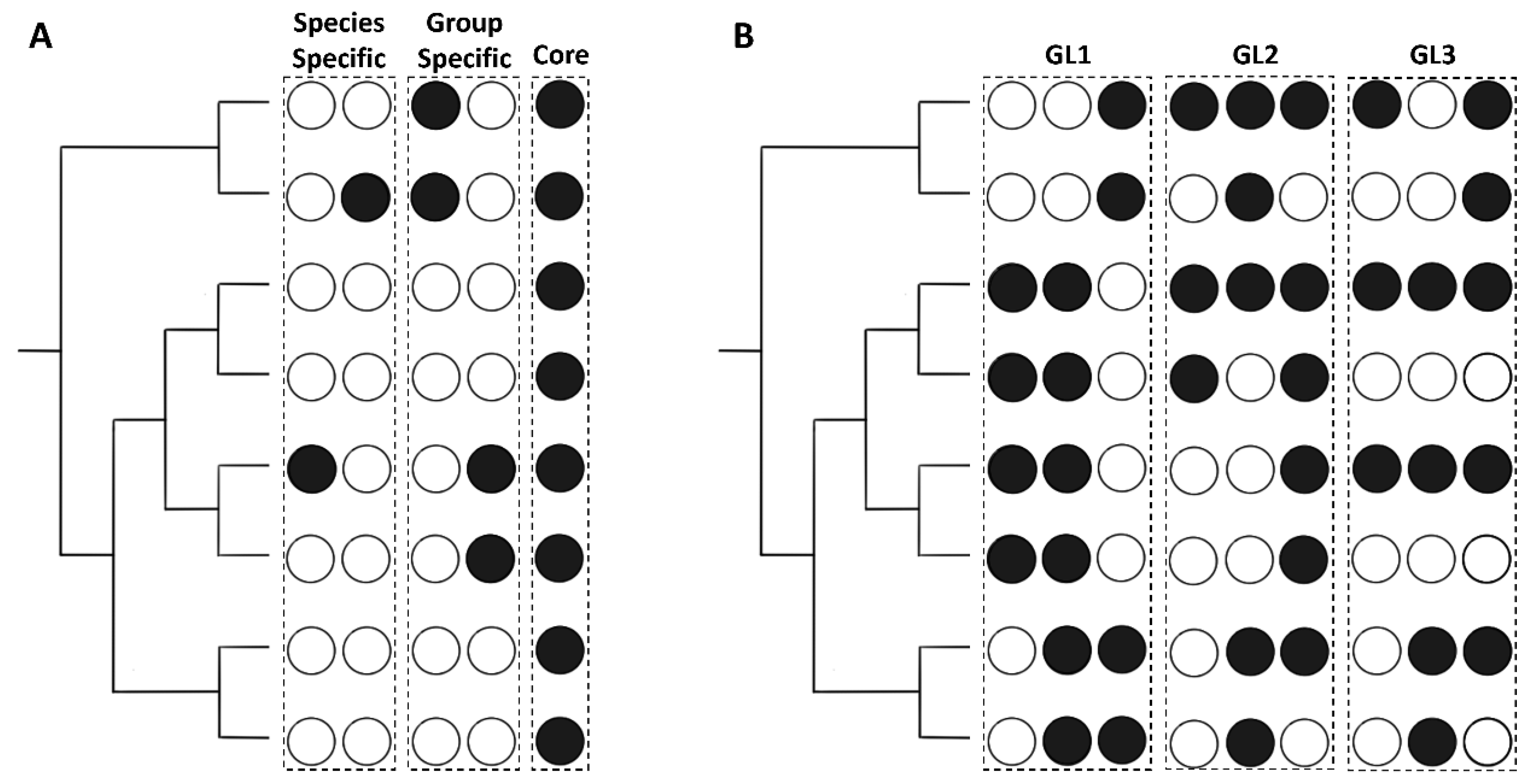
2.5.1. Core Effectors
2.5.2. Group Specific Effectors
2.5.3. Species Specific Effectors
2.5.4. Plasmid-Borne Effectors
2.5.5. Other Effectors
2.6. Effectors in Pathogenc and Non-Pathogec Rickettsia
2.7. Arthropod Host Associated Effectors
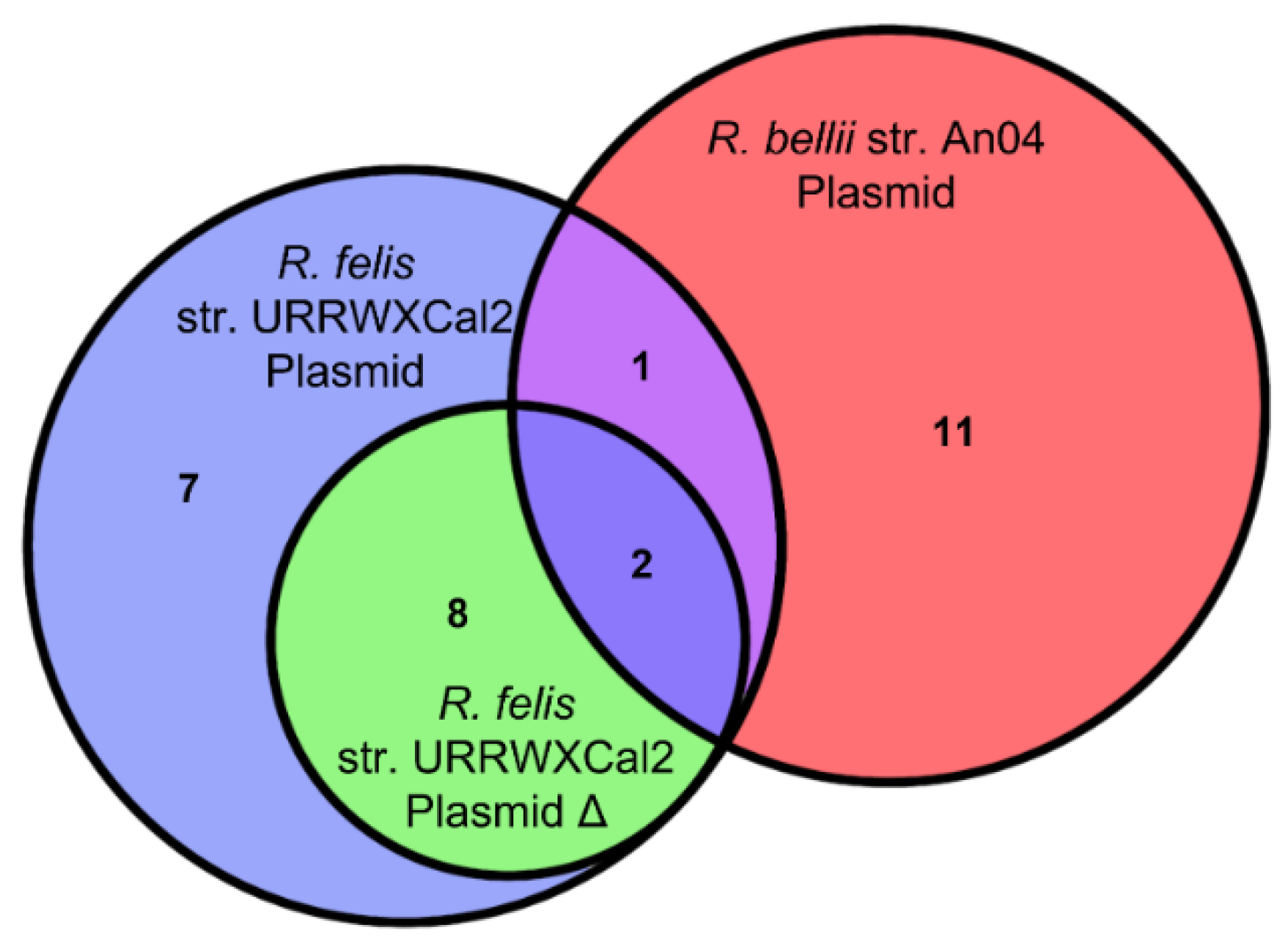
2.8. The Relationship between R. bellii and R. felis
3. Materials and Methods
3.1. Sequences
3.2. Phylogenetic Analysis
3.3. Effector Prediction
3.4. Homolog Identification
3.5. Protein Domain Identification
3.6. Domain of Interest Identification
3.7. Comparison of G+C Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perlman, S.J.; Hunter, M.S.; Zchori-Fein, E. The Emerging Diversity of Rickettsia. Proc. R. Soc. B 2006, 273, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Schön, M.E.; Martijn, J.; Vosseberg, J.; Köstlbacher, S.; Ettema, T.J.G. The Evolutionary Origin of Host Association in the Rickettsiales. Nat. Microbiol. 2022, 7, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Williams, K.; Shukla, M.; Snyder, E.E.; Nordberg, E.K.; Ceraul, S.M.; Dharmanolla, C.; Rainey, D.; Soneja, J.; Shallom, J.M.; et al. Rickettsia Phylogenomics: Unwinding the Intricacies of Obligate Intracellular Life. PLoS ONE 2008, 3, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Bozeman, M.F.; Elisberg, B.L.; Humphries, J.W.; Runcik, K.; Palmer, D.B. Serologic Evidence of Rickettsia canada Infection of Man. J. Infect. Dis. 1970, 121, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.B.; Cunha-Filho, N.A.; Pacheco, R.C.; Ferreira, F.; Pappen, F.G.; Farias, N.A.R.; Larsson, C.E.; Labruna, M.B. Canine Infection by Rickettsiae and Ehrlichiae in Southern Brazil. Am. J. Trop. Med. 2008, 79, 102–108. [Google Scholar] [CrossRef]
- Rao, Q.; Rollat-Farnier, P.-A.; Zhu, D.-T.; Santos-Garcia, D.; Silva, F.J.; Moya, A.; Latorre, A.; Klein, C.C.; Vavre, F.; Sagot, M.-F.; et al. Genome Reduction and Potential Metabolic Complementation of the Dual Endosymbionts in the Whitefly Bemisia tabaci. BMC Genom. 2015, 16, 226:1–226:13. [Google Scholar] [CrossRef]
- Zomorodipour, A.; Andersson, S.G.E. Obligate Intracellular Parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett. 1999, 452, 11–15. [Google Scholar] [CrossRef]
- Sakharkar, K.R.; Sakharkar, M.K.; Verma, C.; Chow, V.T.K. Comparative Study of Overlapping Genes in Bacteria, with Special Reference to Rickettsia prowazekii and Rickettsia conorii. Int. J. Syst. Evol. Microbiol. 2005, 55, 1205–1209. [Google Scholar] [CrossRef]
- Fournier, P.-E.; El Karkouri, K.; Leroy, Q.; Robert, C.; Giumelli, B.; Renesto, P.; Socolovschi, C.; Parola, P.; Audic, S.; Raoult, D. Analysis of the Rickettsia africae Genome Reveals That Virulence Acquisition in Rickettsia Species May Be Explained by Genome Reduction. BMC Genom. 2009, 10, 166:1–166:15. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Joardar, V.; Williams, K.P.; Driscoll, T.; Hostetler, J.B.; Nordberg, E.; Shukla, M.; Walenz, B.; Hill, C.A.; Nene, V.M.; et al. A Rickettsia Genome Overrun by Mobile Genetic Elements Provides Insight into the Acquisition of Genes Characteristic of an Obligate Intracellular Lifestyle. J. Bacteriol. 2012, 194, 376–394. [Google Scholar] [CrossRef]
- Ogata, H.; Renesto, P.; Audic, S.; Robert, C.; Blanc, G.; Fournier, P.-E.; Parinello, H.; Claverie, J.-M.; Raoult, D. The Genome Sequence of Rickettsia felis Identifies the First Putative Conjugative Plasmid in an Obligate Intracellular Parasite. PLoS Biol. 2005, 3, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; La Scola, B.; Audic, S.; Renesto, P.; Blanc, G.; Robert, C.; Fournier, P.-E.; Claverie, J.-M.; Raoult, D. Genome Sequence of Rickettsia bellii Illuminates the Role of Ancestral Protozoa in Gene Exchanges between Intracellular Pathogens. PLoS Genet. 2006, 2, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Phan, I.Q.H.; Driscoll, T.P.; Guillotte, M.L.; Lehman, S.S.; Rennoll-Bankert, K.E.; Subramanian, S.; Beier-Sexton, M.; Myler, P.J.; Rahman, M.S.; et al. The Rickettsia Type IV Secretion System: Unrealized Complexity Mired by Gene Family Expansion. Pathog. Dis. 2016, 74, ftw058:1–ftw058:13. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.S.; Noriea, N.F.; Aistleitner, K.; Clark, T.R.; Dooley, C.A.; Nair, V.; Kaur, S.J.; Rahman, M.S.; Gillespie, J.J.; Azad, A.F.; et al. The Rickettsial Ankyrin Repeat Protein 2 Is a Type IV Secreted Effector That Associates with the Endoplasmic Reticulum. mBio 2018, 9, e00975-18:1–e00975-18:15. [Google Scholar] [CrossRef] [PubMed]
- Rennoll-Bankert, K.E.; Rahman, M.S.; Gillespie, J.J.; Guillotte, M.L.; Kaur, S.J.; Lehman, S.S.; Beier-Sexton, M.; Azad, A.F. Which Way In? The RalF Arf-GEF Orchestrates Rickettsia Host Cell Invasion. PLOS Pathog. 2015, 11, e1005115:1–e1005115:28. [Google Scholar] [CrossRef] [PubMed]
- Voss, O.H.; Gillespie, J.J.; Lehman, S.S.; Rennoll, S.A.; Beier-Sexton, M.; Rahman, M.S.; Azad, A.F. Risk1, a Phosphatidylinositol 3-Kinase Effector, Promotes Rickettsia typhi Intracellular Survival. mBio 2020, 11, e00820-20:1–e00820-20:22. [Google Scholar] [CrossRef]
- Ashari, Z.E.; Dasgupta, N.; Brayton, K.A.; Broschat, S.L. An Optimal Set of Features for Predicting Type IV Secretion System Effector Proteins for a Subset of Species Based on a Multi-Level Feature Selection Approach. PLoS ONE 2018, 13, e0197041:1–e0197041:16. [Google Scholar] [CrossRef]
- Ashari, Z.E.; Brayton, K.A.; Broschat, S.L. Prediction of T4SS Effector Proteins for Anaplasma phagocytophilum Using OPT4e, a New Software Tool. Front. Microbiol. 2019, 10, 1391:1–1391:10. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Beier, M.S.; Rahman, M.S.; Ammerman, N.C.; Shallom, J.M.; Purkayastha, A.; Sobral, B.S.; Azad, A.F. Plasmids and Rickettsial Evolution: Insight from Rickettsia felis. PLoS ONE 2007, 2, e266:1–e266:17. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Burstein, D.; Amaro, F.; Zusman, T.; Lifshitz, Z.; Cohen, O.; Gilbert, J.A.; Pupko, T.; Shuman, H.A.; Segal, G. Genomic Analysis of 38 Legionella Species Identifies Large and Diverse Effector Repertoires. Nat. Genet. 2016, 48, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Noroy, C.; Meyer, D.F. The Super Repertoire of Type IV Effectors in the Pangenome of Ehrlichia Spp. Provides Insights into Host-Specificity and Pathogenesis. PLOS Comput. Biol. 2021, 17, e1008788:1–e1008788:26. [Google Scholar] [CrossRef]
- Bröms, J.E.; Edqvist, P.J.; Forsberg, Å.; Francis, M.S. Tetratricopeptide Repeats Are Essential for PcrH Chaperone Function in Pseudomonas aeruginosa Type III Secretion. FEMS Microbiol. Lett. 2006, 256, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Haraga, A.; Miller, S.I. A Salmonella enterica Serovar Typhimurium Translocated Leucine-Rich Repeat Effector Protein Inhibits NF- κB-Dependent Gene Expression. Infect. Immun. 2003, 71, 4052–4058. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Luhrmann, A.; Satoh, A.; Laskowski-Arce, M.A.; Roy, C.R. Ankyrin Repeat Proteins Comprise a Diverse Family of Bacterial Type IV Effectors. Science 2008, 320, 1651–1654. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Mukherjee, S.; Roy, C.R.; Cherfils, J. Structure of the Legionella Effector AnkX Reveals the Mechanism of Phosphocholine Transfer by the FIC Domain. EMBO J. 2013, 32, 1469–1477. [Google Scholar] [CrossRef]
- Triplett, L.R.; Shidore, T.; Long, J.; Miao, J.; Wu, S.; Han, Q.; Zhou, C.; Ishihara, H.; Li, J.; Zhao, B.; et al. AvrRxo1 Is a Bifunctional Type III Secreted Effector and Toxin-Antitoxin System Component with Homologs in Diverse Environmental Contexts. PLoS ONE 2016, 11, e0158856:1–e0158856:16. [Google Scholar] [CrossRef]
- Eichinger, V.; Nussbaumer, T.; Platzer, A.; Jehl, M.-A.; Arnold, R.; Rattei, T. EffectiveDB—Updates and Novel Features for a Better Annotation of Bacterial Secreted Proteins and Type III, IV, VI Secretion Systems. Nucleic Acids Res. 2015, 44, D669–D674. [Google Scholar] [CrossRef]
- Reed, S.C.O.; Lamason, R.L.; Risca, V.I.; Abernathy, E.; Welch, M.D. Rickettsia Actin-Based Motility Occurs in Distinct Phases Mediated by Different Actin Nucleators. Curr. Biol. 2014, 24, 98–103. [Google Scholar] [CrossRef]
- Colonne, P.M.; Winchell, C.G.; Voth, D.E. Hijacking Host Cell Highways: Manipulation of the Host Actin Cytoskeleton by Obligate Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 107:1–107:8. [Google Scholar] [CrossRef]
- Aistleitner, K.; Clark, T.; Dooley, C.; Hackstadt, T. Selective Fragmentation of the trans-Golgi Apparatus by Rickettsia rickettsii. PLOS Pathog. 2020, 16, e1008582. [Google Scholar] [CrossRef] [PubMed]
- Burstein, D.; Zusman, T.; Degtyar, E.; Viner, R.; Segal, G.; Pupko, T. Genome-Scale Identification of Legionella pneumophila Effectors Using a Machine Learning Approach. PLOS Pathog. 2009, 5, e1000508:1–e1000508:12. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Gillespie, J.J.; Kaur, S.J.; Sears, K.T.; Ceraul, S.M.; Beier-Sexton, M.; Azad, A.F. Rickettsia typhi Possesses Phospholipase A2 Enzymes That Are Involved in Infection of Host Cells. PLOS Pathog. 2013, 9, e1003399:1–e1003399:1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sears, K.T.; Ceraul, S.M.; Gillespie, J.J.; Allen, E.D.; Popov, V.L.; Ammerman, N.C.; Rahman, M.S.; Azad, A.F. Surface Proteome Analysis and Characterization of Surface Cell Antigen (Sca) or Autotransporter Family of Rickettsia typhi. PLOS Pathog. 2012, 8, e1002856:1–e1002856:17. [Google Scholar] [CrossRef]
- Ellison, D.W.; Clark, T.R.; Sturdevant, D.E.; Virtaneva, K.; Hackstadt, T. Limited Transcriptional Responses of Rickettsia rickettsii Exposed to Environmental Stimuli. PLoS ONE 2009, 4, e5612:1–e5612:11. [Google Scholar] [CrossRef]
- Luong, P.; Kinch, L.N.; Brautigam, C.A.; Grishin, N.V.; Tomchick, D.R.; Orth, K. Kinetic and Structural Insights into the Mechanism of AMPylation by VopS Fic Domain. J. Biol. Chem. 2010, 285, 20155–20163. [Google Scholar] [CrossRef]
- Harris, E.K.; Verhoeve, V.I.; Banajee, K.H.; Macaluso, J.A.; Azad, A.F.; Macaluso, K.R. Comparative Vertical Transmission of Rickettsia by Dermacentor variabilis and Amblyomma maculatum. Ticks Tick-borne Dis. 2017, 8, 598–604. [Google Scholar] [CrossRef]
- Roth, T.; Lane, R.S.; Foley, J. A Molecular Survey for Francisella tularensis and Rickettsia Spp. In Haemaphysalis leporispalustris (Acari: Ixodidae) in Northern California. J. Med. Entomol. 2016, 54, 492–495. [Google Scholar] [CrossRef]
- Krawczak, F.S.; Labruna, M.B.; Hecht, J.A.; Paddock, C.D.; Karpathy, S.E. Genotypic Characterization of Rickettsia bellii Reveals Distinct Lineages in the United States and South America. BioMed Res. Int. 2018, 2018, 8505483:1–8505483:8. [Google Scholar] [CrossRef]
- Behar, A.; McCormick, L.J.; Perlman, S.J. Rickettsia felis Infection in a Common Household Insect Pest, Liposcelis bostrychophila (Psocoptera: Liposcelidae). Appl. Environ. Microbiol. 2010, 76, 2280–2285. [Google Scholar] [CrossRef]
- Fongsaran, C.; Jirakanwisal, K.; Tongluan, N.; Latour, A.; Healy, S.; Christofferson, R.C.; Macaluso, K.R. The Role of Cofeeding Arthropods in the Transmission of Rickettsia felis. PLOS Negl. Trop. Dis. 2022, 16, e0010576:1–e0010576:12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Noroy, C.; Lefrançois, T.; Meyer, D.F. Searching Algorithm for Type IV Effector Proteins (S4TE) 2.0: Improved Tools for Type IV Effector Prediction, Analysis and Comparison in Proteobacteria. PLOS Comput. Biol. 2019, 15, e1006847:1–e1006847:12. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421:1–421:9. [Google Scholar] [CrossRef] [PubMed]
- HMMER. Available online: http://hmmer.org/ (accessed on 25 July 2020).


| R. bellii | R. sp. MEAM1 | R. felis | R. akari | R. rickettsii | R. montanensis | R. prowazekii | R. typhi | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Effectors | 345 (25%) | 203 (14%) | 291 (23%) | 171 (17%) | 186 (15%) | 196 (17%) | 101 (12%) | 84 (10%) | 1571 (17%) |
| Coding Genes | 1400 | 1419 * | 1257 | 1034 | 1230 | 1178 | 839 | 815 | 9021 |
| R. bellii | R. sp. MEAM1 | R. felis | R. akari | R. rickettsii | R. montanensis | R. prowazekii | R. typhi | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | Gene Frg | Total | |
| EHSs | 28 (9%) | 296 | 39 (25%) | 158 | 33 (13%) | 250 | 18 (15%) | 117 | 26 (19%) | 134 | 28 (19%) | 144 | 9 (12%) | 75 | 1 (2%) | 66 |
| Non-EHSs | 47 (5%) | 897 | 52 (7%) | 786 | 58 (7%) | 891 | 43 (6%) | 745 | 49 (6%) | 770 | 45 (6%) | 791 | 19 (3%) | 701 | 6 (1%) | 685 |
| >300 bp | 75 (6%) | 1193 | 91 (10%) | 944 | 91 (8%) | 1141 | 61 (7%) | 862 | 75 (8%) | 904 | 73 (8%) | 935 | 28 (4%) | 776 | 7 (1%) | 751 |
| <300 bp | 33 (25%) | 134 | 68 (41%) | 165 | 43 (29%) | 151 | 170 (46%) | 370 | 210 (49%) | 425 | 96 (38%) | 252 | 25 (19%) | 131 | 4 (5%) | 73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aspinwall, J.A.; Brayton, K.A. Analysis of the Type 4 Effectome across the Genus Rickettsia. Int. J. Mol. Sci. 2022, 23, 15513. https://doi.org/10.3390/ijms232415513
Aspinwall JA, Brayton KA. Analysis of the Type 4 Effectome across the Genus Rickettsia. International Journal of Molecular Sciences. 2022; 23(24):15513. https://doi.org/10.3390/ijms232415513
Chicago/Turabian StyleAspinwall, Joseph A., and Kelly A. Brayton. 2022. "Analysis of the Type 4 Effectome across the Genus Rickettsia" International Journal of Molecular Sciences 23, no. 24: 15513. https://doi.org/10.3390/ijms232415513
APA StyleAspinwall, J. A., & Brayton, K. A. (2022). Analysis of the Type 4 Effectome across the Genus Rickettsia. International Journal of Molecular Sciences, 23(24), 15513. https://doi.org/10.3390/ijms232415513





