Efficient De Novo Biosynthesis of Heme by Membrane Engineering in Escherichia coli
Abstract
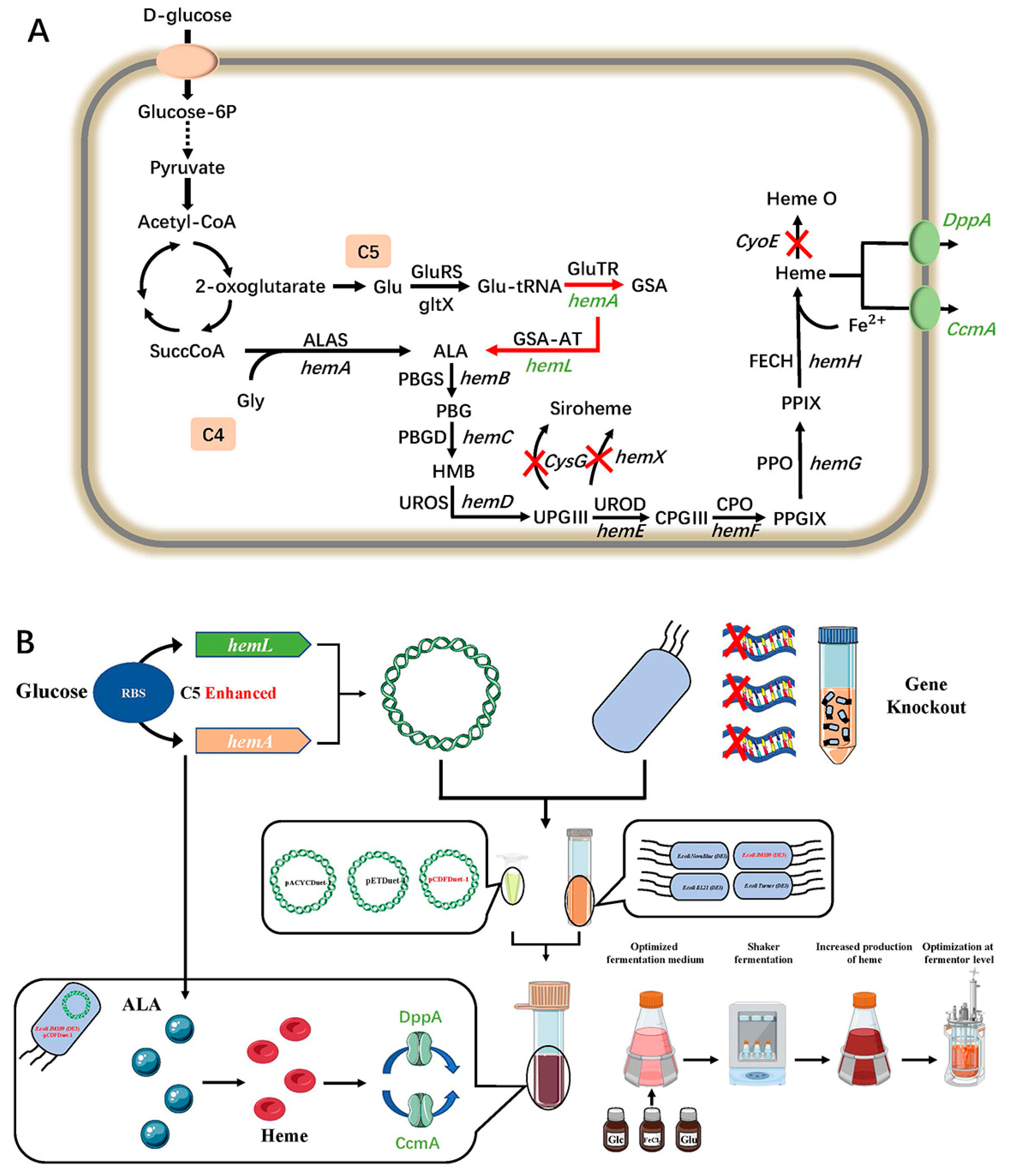
:1. Introduction
2. Results and Discussion
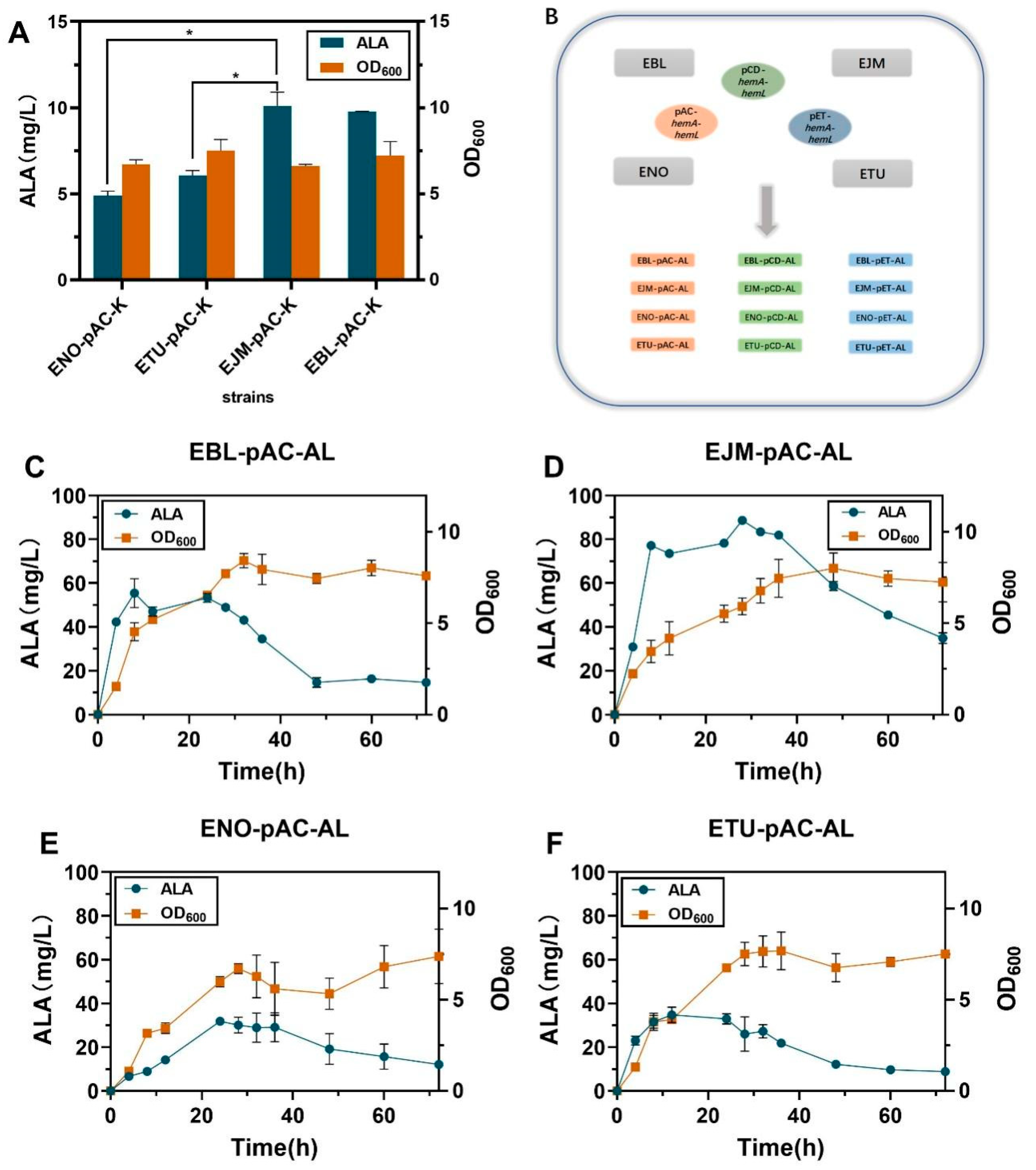
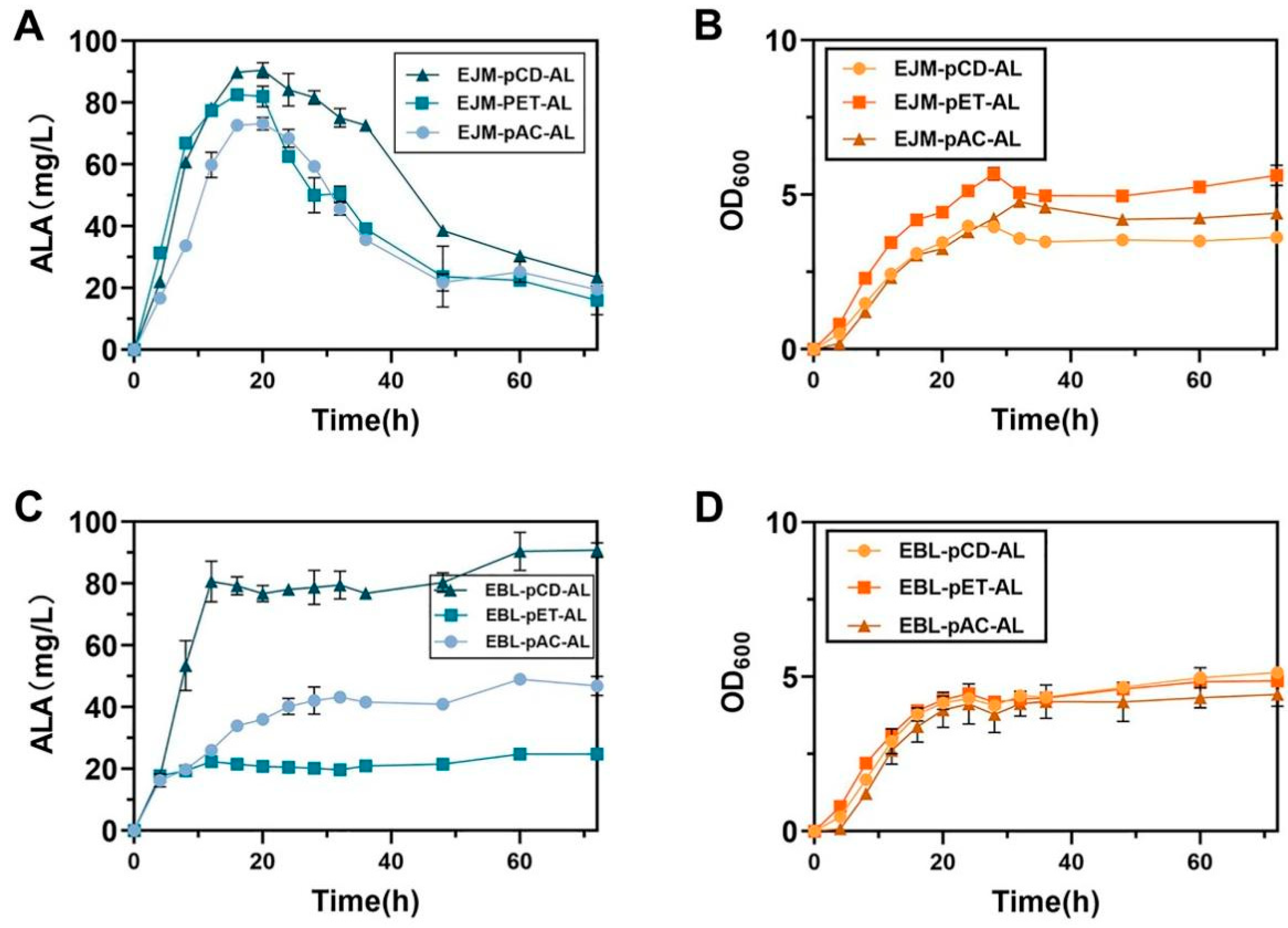
2.1. Improving HemA and HemL Expression in E. coli Host Strains
2.2. Increasing Heme Production by Removing Competitive Pathways
2.3. Overexpression of Heme Exporters to Increases Heme Production
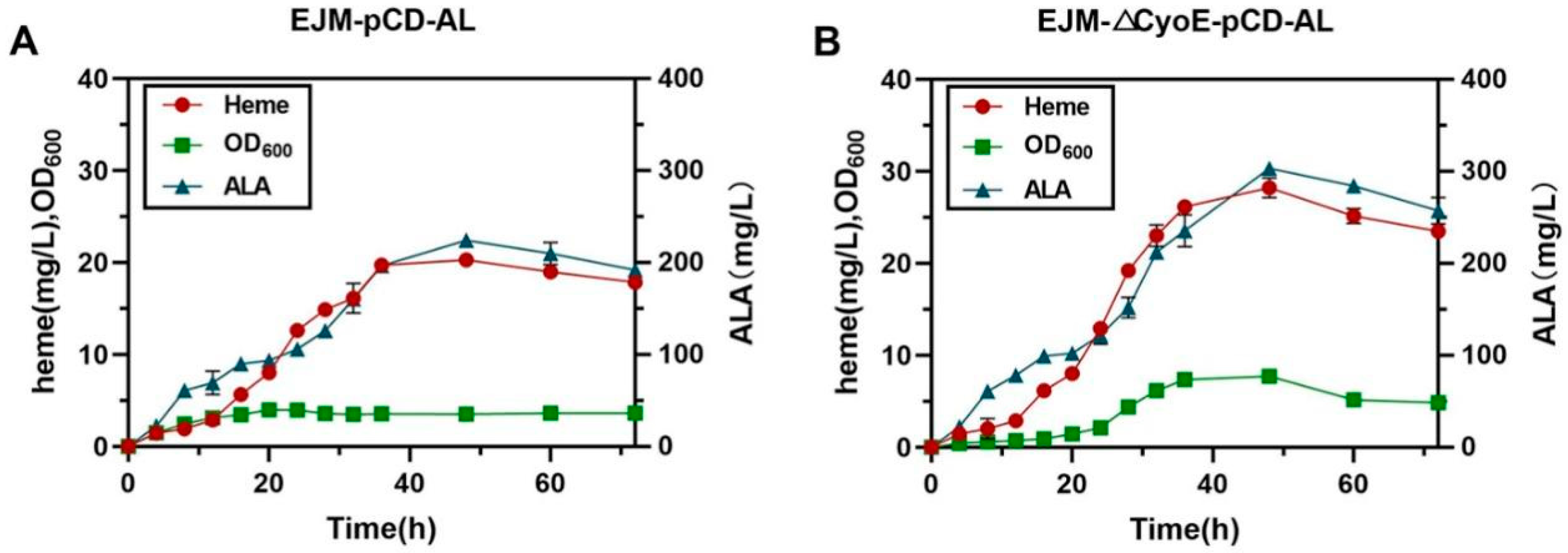
2.4. Improving the Heme Production in Bioreactor
2.4.1. Optimization of Fermentation Conditions
2.4.2. Fed-Batch Fermentation of Heme by Recombinant Strains
3. Materials and Methods
3.1. Strains and Plasmids
3.2. Culture Conditions
3.3. Construction of Recombinant Plasmids and Strains
3.4. Red/ET-Based Recombineering
3.5. Fed-Batch Fermentation
3.6. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mukherjee, M.; Dey, A. Rejigging Electron and Proton Transfer to Transition between Dioxygenase, Monooxygenase, Peroxygenase, and Oxygen Reduction Activity: Insights from Bioinspired Constructs of Heme Enzymes. JACS Au 2021, 1, 1296–1311. [Google Scholar] [CrossRef]
- Zamarreño Beas, J.; Videira, M.A.M.; Saraiva, L.M. Regulation of bacterial haem biosynthesis. Coord. Chem. Rev. 2022, 452. [Google Scholar] [CrossRef]
- Donegan, R.K.; Moore, C.M.; Hanna, D.A.; Reddi, A.R. Handling heme: The mechanisms underlying the movement of heme within and between cells. Free Radic. Biol. Med. 2019, 133, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.G.; Willoughby, M.M.; Hamza, I.; Reddi, A.R. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118881. [Google Scholar] [CrossRef] [PubMed]
- Simsa, R.; Yuen, J.; Stout, A.; Rubio, N.; Fogelstrand, P.; Kaplan, D.L. Extracellular Heme Proteins Influence Bovine Myosatellite Cell Proliferation and the Color of Cell-Based Meat. Foods 2019, 8, 521. [Google Scholar] [CrossRef] [Green Version]
- Gamage, S.M.K.; Dissabandara, L.; Lam, A.K.; Gopalan, V. The role of heme iron molecules derived from red and processed meat in the pathogenesis of colorectal carcinoma. Crit. Rev. Oncol. Hematol. 2018, 126, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Abdelazim, I.A.; Abu-Faza, M.; Shikanova, S.; Zhurabekova, G.; Maghrabi, M.M. Heme-bound iron in treatment of pregnancy-associated iron deficiency anemia. J. Fam. Med. Prim. Care 2018, 7, 1434–1438. [Google Scholar] [CrossRef]
- Buzała, M.; Janicki, B.; Buzała, M.; Słomka, A. Heme iron in meat as the main source of iron in the human diet. J. Elem. 2016, 21, 303–314. [Google Scholar] [CrossRef]
- Shimizu, T.; Huang, D.; Yan, F.; Stranava, M.; Bartosova, M.; Fojtikova, V.; Martinkova, M. Gaseous O2, NO, and CO in signal transduction: Structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 2015, 115, 6491–6533. [Google Scholar] [CrossRef]
- Shimizu, T.; Lengalova, A.; Martinek, V.; Martinkova, M. Heme: Emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 2019, 48, 5624–5657. [Google Scholar] [CrossRef]
- Shisaka, Y.; Shoji, O. Bridging the gap: Unveiling novel functions of a bacterial haem-acquisition protein capturing diverse synthetic porphyrinoids. Coord. Chem. Rev. 2022, 472. [Google Scholar] [CrossRef]
- Wakamatsu, J.; Odagiri, H.; Nishimura, T.; Hattori, A. Quantitative determination of Zn protoporphyrin IX, heme and protoporphyrin IX in Parma ham by HPLC. Meat Sci. 2009, 82, 139–142. [Google Scholar] [CrossRef]
- Ko, Y.J.; Kim, M.; You, S.K.; Shin, S.K.; Chang, J.; Choi, H.J.; Jeong, W.Y.; Lee, M.E.; Hwang, D.H.; Han, S.O. Animal-free heme production for artificial meat in Corynebacterium glutamicum via systems metabolic and membrane engineering. Metab. Eng. 2021, 66, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Wang, Z.; Shen, L.; Xiao, H. Synthetic biology: A new frontier in food production. Trends Biotechnol. 2022, 40, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, Y.; Badar, I.H.; Xia, X.; Kong, B.; Chen, Q. Prospects of artificial meat: Opportunities and challenges around consumer acceptance. Trends Food Sci. Technol. 2021, 116, 434–444. [Google Scholar] [CrossRef]
- Palmer, C. Engineer Dinner—Save the Environment? Engineering 2020, 6, 213–215. [Google Scholar] [CrossRef]
- Ge, B.; Chen, Y.; Yu, Q.; Lin, X.; Li, J.; Qin, S. Regulation of the heme biosynthetic pathway for combinational biosynthesis of phycocyanobilin in Escherichia coli. Process Biochem. 2018, 71, 23–30. [Google Scholar] [CrossRef]
- Celis, A.I.; Choby, J.E.; Kentro, J.; Skaar, E.P.; DuBois, J.L. Control of Metabolite Flux during the Final Steps of Heme b Biosynthesis in Gram-Positive Bacteria. Biochemistry 2019, 58, 5259–5270. [Google Scholar] [CrossRef]
- Feng, C.; Pan, M.; Tang, L. 5-Aminolevulinic acid level and dye-decolorizing peroxidase expression regulate heme synthesis in Escherichia coli. Biotechnol. Lett. 2022, 44, 271–277. [Google Scholar] [CrossRef]
- Ko, Y.J.; Joo, Y.C.; Hyeon, J.E.; Lee, E.; Lee, M.E.; Seok, J.; Kim, S.W.; Park, C.; Han, S.O. Biosynthesis of organic photosensitizer Zn-porphyrin by diphtheria toxin repressor (DtxR)-mediated global upregulation of engineered heme biosynthesis pathway in Corynebacterium glutamicum. Sci. Rep. 2018, 8, 14460. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, J.; Du, G.; Chen, J. Recent Advances in the Microbial Synthesis of Hemoglobin. Trends Biotechnol. 2021, 39, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Richtová, J.; Sheiner, L.; Gruber, A.; Yang, S.-M.; Kořený, L.; Striepen, B.; Oborník, M. Using Diatom and Apicomplexan Models to Study the Heme Pathway of Chromera velia. Int. J. Mol. Sci. 2021, 22, 6495. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qian, X.; Chen, D.; Ge, M. Role of two 5-aminolevulinic acid biosynthetic pathways in heme and secondary metabolite biosynthesis in Amycolatopsis orientalis. J. Basic Microbiol. 2018, 58, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, I.U.; Jahn, M.; Jahn, D. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 2008, 474, 238–251. [Google Scholar] [CrossRef]
- Hubner, I.; Shapiro, J.A.; Hossmann, J.; Drechsel, J.; Hacker, S.M.; Rather, P.N.; Pieper, D.H.; Wuest, W.M.; Sieber, S.A. Broad Spectrum Antibiotic Xanthocillin X Effectively Kills Acinetobacter baumannii via Dysregulation of Heme Biosynthesis. ACS Cent. Sci. 2021, 7, 488–498. [Google Scholar] [CrossRef]
- Steinbrink, J.M.; Myers, R.A.; Hua, K.; Johnson, M.D.; Seidelman, J.L.; Tsalik, E.L.; Henao, R.; Ginsburg, G.S.; Woods, C.W.; Alexander, B.D.; et al. The host transcriptional response to Candidemia is dominated by neutrophil activation and heme biosynthesis and supports novel diagnostic approaches. Genome Med. 2021, 13, 108. [Google Scholar] [CrossRef]
- Phillips, J.D. Heme biosynthesis and the porphyrias. Mol. Genet. Metab. 2019, 128, 164–177. [Google Scholar] [CrossRef]
- Su, T.; Guo, Q.; Zheng, Y.; Liang, Q.; Wang, Q.; Qi, Q. Fine-Tuning of hemB Using CRISPRi for Increasing 5-Aminolevulinic Acid Production in Escherichia coli. Front. Microbiol. 2019, 10, 1731. [Google Scholar] [CrossRef] [Green Version]
- Fiege, K.; Frankenberg-Dinkel, N. Construction of a new T7 promoter compatible Escherichia coli Nissle 1917 strain for recombinant production of heme-dependent proteins. Microb. Cell Factories 2020, 19, 190. [Google Scholar] [CrossRef]
- Yang, P.; Liu, W.; Cheng, X.; Wang, J.; Wang, Q.; Qi, Q. A New Strategy for Production of 5-Aminolevulinic Acid in Recombinant Corynebacterium glutamicum with High Yield. Appl. Environ. Microbiol. 2016, 82, 2709–2717. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, J.; Zhou, J.; Qi, Q.; Du, G.; Chen, J. Recent advances in microbial production of delta-aminolevulinic acid and vitamin B12. Biotechnol. Adv. 2012, 30, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.R.; Choi, K.R.; Lee, S.Y. Metabolic engineering of Escherichia coli for secretory production of free haem. Nat. Catal. 2018, 1, 720–728. [Google Scholar] [CrossRef]
- Cui, Z.; Jiang, Z.; Zhang, J.; Zheng, H.; Jiang, X.; Gong, K.; Liang, Q.; Wang, Q.; Qi, Q. Stable and Efficient Biosynthesis of 5-Aminolevulinic Acid Using Plasmid-Free Escherichia coli. J. Agric. Food Chem. 2019, 67, 1478–1483. [Google Scholar] [CrossRef]
- Duvigneau, J.C.; Esterbauer, H.; Kozlov, A.V. Role of Heme Oxygenase as a Modulator of Heme-Mediated Pathways. Antioxidants 2019, 8, 475. [Google Scholar] [CrossRef] [Green Version]
- Testa, G.; Schaft, J.; van der Hoeven, F.; Glaser, S.; Anastassiadis, K.; Zhang, Y.; Hermann, T.; Stremmel, W.; Stewart, A.F. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 2004, 38, 151–158. [Google Scholar] [CrossRef]
- Rivero-Muller, A.; Lajic, S.; Huhtaniemi, I. Assisted large fragment insertion by Red/ET-recombination (ALFIRE)—An alternative and enhanced method for large fragment recombineering. Nucleic Acids Res. 2007, 35, e78. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Weng, H.; Zhou, Z.; Du, G.; Kang, Z. Engineering of multiple modular pathways for high-yield production of 5-aminolevulinic acid in Escherichia coli. Bioresour. Technol. 2019, 274, 353–360. [Google Scholar] [CrossRef]
- Mauzerall, D.; Granick, S. The Occurrence and Determination of δ-Aminolevulinic Acid and Porphobilinogen in Urine. J. Biol. Chem. 1956, 219, 435–446. [Google Scholar] [CrossRef]
- Espinas, N.A.; Kobayashi, K.; Takahashi, S.; Mochizuki, N.; Masuda, T. Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant Cell Physiol. 2012, 53, 1344–1354. [Google Scholar] [CrossRef]
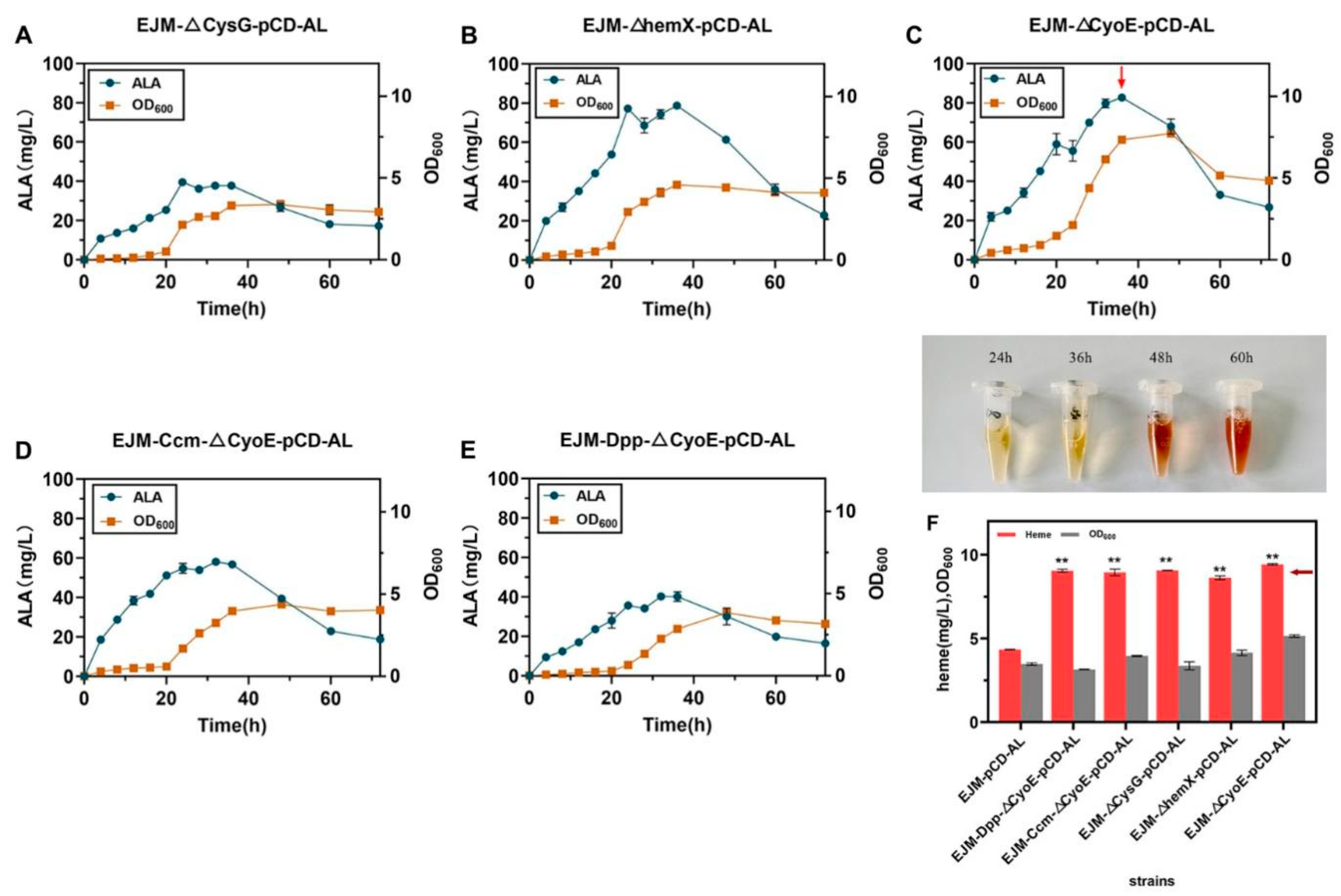

| Strain | Characterization | ALA (mg/L) | Heme (mg/L) |
|---|---|---|---|
| EBL-pCD-AL | E. coli BL21(DE3) contains pCD-hemA-hemL | 90.47 ± 4.37 | 3.82 ± 0.12 |
| EJM-pCD-AL | E. coli JM109(DE3) contains pCD-hemA-hemL | 90.55 ± 1.72 | 4.34 ± 0.02 |
| EJM-Dpp-△CyoE-pCD-AL | Recombinant strains overexpressing the Dpp pathway by gene editing after knockout of gene CyoE | 40.09 ± 1.71 | 9.04 ± 0.06 |
| EJM-Ccm-△CyoE-pCD-AL | Recombinant strains overexpressing the Ccm pathway by gene editing after knockout of gene CyoE | 58.01 ± 1.38 | 8.94 ± 0.14 |
| EJM-△CysG-pCD-AL | Recombinant strain after knockout of gene CysG | 39.53 ± 1.38 | 9.06 ± 0.01 |
| EJM-△hemX-pCD-AL | Recombinant strain after knockout of gene hemX | 78.68 ± 0.83 | 8.62 ± 0.08 |
| EJM-△CyoE-pCD-AL | Recombinant strain after knockout of gene CyoE | 82.73 ± 0.98 | 9.43 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, Z.; Ge, J.; Cui, W.; Zhou, H.; Deng, J.; Xu, B. Efficient De Novo Biosynthesis of Heme by Membrane Engineering in Escherichia coli. Int. J. Mol. Sci. 2022, 23, 15524. https://doi.org/10.3390/ijms232415524
Geng Z, Ge J, Cui W, Zhou H, Deng J, Xu B. Efficient De Novo Biosynthesis of Heme by Membrane Engineering in Escherichia coli. International Journal of Molecular Sciences. 2022; 23(24):15524. https://doi.org/10.3390/ijms232415524
Chicago/Turabian StyleGeng, Zhexian, Jinxia Ge, Wei Cui, Hui Zhou, Jieying Deng, and Baocai Xu. 2022. "Efficient De Novo Biosynthesis of Heme by Membrane Engineering in Escherichia coli" International Journal of Molecular Sciences 23, no. 24: 15524. https://doi.org/10.3390/ijms232415524
APA StyleGeng, Z., Ge, J., Cui, W., Zhou, H., Deng, J., & Xu, B. (2022). Efficient De Novo Biosynthesis of Heme by Membrane Engineering in Escherichia coli. International Journal of Molecular Sciences, 23(24), 15524. https://doi.org/10.3390/ijms232415524






