Comprehensive Analysis of FASN in Tumor Immune Infiltration and Prognostic Value for Immunotherapy and Promoter DNA Methylation
Abstract
:1. Introduction
2. Results
2.1. Differential Expression of FASN between Tumor and Normal Tissue Samples
2.2. The Correlation of FASN Expression and Tumor Grade or Stage
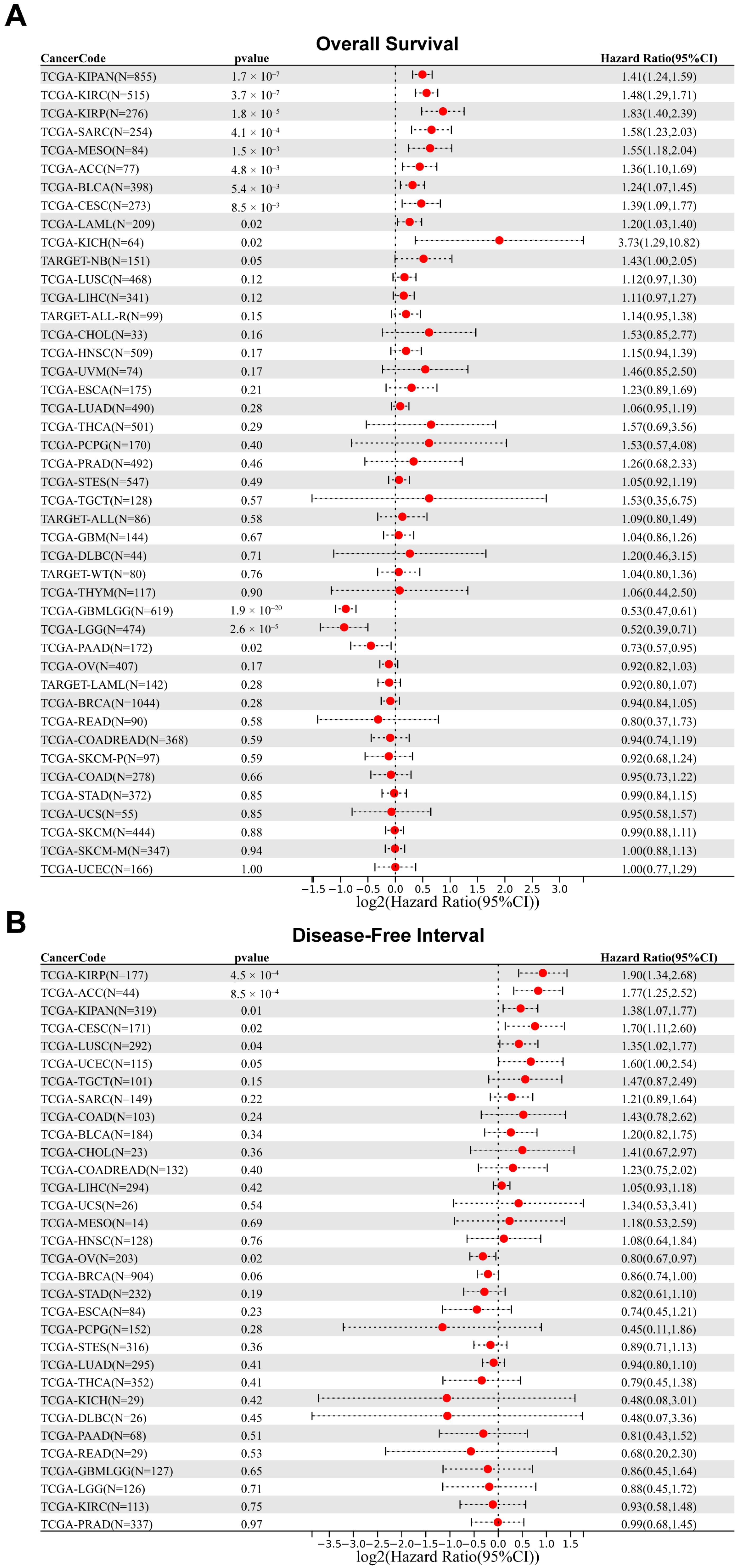
2.3. Prognostic Analysis of FASN in Patients with Different Cancers
2.4. FASN Expression Regulates Tumor Infiltration of Immune Cells in Multiple Cancers
2.5. FASN Offers Significant Prognostic Value for Immunotherapy
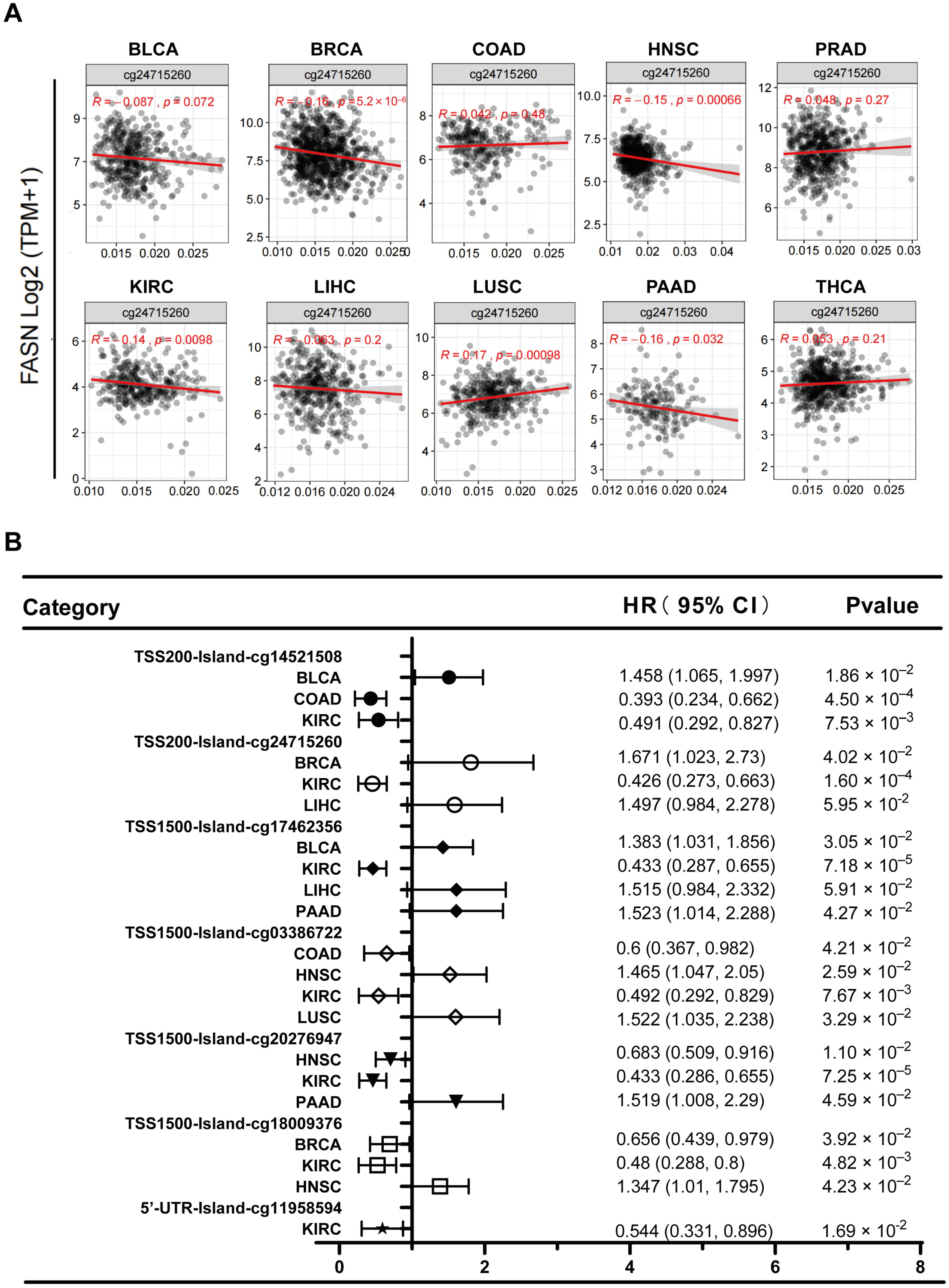
2.6. Analysis of DNA Methylation at the FASN Promoter
2.7. MiRNA-Targeting FASN Prediction and Tumor Suppressor Function Verification
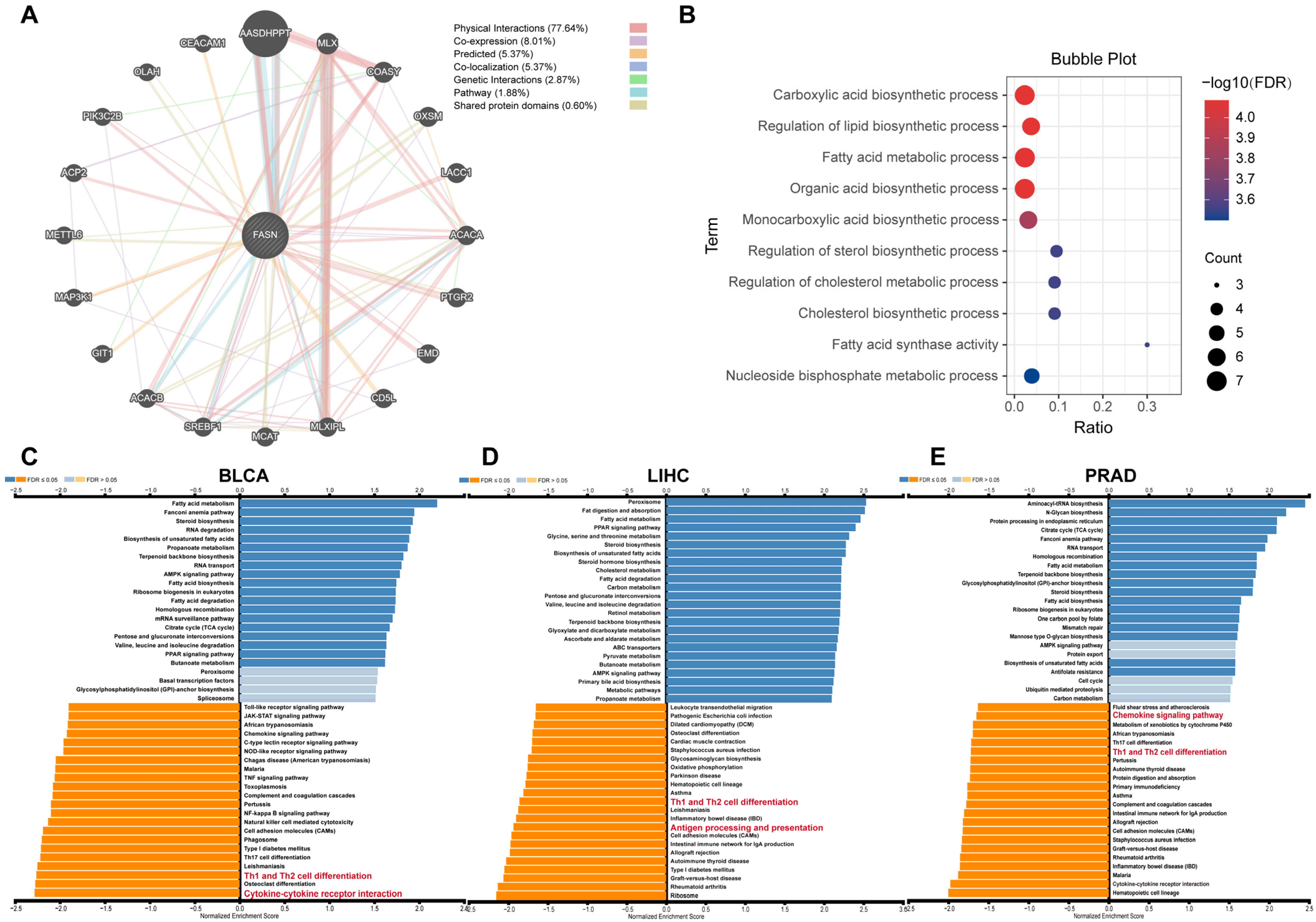
2.8. FASN Functional Enrichment
3. Discussion
4. Materials and Methods
4.1. FASN mRNA and Protein Expression Profiles
4.2. Survival Analysis and Relationship with Clinical Grade and Stage
4.3. Relationship between FASN Expression and Immunity
4.4. Analysis of Mutations in FASN and Immunotherapeutic Response
4.5. Analysis of miRNAs Targeting FASN
4.6. Correlation between FASN Expression and DNA Methylation
4.7. Cell Lines and Cell Culture
4.8. Tissue Sample Sources
4.9. MiRNA Detection and Transfection
4.10. Cell Proliferation Assay
4.11. Clone Formation Assay
4.12. Transwell Assay
4.13. Statistical Analysis
4.14. FASN Functional Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Gong, J.; Shen, S.; Yang, Y.; Qin, S.; Huang, L.; Zhang, H.; Chen, L.; Chen, Y.; Li, S.; She, S.; et al. Inhibition of FASN Suppresses Migration, Invasion and Growth in Hepatoma Carcinoma Cells by Deregulating the HIF-1α/IGFBP1 Pathway. Int. J. Oncol. 2017, 50, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Zaytseva, Y.Y.; Elliott, V.A.; Rychahou, P.; Mustain, W.C.; Kim, J.T.; Valentino, J.; Gao, T.; O’Connor, K.L.; Neltner, J.M.; Lee, E.Y.; et al. Cancer Cell-Associated Fatty Acid Synthase Activates Endothelial Cells and Promotes Angiogenesis in Colorectal Cancer. Carcinogenesis 2014, 35, 1341–1351. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Zhong, X.; Song, H.; Liu, J.; Li, J.; Leung, F.; Lu, W.W.; Liu, Z.-L. Anoikis Resistant Mediated by FASN Promoted Growth and Metastasis of Osteosarcoma. Cell Death Dis. 2019, 10, 298. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.A.; Wei, J.; Nguyen, T.-L.M.; Shi, H.; Su, W.; Palacios, G.; Dhungana, Y.; Chapman, N.M.; Long, L.; Saravia, J.; et al. Lipid Signalling Enforces Functional Specialization of Treg Cells in Tumours. Nature 2021, 591, 306–311. [Google Scholar] [CrossRef]
- Jiang, L.; Fang, X.; Wang, H.; Li, D.; Wang, X. Ovarian Cancer-Intrinsic Fatty Acid Synthase Prevents Anti-Tumor Immunity by Disrupting Tumor-Infiltrating Dendritic Cells. Front. Immunol. 2018, 9, 2927. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Tsai, Y.; Zhu, R.; Keng, P.C.; Chen, Y.; Chen, Y.; Lee, S.O. Retraction Notice to “FASN-TGF-Β1-PD-L1 Axis Contributes to the Development of Resistance to NK Cell Cytotoxicity of Cisplatin-Resistant Lung Cancer Cells” [BBA—Molecular and Cell Biology of Lipids 1863/3 (March 2018) 313-322]. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158946. [Google Scholar] [CrossRef]
- Gouw, A.M.; Eberlin, L.S.; Margulis, K.; Sullivan, D.K.; Toal, G.G.; Tong, L.; Zare, R.N.; Felsher, D.W. Oncogene KRAS Activates Fatty Acid Synthase, Resulting in Specific ERK and Lipid Signatures Associated with Lung Adenocarcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 4300–4305. [Google Scholar] [CrossRef] [Green Version]
- Shiragami, R.; Murata, S.; Kosugi, C.; Tezuka, T.; Yamazaki, M.; Hirano, A.; Yoshimura, Y.; Suzuki, M.; Shuto, K.; Koda, K. Enhanced Antitumor Activity of Cerulenin Combined with Oxaliplatin in Human Colon Cancer Cells. Int. J. Oncol. 2013, 43, 431–438. [Google Scholar] [CrossRef]
- Relat, J.; Blancafort, A.; Oliveras, G.; Cufí, S.; Haro, D.; Marrero, P.F.; Puig, T. Different Fatty Acid Metabolism Effects of (-)-Epigallocatechin-3-Gallate and C75 in Adenocarcinoma Lung Cancer. BMC Cancer 2012, 12, 280. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Zhang, X.; Wang, R.-J.; Ma, Q.-Y.; Xu, L.; Wang, Y.; Liao, H.-P.; Wang, H.-L.; Hu, L.-D.; Kong, X.; et al. PI3Kα Inhibitor CYH33 Triggers Antitumor Immunity in Murine Breast Cancer by Activating CD8+T Cells and Promoting Fatty Acid Metabolism. J. Immunother. Cancer 2021, 9, e003093. [Google Scholar] [CrossRef]
- de Almeida, L.Y.; Mariano, F.S.; Bastos, D.C.; Cavassani, K.A.; Raphelson, J.; Mariano, V.S.; Agostini, M.; Moreira, F.S.; Coletta, R.D.; Mattos-Graner, R.O.; et al. The Antimetastatic Activity of Orlistat Is Accompanied by an Antitumoral Immune Response in Mouse Melanoma. Cancer Chemother. Pharmacol. 2020, 85, 321–330. [Google Scholar] [CrossRef]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic Dysregulation and Emerging Therapeutical Targets for Hepatocellular Carcinoma. Acta Pharm. Sin. B 2022, 12, 558–580. [Google Scholar] [CrossRef]
- Sternby, B.; Hartmann, D.; Borgström, B.; Nilsson, A. Degree of in Vivo Inhibition of Human Gastric and Pancreatic Lipases by Orlistat (Tetrahydrolipstatin, THL) in the Stomach and Small Intestine. Clin. Nutr. 2002, 21, 395–402. [Google Scholar] [CrossRef]
- Agostini, M.; Almeida, L.Y.; Bastos, D.C.; Ortega, R.M.; Moreira, F.S.; Seguin, F.; Zecchin, K.G.; Raposo, H.F.; Oliveira, H.C.F.; Amoêdo, N.D.; et al. The Fatty Acid Synthase Inhibitor Orlistat Reduces the Growth and Metastasis of Orthotopic Tongue Oral Squamous Cell Carcinomas. Mol. Cancer Ther. 2014, 13, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty Acid Synthase (FASN) as a Therapeutic Target in Breast Cancer. Expert Opin. Ther. Targets 2017, 21, 1001–1016. [Google Scholar] [CrossRef]
- Ates, G.; Goldberg, J.; Currais, A.; Maher, P. CMS121, a Fatty Acid Synthase Inhibitor, Protects against Excess Lipid Peroxidation and Inflammation and Alleviates Cognitive Loss in a Transgenic Mouse Model of Alzheimer’s Disease. Redox Biol. 2020, 36, 101648. [Google Scholar] [CrossRef]
- Hu, J.; Wang, H.; Li, X.; Liu, Y.; Mi, Y.; Kong, H.; Xi, D.; Yan, W.; Luo, X.; Ning, Q.; et al. Fibrinogen-like Protein 2 Aggravates Nonalcoholic Steatohepatitis via Interaction with TLR4, Eliciting Inflammation in Macrophages and Inducing Hepatic Lipid Metabolism Disorder. Theranostics 2020, 10, 9702–9720. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Shang, C.; Li, Y.; He, T.; Liao, Y.; Du, Q.; Wang, P.; Qiao, J.; Guo, H. The Prognostic MiR-532-5p-Correlated CeRNA-Mediated Lipid Droplet Accumulation Drives Nodal Metastasis of Cervical Cancer. J. Adv. Res. 2021, 37, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Liu, P.; Zhang, C.; Liu, T.; Wang, W.; Shang, C.; Wu, J.; Liao, Y.; Chen, Y.; Huang, J.; et al. FASN Promotes Lymph Node Metastasis in Cervical Cancer via Cholesterol Reprogramming and Lymphangiogenesis. Cell Death Dis. 2022, 13, 488. [Google Scholar] [CrossRef] [PubMed]
- Man, S.; Yao, J.; Lv, P.; Liu, Y.; Yang, L.; Ma, L. Curcumin-Enhanced Antitumor Effects of Sorafenib via Regulating the Metabolism and Tumor Microenvironment. Food Funct. 2020, 11, 6422–6432. [Google Scholar] [CrossRef]
- Yang, Y. Cancer Immunotherapy: Harnessing the Immune System to Battle Cancer. J. Clin. Investig. 2015, 125, 3335–3337. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Zhang, J.-G.; Deng, H.-W.; Wang, Y.-P. Joint Detection of Associations between DNA Methylation and Gene Expression from Multiple Cancers. IEEE J. Biomed. Health Inform. 2018, 22, 1960–1969. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA Methylation in Cancer: Location Revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a Molecular Understanding of MicroRNA-Mediated Gene Silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on Fat: The Role of Lipid Synthesis in Cancer Metabolism and Tumour Development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef]
- Medes, G.; Thomas, A.; Weinhouse, S. Metabolism of Neoplastic Tissue. IV. A Study of Lipid Synthesis in Neoplastic Tissue Slices in Vitro. Cancer Res. 1953, 13, 27–29. [Google Scholar] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Dellorusso, P.V.; Olson, O.C.; Passegué, E. Dysregulated Haematopoietic Stem Cell Behaviour in Myeloid Leukaemogenesis. Nat. Rev. Cancer 2020, 20, 365–382. [Google Scholar] [CrossRef]
- Shafat, M.S.; Oellerich, T.; Mohr, S.; Robinson, S.D.; Edwards, D.R.; Marlein, C.R.; Piddock, R.E.; Fenech, M.; Zaitseva, L.; Abdul-Aziz, A.; et al. Leukemic Blasts Program Bone Marrow Adipocytes to Generate a Protumoral Microenvironment. Blood 2017, 129, 1320–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Adane, B.; Khan, N.; Sullivan, T.; Minhajuddin, M.; Gasparetto, M.; Stevens, B.; Pei, S.; Balys, M.; Ashton, J.M.; et al. Leukemic Stem Cells Evade Chemotherapy by Metabolic Adaptation to an Adipose Tissue Niche. Cell Stem Cell 2016, 19, 23–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, N.; Li, B.; Xu, X.; Xu, J.; Hu, S. Inhibition of FASN Expression Enhances Radiosensitivity in Human Non-Small Cell Lung Cancer. Oncol. Lett. 2018, 15, 4578–4584. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Xu, Y.; Guo, F.; Chen, X.; Ji, J.; Xu, H.; He, J.; Yu, Y.; Sun, Y.; Lu, X.; et al. FASN Protein Overexpression Indicates Poor Biochemical Recurrence-Free Survival in Prostate Cancer. Dis. Markers 2020, 2020, 3904947. [Google Scholar] [CrossRef]
- Zhou, Y.; Su, W.; Liu, H.; Chen, T.; Höti, N.; Pei, H.; Zhu, H. Fatty Acid Synthase Is a Prognostic Marker and Associated with Immune Infiltrating in Gastric Cancers Precision Medicine. Biomark. Med. 2020, 14, 185–199. [Google Scholar] [CrossRef]
- Lv, M.; Chen, M.; Zhang, R.; Zhang, W.; Wang, C.; Zhang, Y.; Wei, X.; Guan, Y.; Liu, J.; Feng, K.; et al. Manganese Is Critical for Antitumor Immune Responses via CGAS-STING and Improves the Efficacy of Clinical Immunotherapy. Cell Res. 2020, 30, 966–979. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Wang, Z.; Li, G.-Z.; Wang, Q.-W.; Bao, Z.-S.; Zhang, C.-B.; Jiang, T. Immune Cytolytic Activity Is Associated With Genetic and Clinical Properties of Glioma. Front. Immunol. 2019, 10, 1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8+ T Cell States in Human Cancer: Insights from Single-Cell Analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Ahrends, T.; Borst, J. The Opposing Roles of CD4+ T Cells in Anti-Tumour Immunity. Immunology 2018, 154, 582–592. [Google Scholar] [CrossRef] [Green Version]
- Kershaw, M.H.; Westwood, J.A.; Darcy, P.K. Gene-Engineered T Cells for Cancer Therapy. Nat. Rev. Cancer 2013, 13, 525–541. [Google Scholar] [CrossRef]
- Houghton, A.M.; Rzymkiewicz, D.M.; Ji, H.; Gregory, A.D.; Egea, E.E.; Metz, H.E.; Stolz, D.B.; Land, S.R.; Marconcini, L.A.; Kliment, C.R.; et al. Neutrophil Elastase-Mediated Degradation of IRS-1 Accelerates Lung Tumor Growth. Nat. Med. 2010, 16, 219–223. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, A.; Brooks, M.W.; Houshyar, S.; Reinhardt, F.; Ardolino, M.; Fessler, E.; Chen, M.B.; Krall, J.A.; DeCock, J.; Zervantonakis, I.K.; et al. Neutrophils Suppress Intraluminal NK Cell-Mediated Tumor Cell Clearance and Enhance Extravasation of Disseminated Carcinoma Cells. Cancer Discov. 2016, 6, 630–649. [Google Scholar] [CrossRef] [Green Version]
- Allavena, P.; Sica, A.; Solinas, G.; Porta, C.; Mantovani, A. The Inflammatory Micro-Environment in Tumor Progression: The Role of Tumor-Associated Macrophages. Crit. Rev. Oncol. Hematol. 2008, 66, 1–9. [Google Scholar] [CrossRef]
- Jackute, J.; Zemaitis, M.; Pranys, D.; Sitkauskiene, B.; Miliauskas, S.; Vaitkiene, S.; Sakalauskas, R. Distribution of M1 and M2 Macrophages in Tumor Islets and Stroma in Relation to Prognosis of Non-Small Cell Lung Cancer. BMC Immunol. 2018, 19, 3. [Google Scholar] [CrossRef] [Green Version]
- Edin, S.; Wikberg, M.L.; Oldenborg, P.-A.; Palmqvist, R. Macrophages: Good Guys in Colorectal Cancer. Oncoimmunology 2013, 2, e23038. [Google Scholar] [CrossRef] [Green Version]
- Aj, B.; Sf, E. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef]
- Talmadge, J.E.; Donkor, M.; Scholar, E. Inflammatory Cell Infiltration of Tumors: Jekyll or Hyde. Cancer Metastasis Rev. 2007, 26, 373–400. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage Plasticity and Polarization: In Vivo Veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.; Meier, B.; Otsuka, A.; Fenini, G.; Satoh, T.; Gehrke, S.; Widmer, D.; Levesque, M.P.; Mangana, J.; Kerl, K.; et al. Tumour Hypoxia Promotes Melanoma Growth and Metastasis via High Mobility Group Box-1 and M2-like Macrophages. Sci. Rep. 2016, 6, 29914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; He, Y.; Sun, X.; Li, Q.; Wang, W.; Zhao, A.; Di, W. A High M1/M2 Ratio of Tumor-Associated Macrophages Is Associated with Extended Survival in Ovarian Cancer Patients. J. Ovarian Res. 2014, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gryfe, R.; Kim, H.; Hsieh, E.T.; Aronson, M.D.; Holowaty, E.J.; Bull, S.B.; Redston, M.; Gallinger, S. Tumor Microsatellite Instability and Clinical Outcome in Young Patients with Colorectal Cancer. N. Engl. J. Med. 2000, 342, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Fumet, J.-D.; Truntzer, C.; Yarchoan, M.; Ghiringhelli, F. Tumour Mutational Burden as a Biomarker for Immunotherapy: Current Data and Emerging Concepts. Eur. J. Cancer 2020, 131, 40–50. [Google Scholar] [CrossRef]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Dersh, D.; Hollý, J.; Yewdell, J.W. A Few Good Peptides: MHC Class I-Based Cancer Immunosurveillance and Immunoevasion. Nat. Rev. Immunol. 2021, 21, 116–128. [Google Scholar] [CrossRef]
- Yang, S.; Wei, W.; Zhao, Q. B7-H3, a Checkpoint Molecule, as a Target for Cancer Immunotherapy. Int. J. Biol. Sci. 2020, 16, 1767–1773. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zhao, S.; Zhang, Y.; Wang, Y.; Zhang, Z.; Yang, M.; Zhu, Y.; Zhang, G.; Guo, G.; Tong, A.; et al. B7-H3 as a Novel CAR-T Therapeutic Target for Glioblastoma. Mol. Ther. Oncolytics 2019, 14, 279–287. [Google Scholar] [CrossRef]
- Ramjiawan, R.R.; Griffioen, A.W.; Duda, D.G. Anti-Angiogenesis for Cancer Revisited: Is There a Role for Combinations with Immunotherapy? Angiogenesis 2017, 20, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Hurwitz, H.I.; Sandler, A.B.; Miles, D.; Coleman, R.L.; Deurloo, R.; Chinot, O.L. Bevacizumab (Avastin®) in Cancer Treatment: A Review of 15 Years of Clinical Experience and Future Outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A Review of Its Role in Cancers. OncoTargets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Wang, J.; Zhang, D.; Yuan, Y.; Tang, L.; Zhou, Y.; Jiang, X. Immune-Related MiRNA-195-5p Inhibits the Progression of Lung Adenocarcinoma by Targeting Polypyrimidine Tract-Binding Protein 1. Front. Oncol. 2022, 12, 862564. [Google Scholar] [CrossRef] [PubMed]
- Dobosz, P.; Stempor, P.A.; Ramírez Moreno, M.; Bulgakova, N.A. Transcriptional and Post-Transcriptional Regulation of Checkpoint Genes on the Tumour Side of the Immunological Synapse. Heredity 2022, 129, 64–74. [Google Scholar] [CrossRef]
- Xia, M.; Liu, C.J.; Zhang, Q.; Guo, A.Y. GEDS: A Gene Expression Display Server for mRNAs, miRNAs and Proteins. Available online: http://bioinfo.life.hust.edu.cn/web/GEDS/ (accessed on 3 July 2019).
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A Comprehensive, Interaction-Friendly Clinical Bioinformatics Analysis Platform. Available online: http://vip.sangerbox.com/login.html (accessed on 8 July 2022).
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An Integrated Repository Portal for Tumor–Immune System Interactions. Available online: http://cis.hku.hk/TISIDB/index.php (accessed on 15 October 2019).
- Lin, A.; Qi, C.; Wei, T.; Li, M.; Cheng, Q.; Liu, Z.; Luo, P.; Zhang, J. CAMOIP: A Web Server for Comprehensive Analysis on Multi-Omics of Immunotherapy in Pan-Cancer. Available online: http://220.189.241.246:13838/ (accessed on 13 May 2022).
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yu, L.; Sun, Y.; Hao, L.; Bai, J.; Yuan, X.; Wu, R.; Hong, M.; Liu, P.; Duan, X.; et al. Comprehensive Analysis of FASN in Tumor Immune Infiltration and Prognostic Value for Immunotherapy and Promoter DNA Methylation. Int. J. Mol. Sci. 2022, 23, 15603. https://doi.org/10.3390/ijms232415603
Zhang M, Yu L, Sun Y, Hao L, Bai J, Yuan X, Wu R, Hong M, Liu P, Duan X, et al. Comprehensive Analysis of FASN in Tumor Immune Infiltration and Prognostic Value for Immunotherapy and Promoter DNA Methylation. International Journal of Molecular Sciences. 2022; 23(24):15603. https://doi.org/10.3390/ijms232415603
Chicago/Turabian StyleZhang, Mingyang, Lei Yu, Yannan Sun, Li Hao, Jing Bai, Xinyu Yuan, Rihan Wu, Mei Hong, Pengxia Liu, Xiaojun Duan, and et al. 2022. "Comprehensive Analysis of FASN in Tumor Immune Infiltration and Prognostic Value for Immunotherapy and Promoter DNA Methylation" International Journal of Molecular Sciences 23, no. 24: 15603. https://doi.org/10.3390/ijms232415603




