Obesity as a Risk Factor for Breast Cancer—The Role of miRNA
Abstract
1. Introduction
1.1. Epidemiology
1.2. Pathogenesis of Breast Cancer
2. microRNAs (miRNAs) in Breast Cancer
2.1. miRNAs and Obesity
2.2. miRNA and Inflammation
2.3. miRNA and Estrogens
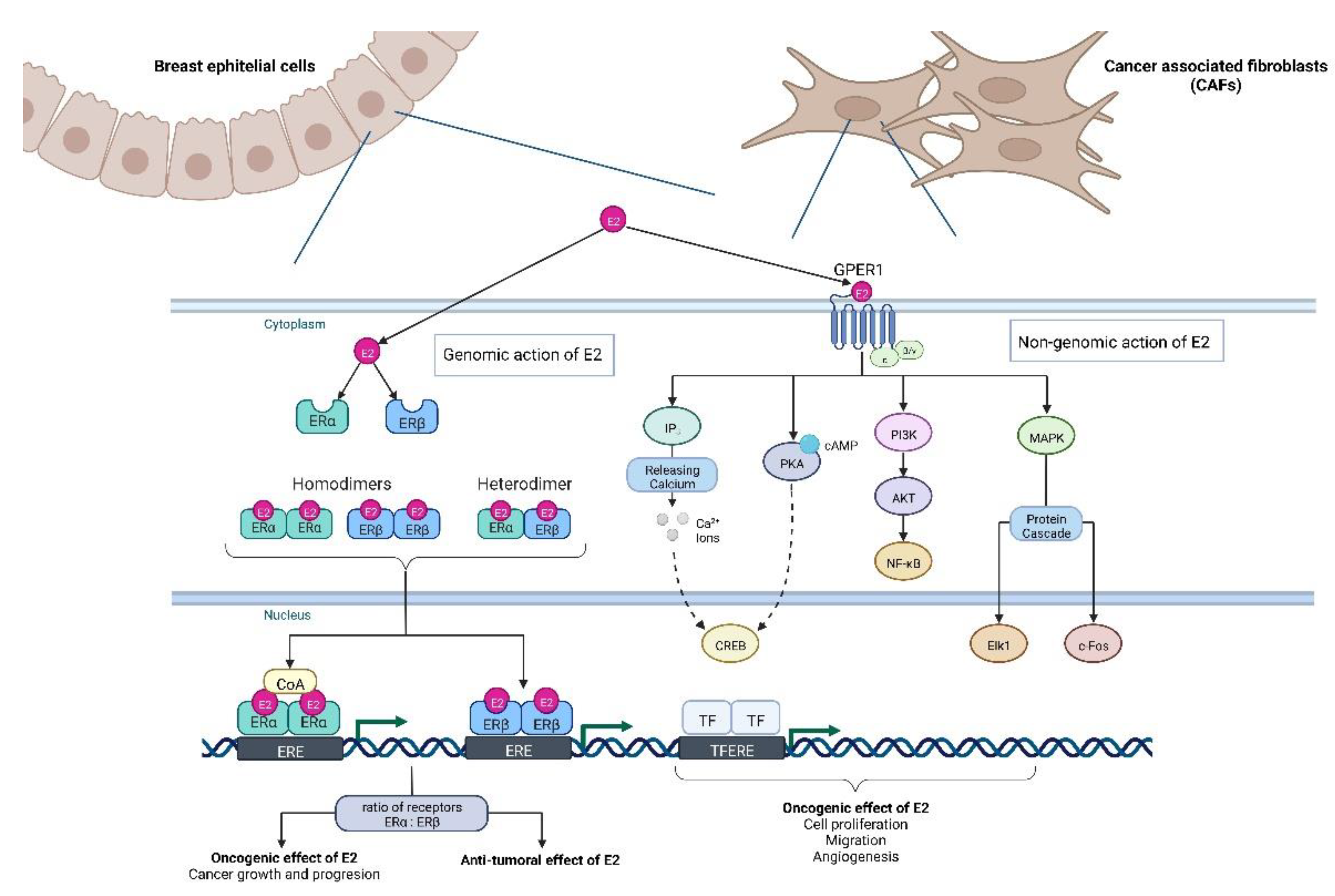

3. Clinical Significance—Diagnosis and Therapy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| miRNAs | microRNAs |
| Her2 | human epidermal growth factor receptor 2 |
| TNBC | triple-negative breast cancer |
| BMI | body mass index |
| WHR | waist-hip ratio |
| AT | adipose tissue |
| IGF-1 | insulin-like growth factor |
| TBARS | thiobarbituric reactive acid substances |
| MDA | malondialdehyde |
| LOOH | lipid hydroperoxides |
| AMPK | AMP-activated protein kinase |
| ERK | signal-regulated kinase |
| JAK/STAT | signaling pathways, Janus kinase |
| PI3K | phosphatidylinositol 3-kinase |
| MAPK | mitogen-activated protein kinase |
| MA | mature adipocytes |
| FOXP4 | forkhead box protein P4 () |
| PPARα | peroxisome proliferator-activated receptor alpha |
| MSCs | marrow stromal stem cells |
| ADIPOQ | adiponectin |
| ADIPOR2 | adiponectin receptor 2 |
| BECN1 | beclin 1 |
| GLUT4 | glucose transporter 4 |
| SCD1 | stearoyl-CoA desaturase-1 |
| DGAT1/2 | diacyl glycerol acyl transferase1/2 |
| FAS | fatty acid synthase |
| AdipoQ | adiponectin |
| ATGL | adipocyte triglyceride lipase |
| AP2 | adipocyte fatty acid binding protein |
| IRS1 | insulin receptor substrate 1 |
| phosphor-AMPKα | Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 |
| ATM | adipose tissue macrophages |
| GLUTs | glucose transporters |
| EVs | extracellular vesicles |
| TDO2 | tryptophan-2,3-dioxygenase |
| LA | linoleic acid |
| DHA | docosahexaenoic acid |
| HIF-1α | hypoxia-inducible factor |
| VEGF | vascular endothelial growth factor |
| ADM | adrenomedullin |
| TGF-β3 | transforming growth factor-β3 |
| HK1, HK2 | hexokinase |
| IGF2 | insulin-like growth factor 2 |
| C-MYC | myelocytomatosis virus oncogene cellular homolog |
| TGFβ | transforming growth factor-β |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| NF-kB | nuclear factor kappa B |
| STAT3 | signal transducer and activator of transcription 3 |
| TLR | toll-like receptor |
| CAFs | cancer-associated fibroblasts |
| TAMs | tumor-associated macrophages |
| ROS | reactive oxygen species |
| DHEA | dehydroepiandrosterone |
| DHEA-S | DHEA sulfate () |
| 17βHSDs | 17β-hydroxysteroid dehydrogenases |
| CYP19A1 | Cytochrome P450 Family 19 Subfamily A Member 1 |
| ASCs | adipose stromal cells |
| GPER1G | protein-coupled estrogen receptor 1 |
| EMT | epithelial-mesenchymal transition |
| SERM | selective estrogen receptor modulators |
| SERD | selective estrogen receptor down regulators). |
| DNMT1 | DNA methyltransferase 1 |
| Ais | aromatase inhibitors |
References
- Chen, D.; Huang, M.; Li, W. Knowledge-Powered Deep Breast Tumor Classification With Multiple Medical Reports. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 891–901. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast Cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Now Most Common Form of Cancer: WHO Taking Action. Available online: https://www.who.int/news/item/03-02-2021-breast-cancer-now-most-common-form-of-cancer-who-taking-action (accessed on 3 February 2021).
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Rosen, J.M. Breast Cancer Heterogeneity through the Lens of Single-Cell Analysis and Spatial Pathologies. Semin. Cancer Biol. 2021, 82, 3–10. [Google Scholar] [CrossRef]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef]
- Berger, A.C.; Korkut, A.; Kanchi, R.S.; Hegde, A.M.; Lenoir, W.; Liu, W.; Liu, Y.; Fan, H.; Shen, H.; Ravikumar, V.; et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018, 33, 690–705.e9. [Google Scholar] [CrossRef]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast Cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Crimini, E.; Repetto, M.; Aftimos, P.; Botticelli, A.; Marchetti, P.; Curigliano, G. Precision Medicine in Breast Cancer: From Clinical Trials to Clinical Practice. Cancer Treat. Rev. 2021, 98, 102223. [Google Scholar] [CrossRef]
- Cserni, G.; Quinn, C.M.; Foschini, M.P.; Bianchi, S.; Callagy, G.; Chmielik, E.; Decker, T.; Fend, F.; Kovács, A.; van Diest, P.J.; et al. Triple-Negative Breast Cancer Histological Subtypes with a Favourable Prognosis. Cancers 2021, 13, 5694. [Google Scholar] [CrossRef]
- Bhushan, A.; Gonsalves, A.; Menon, J.U. Current State of Breast Cancer Diagnosis, Treatment, and Theranostics. Pharmaceutics 2021, 13, 723. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, M.; Focaccetti, C.; Izzi, V.; Masuelli, L.; Modesti, A.; Bei, R. Tumor Antigens Heterogeneity and Immune Response-Targeting Neoantigens in Breast Cancer. Semin. Cancer Biol. 2021, 72, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Fumagalli, C.; Barberis, M. Breast Cancer Heterogeneity. Diagnostics 2021, 11, 1555. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, J.; Gao, Y.; Yang, K.; He, J.; Li, N.; Tian, J. A Systematic Review of Recommendations on Screening Strategies for Breast Cancer Due to Hereditary Predisposition: Who, When, and How? Cancer Med. 2021, 10, 3437–3448. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Benada, J.; Thatte, J.V.; Kinalis, S.; Ejlertsen, B.; Nielsen, F.C.; Sørensen, C.S.; Rossing, M. A Catalog of Curated Breast Cancer Genes. Breast Cancer Res. Treat. 2022, 191, 431–441. [Google Scholar] [CrossRef]
- Liu, H.; Shi, W.; Jin, Z.; Zhuo, R.; Dong, J.; Lao, Q.; Li, S.; Pang, W. Global, Regional, and National Mortality Trends of Female Breast Cancer by Risk Factor, 1990-2017. BMC Cancer 2021, 21, 459. [Google Scholar] [CrossRef]
- Stefan, N. Metabolically Healthy and Unhealthy Normal Weight and Obesity. Endocrinol. Metab. 2020, 35, 487–493. [Google Scholar] [CrossRef]
- Goossens, G.H. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes. Facts 2017, 10, 207–215. [Google Scholar] [CrossRef]
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 9 June 2021).
- Centers for Disease Control and Prevention. Defining Adult Overweight & Obesity. Available online: https://www.cdc.gov/obesity/basics/adult-defining.html (accessed on 3 June 2022).
- Centers for Disease Control and Prevention. National Center for Chronic Disease Prevention and Health Promotion, Division of Nutrition, Physical Activity, and Obesity. Data, Trend and Maps. Available online: https://www.cdc.gov/nccdphp/dnpao/data-trends-maps/index.html (accessed on 25 November 2022).
- Hannah Ritchie and Max Roser—“Obesity”. Published Online at Our-WorldInData.Org. 2017. Available online: https://ourworldindata.org/obesity (accessed on 11 August 2017).
- Zhang, H.; Zhao, X.; Guo, Y.; Chen, R.; He, J.; Li, L.; Qiang, Z.; Yang, Q.; Liu, X.; Huang, C.; et al. Hypoxia Regulates Overall MRNA Homeostasis by Inducing Met1-Linked Linear Ubiquitination of AGO2 in Cancer Cells. Nat. Commun. 2021, 12, 5416. [Google Scholar] [CrossRef] [PubMed]
- Gajeton, J.; Krukovets, I.; Muppala, S.; Verbovetskiy, D.; Zhang, J.; Stenina-Adognravi, O. Hyperglycemia-Induced MiR-467 Drives Tumor Inflammation and Growth in Breast Cancer. Cancers 2021, 13, 1346. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Siegel, R.L.; Torre, L.A.; Pearson-Stuttard, J.; Islami, F.; Fedewa, S.A.; Goding Sauer, A.; Shuval, K.; Gapstur, S.M.; Jacobs, E.J.; et al. Global Patterns in Excess Body Weight and the Associated Cancer Burden. CA Cancer J. Clin. 2018, 69, 88–112. [Google Scholar] [CrossRef]
- Kolb, R.; Zhang, W. Obesity and Breast Cancer: A Case of Inflamed Adipose Tissue. Cancers 2020, 12, 1686. [Google Scholar] [CrossRef]
- Protani, M.; Coory, M.; Martin, J.H. Effect of Obesity on Survival of Women with Breast Cancer: Systematic Review and Meta-Analysis. Breast Cancer Res. Treat. 2010, 123, 627–635. [Google Scholar] [CrossRef]
- Liu, Y.L.; Saraf, A.; Catanese, B.; Lee, S.M.; Zhang, Y.; Connolly, E.P.; Kalinsky, K. Obesity and Survival in the Neoadjuvant Breast Cancer Setting: Role of Tumor Subtype in an Ethnically Diverse Population. Breast Cancer Res. Treat. 2018, 167, 277–288. [Google Scholar] [CrossRef]
- Zhao, X.-B.; Ren, G.-S. Diabetes Mellitus and Prognosis in Women with Breast Cancer: A Systematic Review and Meta-Analysis. Medicine 2016, 95, e5602. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cai, Y.; Yu, F.; Ping, Z.; Liu, L. Body Mass Index Increases the Lymph Node Metastasis Risk of Breast Cancer: A Dose-Response Meta-Analysis with 52904 Subjects from 20 Cohort Studies. BMC Cancer 2020, 20, 601. [Google Scholar] [CrossRef] [PubMed]
- Ewertz, M.; Jensen, M.-B.; Gunnarsdóttir, K.Á.; Højris, I.; Jakobsen, E.H.; Nielsen, D.; Stenbygaard, L.E.; Tange, U.B.; Cold, S. Effect of Obesity on Prognosis after Early-Stage Breast Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 25–31. [Google Scholar] [CrossRef]
- Lenz, M.; Arts, I.C.W.; Peeters, R.L.M.; de Kok, T.M.; Ertaylan, G. Adipose Tissue in Health and Disease through the Lens of Its Building Blocks. Sci. Rep. 2020, 10, 10433. [Google Scholar] [CrossRef] [PubMed]
- Agurs-Collins, T.; Ross, S.A.; Dunn, B.K. The Many Faces of Obesity and Its Influence on Breast Cancer Risk. Front. Oncol. 2019, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Blüher, M.; Bashan, N.; Shai, I.; Harman-Boehm, I.; Tarnovscki, T.; Avinaoch, E.; Stumvoll, M.; Dietrich, A.; Klöting, N.; Rudich, A. Activated Ask1-MKK4-P38MAPK/JNK Stress Signaling Pathway in Human Omental Fat Tissue May Link Macrophage Infiltration to Whole-Body Insulin Sensitivity. J. Clin. Endocrinol. Metab. 2009, 94, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Haim, Y.; Blüher, M.; Konrad, D.; Goldstein, N.; Klöting, N.; Harman-Boehm, I.; Kirshtein, B.; Ginsberg, D.; Tarnovscki, T.; Gepner, Y.; et al. ASK1 (MAP3K5) Is Transcriptionally Upregulated by E2F1 in Adipose Tissue in Obesity, Molecularly Defining a Human Dys-Metabolic Obese Phenotype. Mol. Metab. 2017, 6, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Nono Nankam, P.A.; Nguelefack, T.B.; Goedecke, J.H.; Blüher, M. Contribution of Adipose Tissue Oxidative Stress to Obesity-Associated Diabetes Risk and Ethnic Differences: Focus on Women of African Ancestry. Antioxidants 2021, 10, 622. [Google Scholar] [CrossRef]
- Pham, D.-V.; Park, P.-H. Tumor Metabolic Reprogramming by Adipokines as a Critical Driver of Obesity-Associated Cancer Progression. Int. J. Mol. Sci. 2021, 22, 1444. [Google Scholar] [CrossRef]
- Nam, G.E.; Zhang, Z.-F.; Rao, J.; Zhou, H.; Jung, S.Y. Interactions Between Adiponectin-Pathway Polymorphisms and Obesity on Postmenopausal Breast Cancer Risk Among African American Women: The WHI SHARe Study. Front. Oncol. 2021, 11, 698198. [Google Scholar] [CrossRef]
- Barone, I.; Caruso, A.; Gelsomino, L.; Giordano, C.; Bonofiglio, D.; Catalano, S.; Andò, S. Obesity and Endocrine Therapy Resistance in Breast Cancer: Mechanistic Insights and Perspectives. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2022, 23, e13358. [Google Scholar] [CrossRef]
- Bjune, J.-I.; Strømland, P.P.; Jersin, R.Å.; Mellgren, G.; Dankel, S.N. Metabolic and Epigenetic Regulation by Estrogen in Adipocytes. Front. Endocrinol. 2022, 13, 828780. [Google Scholar] [CrossRef]
- Rubinstein, M.M.; Brown, K.A.; Iyengar, N.M. Targeting Obesity-Related Dysfunction in Hormonally Driven Cancers. Br. J. Cancer 2021, 125, 495–509. [Google Scholar] [CrossRef]
- Gérard, C.; Brown, K.A. Obesity and Breast Cancer—Role of Estrogens and the Molecular Underpinnings of Aromatase Regulation in Breast Adipose Tissue. Mol. Cell. Endocrinol. 2018, 466, 15–30. [Google Scholar] [CrossRef]
- Kiran, S.; Kumar, V.; Kumar, S.; Price, R.L.; Singh, U.P. Adipocyte, Immune Cells, and MiRNA Crosstalk: A Novel Regulator of Metabolic Dysfunction and Obesity. Cells 2021, 10, 1004. [Google Scholar] [CrossRef] [PubMed]
- Danforth, D.N. The Role of Chronic Inflammation in the Development of Breast Cancer. Cancers 2021, 13, 3918. [Google Scholar] [CrossRef] [PubMed]
- Pérez, S.; Rius-Pérez, S. Macrophage Polarization and Reprogramming in Acute Inflammation: A Redox Perspective. Antioxidants 2022, 11, 1394. [Google Scholar] [CrossRef] [PubMed]
- Artemniak-Wojtowicz, D.; Kucharska, A.M.; Pyrżak, B. Obesity and Chronic Inflammation Crosslinking. Cent.-Eur. J. Immunol. 2020, 45, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Pirola, L.; Ferraz, J.C. Role of Pro- and Anti-Inflammatory Phenomena in the Physiopathology of Type 2 Diabetes and Obesity. World J. Biol. Chem. 2017, 8, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity Is Associated with Macrophage Accumulation in Adipose Tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Ruggiero, A.D.; Vemuri, R.; Block, M.; DeStephanis, D.; Davis, M.; Chou, J.; Williams, A.; Brock, A.; Das, S.K.; Kavanagh, K. Macrophage Phenotypes and Gene Expression Patterns Are Unique in Naturally Occurring Metabolically Healthy Obesity. Int. J. Mol. Sci. 2022, 23, 12680. [Google Scholar] [CrossRef]
- Springer, N.L.; Iyengar, N.M.; Bareja, R.; Verma, A.; Jochelson, M.S.; Giri, D.D.; Zhou, X.K.; Elemento, O.; Dannenberg, A.J.; Fischbach, C. Obesity-Associated Extracellular Matrix Remodeling Promotes a Macrophage Phenotype Similar to Tumor-Associated Macrophages. Am. J. Pathol. 2019, 189, 2019–2035. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-Associated Macrophages in Tumor Metastasis: Biological Roles and Clinical Therapeutic Applications. J. Hematol. Oncol. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Munir, M.T.; Kay, M.K.; Kang, M.H.; Rahman, M.M.; Al-Harrasi, A.; Choudhury, M.; Moustaid-Moussa, N.; Hussain, F.; Rahman, S.M. Tumor-Associated Macrophages as Multifaceted Regulators of Breast Tumor Growth. Int. J. Mol. Sci. 2021, 22, 6526. [Google Scholar] [CrossRef]
- Larionova, I.; Tuguzbaeva, G.; Ponomaryova, A.; Stakheyeva, M.; Cherdyntseva, N.; Pavlov, V.; Choinzonov, E.; Kzhyshkowska, J. Tumor-Associated Macrophages in Human Breast, Colorectal, Lung, Ovarian and Prostate Cancers. Front. Oncol. 2020, 10, 566511. [Google Scholar] [CrossRef] [PubMed]
- Muppala, S.; Xiao, R.; Gajeton, J.; Krukovets, I.; Verbovetskiy, D.; Stenina-Adognravi, O. Thrombospondin-4 Mediates Hyperglycemia- and TGF-Beta-Induced Inflammation in Breast Cancer. Int. J. Cancer 2021, 148, 2010–2022. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues Mantuano, N.; Stanczak, M.A.; Oliveira, I.d.A.; Kirchhammer, N.; Filardy, A.A.; Monaco, G.; Santos, R.C.; Fonseca, A.C.; Fontes, M.; Bastos, C.D.S.; et al. Hyperglycemia Enhances Cancer Immune Evasion by Inducing Alternative Macrophage Polarization through Increased O-GlcNAcylation. Cancer Immunol. Res. 2020, 8, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Lowe, S.W. The Microcosmos of Cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Arun, R.P.; Cahill, H.F.; Marcato, P. Breast Cancer Subtype-Specific MiRNAs: Networks, Impacts, and the Potential for Intervention. Biomedicines 2022, 10, 651. [Google Scholar] [CrossRef] [PubMed]
- Ahonen, M.A.; Asghar, M.Y.; Parviainen, S.J.; Liebisch, G.; Höring, M.; Leidenius, M.; Fischer-Posovszky, P.; Wabitsch, M.; Mikkola, T.S.; Törnquist, K.; et al. Human Adipocyte Differentiation and Composition of Disease-Relevant Lipids Are Regulated by MiR-221-3p. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158841. [Google Scholar] [CrossRef] [PubMed]
- Rajarajan, D.; Selvarajan, S.; Charan Raja, M.R.; Kar Mahapatra, S.; Kasiappan, R. Genome-Wide Analysis Reveals MiR-3184-5p and MiR-181c-3p as a Critical Regulator for Adipocytes-Associated Breast Cancer. J. Cell. Physiol. 2019, 234, 17959–17974. [Google Scholar] [CrossRef]
- Zhang, H.-G.; Wang, X.-B.; Zhao, H.; Zhou, C.-N. MicroRNA-9-5p Promotes Osteoporosis Development through Inhibiting Osteogenesis and Promoting Adipogenesis via Targeting Wnt3a. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 456–463. [Google Scholar] [CrossRef]
- Chung, S.J.; Nagaraju, G.P.; Nagalingam, A.; Muniraj, N.; Kuppusamy, P.; Walker, A.; Woo, J.; Győrffy, B.; Gabrielson, E.; Saxena, N.K.; et al. ADIPOQ/Adiponectin Induces Cytotoxic Autophagy in Breast Cancer Cells through STK11/LKB1-Mediated Activation of the AMPK-ULK1 Axis. Autophagy 2017, 13, 1386–1403. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, S.; Tang, W.; Huang, Q.; Mei, Y.; Yang, H. Exosomes from Tamoxifen-Resistant Breast Cancer Cells Transmit Drug Resistance Partly by Delivering MiR-9-5p. Cancer Cell Int. 2021, 21, 55. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Zhang, D.; Katayama, A.; Kurooka, N.; Sugawara, R.; Albuayjan, H.H.H.; Nakatsuka, A.; Eguchi, J.; Wada, J. Adipocyte-Specific Inhibition of Mir221/222 Ameliorates Diet-Induced Obesity Through Targeting Ddit4. Front. Endocrinol. 2021, 12, 750261. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Shen, L.; Wang, S.; Guo, Z.; Zheng, T.; Tan, Y.; Fan, Y.; Liu, L.; Chen, L.; Jiang, A.; et al. Genistein Inhibits High Fat Diet-Induced Obesity through MiR-222 by Targeting BTG2 and Adipor1. Food Funct. 2020, 11, 2418–2426. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Park, J.-Y. Emerging Roles of 14-3-3γ in the Brain Disorder. BMB Rep. 2020, 53, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Li, Z.; Sun, S.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Zhang, Y.; Sun, S.; et al. Exosomes from the Tumour-Adipocyte Interplay Stimulate Beige/Brown Differentiation and Reprogram Metabolism in Stromal Adipocytes to Promote Tumour Progression. J. Exp. Clin. Cancer Res. 2019, 38, 223. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal MiRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage-Derived MicroRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, C.-J.; Guo, G.-L. MiR-155 Promotes the Proliferation and Migration of Breast Cancer Cells via Targeting SOCS1 and MMP16. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7323–7332. [Google Scholar] [CrossRef] [PubMed]
- Itani, M.M.; Nassar, F.J.; Tfayli, A.H.; Talhouk, R.S.; Chamandi, G.K.; Itani, A.R.S.; Makoukji, J.; Boustany, R.-M.N.; Hou, L.; Zgheib, N.K.; et al. A Signature of Four Circulating MicroRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021, 22, 6121. [Google Scholar] [CrossRef]
- Chen, B.; Li, H.; Zeng, X.; Yang, P.; Liu, X.; Zhao, X.; Liang, S. Roles of MicroRNA on Cancer Cell Metabolism. J. Transl. Med. 2012, 10, 228. [Google Scholar] [CrossRef]
- Wu, W.; Zhao, S. Metabolic Changes in Cancer: Beyond the Warburg Effect. Acta Biochim. Biophys. Sin. 2013, 45, 18–26. [Google Scholar] [CrossRef]
- Thorens, B.; Mueckler, M. Glucose Transporters in the 21st Century. Am. J. Physiol.-Endocrinol. Metab. 2010, 298, E141–E145. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Kim, H.; Nam, K.; Shin, I. Glut1 Promotes Cell Proliferation, Migration and Invasion by Regulating Epidermal Growth Factor Receptor and Integrin Signaling in Triple-Negative Breast Cancer Cells. BMB Rep. 2017, 50, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tang, H.; Liu, X.; Liu, P.; Yang, L.; Xie, X.; Ye, F.; Song, C.; Xie, X.; Wei, W. MiR-22 as a Prognostic Factor Targets Glucose Transporter Protein Type 1 in Breast Cancer. Cancer Lett. 2015, 356, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-Cancer-Secreted MiR-122 Reprograms Glucose Metabolism in Premetastatic Niche to Promote Metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Huang, X.; Ye, F.; Chen, B.; Song, C.; Wen, J.; Zhang, Z.; Zheng, G.; Tang, H.; Xie, X. The MiR-34a-LDHA Axis Regulates Glucose Metabolism and Tumor Growth in Breast Cancer. Sci. Rep. 2016, 6, 21735. [Google Scholar] [CrossRef]
- Li, L.; Kang, L.; Zhao, W.; Feng, Y.; Liu, W.; Wang, T.; Mai, H.; Huang, J.; Chen, S.; Liang, Y.; et al. MiR-30a-5p Suppresses Breast Tumor Growth and Metastasis through Inhibition of LDHA-Mediated Warburg Effect. Cancer Lett. 2017, 400, 89–98. [Google Scholar] [CrossRef]
- Chen, C.; Bai, L.; Cao, F.; Wang, S.; He, H.; Song, M.; Chen, H.; Liu, Y.; Guo, J.; Si, Q.; et al. Targeting LIN28B Reprograms Tumor Glucose Metabolism and Acidic Microenvironment to Suppress Cancer Stemness and Metastasis. Oncogene 2019, 38, 4527–4539. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine Reliance in Cell Metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, D.; Zhao, Z.; Ren, P.; Li, C.; Hu, Y.; Shi, J.; Su, H.; Wang, L.; Liu, H.; et al. Epigenetic Silencing of MicroRNA-137 Enhances ASCT2 Expression and Tumor Glutamine Metabolism. Oncogenesis 2017, 6, e356. [Google Scholar] [CrossRef]
- Liu, H.; Huang, X.; Ye, T. MiR-22 down-Regulates the Proto-Oncogene ATP Citrate Lyase to Inhibit the Growth and Metastasis of Breast Cancer. Am. J. Transl. Res. 2018, 10, 659–669. [Google Scholar]
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.-H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-Cell-Secreted Exosomal MiR-105 Promotes Tumour Growth through the MYC-Dependent Metabolic Reprogramming of Stromal Cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, N.C.; Rogers, T.J.; Gordon, M.A.; Greene, L.I.; Cochrane, D.R.; Spoelstra, N.S.; Nemkov, T.G.; D’Alessandro, A.; Hansen, K.C.; Richer, J.K. A TDO2-AhR Signaling Axis Facilitates Anoikis Resistance and Metastasis in Triple-Negative Breast Cancer. Cancer Res. 2015, 75, 4651–4664. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, P.; Fallarino, F.; Italiano, A.; Soubeyran, I.; MacGrogan, G.; Debled, M.; Velasco, V.; Bodet, D.; Eimer, S.; Veldhoen, M.; et al. Accumulation of an Endogenous Tryptophan-Derived Metabolite in Colorectal and Breast Cancers. PLoS ONE 2015, 10, e0122046. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.J.; Christenson, J.L.; Greene, L.I.; O’Neill, K.I.; Williams, M.M.; Gordon, M.A.; Nemkov, T.; D’Alessandro, A.; Degala, G.D.; Shin, J.; et al. Reversal of Triple-Negative Breast Cancer EMT by MiR-200c Decreases Tryptophan Catabolism and a Program of Immunosuppression. Mol. Cancer Res. 2019, 17, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Li, X.; Chou, J.; Xiang, C.; Guo, Q.; Zhang, Z.; Guo, X.; Gao, L.; Xing, Y.; Xi, T. StarD13 3’-Untranslated Region Functions as a CeRNA for TP53INP1 in Prohibiting Migration and Invasion of Breast Cancer Cells by Regulating MiR-125b Activity. Eur. J. Cell Biol. 2018, 97, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Zhang, R.; He, Y.; Zou, M.; Guo, L.; Xi, T. MicroRNA-125b Induces Metastasis by Targeting STARD13 in MCF-7 and MDA-MB-231 Breast Cancer Cells. PLoS ONE 2012, 7, e35435. [Google Scholar] [CrossRef] [PubMed]
- Klopotowska, D.; Matuszyk, J.; Wietrzyk, J. Steroid Hormone Calcitriol and Its Analog Tacalcitol Inhibit MiR-125b Expression in a Human Breast Cancer MCF-7 Cell Line. Steroids 2019, 141, 70–75. [Google Scholar] [CrossRef]
- Dierssen-Sotos, T.; Gómez-Acebo, I.; Palazuelos, C.; Gracia-Lavedan, E.; Pérez-Gómez, B.; Oribe, M.; Martín, V.; Guevara, M.; Rodríguez-Cundín, P.; Fernández-Tardón, G.; et al. Fatty Acid Intake and Breast Cancer in the Spanish Multicase–Control Study on Cancer (MCC-Spain). Eur. J. Nutr. 2020, 59, 1171–1179. [Google Scholar] [CrossRef]
- Gucalp, A.; Zhou, X.K.; Cook, E.D.; Garber, J.E.; Crew, K.D.; Nangia, J.R.; Bhardwaj, P.; Giri, D.D.; Elemento, O.; Verma, A.; et al. A Randomized Multicenter Phase II Study of Docosahexaenoic Acid in Patients with a History of Breast Cancer, Premalignant Lesions, or Benign Breast Disease. Cancer Prev. Res. 2018, 11, 203–214. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, T.; Zhai, S.; Li, W.; Meng, Q. Linoleic Acid and Breast Cancer Risk: A Meta-Analysis. Public Health Nutr. 2016, 19, 1457–1463. [Google Scholar] [CrossRef]
- Gonzalez-Reyes, C.; Marcial-Medina, C.; Cervantes-Anaya, N.; Cortes-Reynosa, P.; Salazar, E.P. Migration and Invasion Induced by Linoleic Acid Are Mediated through Fascin in MDA-MB-231 Breast Cancer Cells. Mol. Cell. Biochem. 2018, 443, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Elieh Ali Komi, D.; Shekari, N.; Soofian-kordkandi, P.; Javadian, M.; Shanehbandi, D.; Baradaran, B.; Kazemi, T. Docosahexaenoic Acid (DHA) and Linoleic Acid (LA) Modulate the Expression of Breast Cancer Involved MiRNAs in MDA-MB-231 Cell Line. Clin. Nutr. ESPEN 2021, 46, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Luengo-Gil, G.; Gonzalez-Billalabeitia, E.; Perez-Henarejos, S.A.; Navarro Manzano, E.; Chaves-Benito, A.; Garcia-Martinez, E.; Garcia-Garre, E.; Vicente, V.; Ayala de la Peña, F. Angiogenic Role of MiR-20a in Breast Cancer. PLoS ONE 2018, 13, e0194638. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Mudduluru, G.; Ceppi, P.; Muppala, S.; Kozlowski, M.; Niklinski, J.; Papotti, M.; Allgayer, H. MicroRNA-30a Inhibits Epithelial-to-Mesenchymal Transition by Targeting Snai1 and Is Downregulated in Non-Small Cell Lung Cancer. Int. J. Cancer 2012, 130, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Pan, L.; Gao, J.; Ye, X.; Chen, L.; Zhang, X.; Tang, W.; Zheng, W. Prognostic Value of MiR-106b Expression in Breast Cancer Patients. J. Surg. Res. 2015, 195, 158–165. [Google Scholar] [CrossRef]
- Jiang, N.; Zou, C.; Zhu, Y.; Luo, Y.; Chen, L.; Lei, Y.; Tang, K.; Sun, Y.; Zhang, W.; Li, S.; et al. HIF-1α-Regulated MiR-1275 Maintains Stem Cell-like Phenotypes and Promotes the Progression of LUAD by Simultaneously Activating Wnt/β-Catenin and Notch Signaling. Theranostics 2020, 10, 2553–2570. [Google Scholar] [CrossRef]
- Tang, T.; Yang, Z.; Zhu, Q.; Wu, Y.; Sun, K.; Alahdal, M.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. Up-Regulation of MiR-210 Induced by a Hypoxic Microenvironment Promotes Breast Cancer Stem Cells Metastasis, Proliferation, and Self-Renewal by Targeting E-Cadherin. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 6965–6981. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, J.; Wang, L.; Dai, H.; Li, N.; Hu, W.; Cai, H. HIF-1α Promotes Breast Cancer Cell MCF-7 Proliferation and Invasion Through Regulating MiR-210. Cancer Biother. Radiopharm. 2017, 32, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Xiao, J.; Zhou, Z.; Wu, J.; Ge, F.; Li, Z.; Zhang, H.; Sun, J.; Li, F.; Liu, R.; et al. Hypoxia Induces MiR-153 through the IRE1α-XBP1 Pathway to Fine Tune the HIF1α/VEGFA Axis in Breast Cancer Angiogenesis. Oncogene 2018, 37, 1961–1975. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and Tumor Progression: Signaling Pathways and Targeted Intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Danenberg, E.; Bardwell, H.; Zanotelli, V.R.T.; Provenzano, E.; Chin, S.-F.; Rueda, O.M.; Green, A.; Rakha, E.; Aparicio, S.; Ellis, I.O.; et al. Breast Tumor Microenvironment Structures Are Associated with Genomic Features and Clinical Outcome. Nat. Genet. 2022, 54, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, S.; Hu, Y.; Huang, W. Landscape of Immune Microenvironment Under Immune Cell Infiltration Pattern in Breast Cancer. Front. Immunol. 2021, 12, 711433. [Google Scholar] [CrossRef]
- Song, P.; Li, Y.; Dong, Y.; Liang, Y.; Qu, H.; Qi, D.; Lu, Y.; Jin, X.; Guo, Y.; Jia, Y.; et al. Estrogen Receptor β Inhibits Breast Cancer Cells Migration and Invasion through CLDN6-Mediated Autophagy. J. Exp. Clin. Cancer Res. 2019, 38, 354. [Google Scholar] [CrossRef] [PubMed]
- Schüler-Toprak, S.; Häring, J.; Inwald, E.C.; Moehle, C.; Ortmann, O.; Treeck, O. Agonists and Knockdown of Estrogen Receptor β Differentially Affect Invasion of Triple-Negative Breast Cancer Cells in Vitro. BMC Cancer 2016, 16, 951. [Google Scholar] [CrossRef]
- Grober, O.M.; Mutarelli, M.; Giurato, G.; Ravo, M.; Cicatiello, L.; De Filippo, M.R.; Ferraro, L.; Nassa, G.; Papa, M.F.; Paris, O.; et al. Global Analysis of Estrogen Receptor Beta Binding to Breast Cancer Cell Genome Reveals an Extensive Interplay with Estrogen Receptor Alpha for Target Gene Regulation. BMC Genomics 2011, 12, 36. [Google Scholar] [CrossRef]
- Taroeno-Hariadi, K.W.; Hardianti, M.S.; Sinorita, H.; Aryandono, T. Obesity, Leptin, and Deregulation of MicroRNA in Lipid Metabolisms: Their Contribution to Breast Cancer Prognosis. Diabetol. Metab. Syndr. 2021, 13, 10. [Google Scholar] [CrossRef]
- Duggan, C.; Tapsoba, J.d.D.; Scheel, J.; Wang, C.-Y.; McTiernan, A. Weight Loss Reduces Circulating Micro-RNA Related to Obesity and Breast Cancer in Postmenopausal Women. Epigenetics 2022, 17, 1–14. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Kielstein, H. Adipokines Regulate the Expression of Tumor-Relevant MicroRNAs. Obes. Facts 2019, 12, 211–225. [Google Scholar] [CrossRef]
- Martínez-Gutierrez, A.; Carbajal-Lopez, B.; Bui, T.M.; Mendoza-Rodriguez, M.; Campos-Parra, A.D.; Calderillo-Ruiz, G.; Cantú-De Leon, D.; Madrigal-Santillán, E.-O.; Sumagin, R.; Pérez-Plasencia, C.; et al. A MicroRNA Panel That Regulates Proinflammatory Cytokines as Diagnostic and Prognosis Biomarkers in Colon Cancer. Biochem. Biophys. Rep. 2022, 30, 101252. [Google Scholar] [CrossRef]
- Ebright, R.Y.; Zachariah, M.A.; Micalizzi, D.S.; Wittner, B.S.; Niederhoffer, K.L.; Nieman, L.T.; Chirn, B.; Wiley, D.F.; Wesley, B.; Shaw, B.; et al. HIF1A Signaling Selectively Supports Proliferation of Breast Cancer in the Brain. Nat. Commun. 2020, 11, 6311. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-N.; Chen, P.-S.; Chiu, C.-F.; Lyu, Y.-J.; Lo, C.; Tsai, L.-W.; Wang, M.-Y. TARBP2 Suppresses Ubiquitin-Proteasomal Degradation of HIF-1α in Breast Cancer. Int. J. Mol. Sci. 2021, 23, 208. [Google Scholar] [CrossRef] [PubMed]
- Menikdiwela, K.R.; Kahathuduwa, C.; Bolner, M.L.; Rahman, R.L.; Moustaid-Moussa, N. Association between Obesity, Race or Ethnicity, and Luminal Subtypes of Breast Cancer. Biomedicines 2022, 10, 2931. [Google Scholar] [CrossRef] [PubMed]
- Scabia, V.; Ayyanan, A.; De Martino, F.; Agnoletto, A.; Battista, L.; Laszlo, C.; Treboux, A.; Zaman, K.; Stravodimou, A.; Jallut, D.; et al. Estrogen Receptor Positive Breast Cancers Have Patient Specific Hormone Sensitivities and Rely on Progesterone Receptor. Nat. Commun. 2022, 13, 3127. [Google Scholar] [CrossRef]
- Okoh, V.; Deoraj, A.; Roy, D. Estrogen-Induced Reactive Oxygen Species-Mediated Signalings Contribute to Breast Cancer. Biochim. Biophys. Acta 2011, 1815, 115–133. [Google Scholar] [CrossRef]
- Dall, G.V.; Britt, K.L. Estrogen Effects on the Mammary Gland in Early and Late Life and Breast Cancer Risk. Front. Oncol. 2017, 7, 110. [Google Scholar] [CrossRef]
- Mair, K.M.; Gaw, R.; MacLean, M.R. Obesity, Estrogens and Adipose Tissue Dysfunction—Implications for Pulmonary Arterial Hypertension. Pulm. Circ. 2020, 10, 2045894020952019. [Google Scholar] [CrossRef]
- Kakugawa, Y.; Tada, H.; Kawai, M.; Suzuki, T.; Nishino, Y.; Kanemura, S.; Ishida, T.; Ohuchi, N.; Minami, Y. Associations of Obesity and Physical Activity with Serum and Intratumoral Sex Steroid Hormone Levels among Postmenopausal Women with Breast Cancer: Analysis of Paired Serum and Tumor Tissue Samples. Breast Cancer Res. Treat. 2017, 162, 115–125. [Google Scholar] [CrossRef]
- Schairer, C.; Fuhrman, B.J.; Boyd-Morin, J.; Genkinger, J.M.; Gail, M.H.; Hoover, R.N.; Ziegler, R.G. Quantifying the Role of Circulating Unconjugated Estradiol in Mediating the Body Mass Index-Breast Cancer Association. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2016, 25, 105–113. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Au, C.C.; Benito-Martin, A.; Ladumor, H.; Oshchepkova, S.; Moges, R.; Brown, K.A. Estrogens and Breast Cancer: Mechanisms Involved in Obesity-Related Development, Growth and Progression. J. Steroid Biochem. Mol. Biol. 2019, 189, 161–170. [Google Scholar] [CrossRef]
- Chan, H.J.; Petrossian, K.; Chen, S. Structural and Functional Characterization of Aromatase, Estrogen Receptor, and Their Genes in Endocrine-Responsive and -Resistant Breast Cancer Cells. J. Steroid Biochem. Mol. Biol. 2016, 161, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Zahid, H.; Subbaramaiah, K.; Iyengar, N.M.; Zhou, X.K.; Chen, I.-C.; Bhardwaj, P.; Gucalp, A.; Morrow, M.; Hudis, C.A.; Dannenberg, A.J.; et al. Leptin Regulation of the P53-HIF1α/PKM2-Aromatase Axis in Breast Adipose Stromal Cells: A Novel Mechanism for the Obesity-Breast Cancer Link. Int. J. Obes. 2018, 42, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Xu, Y.; Li, J.; Zheng, S.; Sun, P.; Wang, T. Prognostic Role of GPER/Ezrin in Triple-Negative Breast Cancer Is Associated with Menopausal Status. Endocr. Connect. 2019, 8, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Bouris, P.; Skandalis, S.S.; Piperigkou, Z.; Afratis, N.; Karamanou, K.; Aletras, A.J.; Moustakas, A.; Theocharis, A.D.; Karamanos, N.K. Estrogen Receptor Alpha Mediates Epithelial to Mesenchymal Transition, Expression of Specific Matrix Effectors and Functional Properties of Breast Cancer Cells. Matrix Biol. J. Int. Soc. Matrix Biol. 2015, 43, 42–60. [Google Scholar] [CrossRef]
- Warner, M.; Wu, W.; Montanholi, L.; Nalvarte, I.; Antonson, P.; Gustafsson, J.-A. Ventral Prostate and Mammary Gland Phenotype in Mice with Complete Deletion of the ERβ Gene. Proc. Natl. Acad. Sci. USA 2020, 117, 4902–4909. [Google Scholar] [CrossRef]
- Alexander, S.P.H.; Mathie, A.; Peters, J.A. Guide to Receptors and Channels (GRAC), 5th Edition. Br. J. Pharmacol. 2011, 164 (Suppl. 1), S1–S324. [Google Scholar] [CrossRef]
- Cadenas, C.; Bolt, H.M. Estrogen Receptors in Human Disease. Arch. Toxicol. 2012, 86, 1489–1490. [Google Scholar] [CrossRef][Green Version]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef]
- George, A.L.; Rajoria, S.; Suriano, R.; Mittleman, A.; Tiwari, R.K. Hypoxia and Estrogen Are Functionally Equivalent in Breast Cancer-Endothelial Cell Interdependence. Mol. Cancer 2012, 11, 80. [Google Scholar] [CrossRef]
- Wang, C.; Mayer, J.A.; Mazumdar, A.; Fertuck, K.; Kim, H.; Brown, M.; Brown, P.H. Estrogen Induces C-Myc Gene Expression via an Upstream Enhancer Activated by the Estrogen Receptor and the AP-1 Transcription Factor. Mol. Endocrinol. 2011, 25, 1527–1538. [Google Scholar] [CrossRef]
- Xu, D.; Takeshita, F.; Hino, Y.; Fukunaga, S.; Kudo, Y.; Tamaki, A.; Matsunaga, J.; Takahashi, R.-U.; Takata, T.; Shimamoto, A.; et al. MiR-22 Represses Cancer Progression by Inducing Cellular Senescence. J. Cell Biol. 2011, 193, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, C.; Liao, H.; Wang, Q. Activation of GPER by E2 Promotes Proliferation, Invasion and Migration of Breast Cancer Cells by Regulating the MiR-124/CD151 Pathway. Oncol. Lett. 2021, 21, 432. [Google Scholar] [CrossRef] [PubMed]
- Segura-Bautista, D.; Olivares, A.; Casas-González, P.; Bonilla, E.; Salazar, Z.; Pérez-Solis, M.A. GPR30 Expression and Function in Breast Cancer Cells Are Induced through a Cis-acting Element Targeted by ETS Factors. Oncol. Rep. 2020, 43, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Sandén, C.; Broselid, S.; Cornmark, L.; Andersson, K.; Daszkiewicz-Nilsson, J.; Mårtensson, U.E.A.; Olde, B.; Leeb-Lundberg, L.M.F. G Protein-Coupled Estrogen Receptor 1/G Protein-Coupled Receptor 30 Localizes in the Plasma Membrane and Traffics Intracellularly on Cytokeratin Intermediate Filaments. Mol. Pharmacol. 2011, 79, 400–410. [Google Scholar] [CrossRef]
- Lei, B.; Sun, S.; Zhang, X.; Feng, C.; Xu, J.; Wen, Y.; Huang, Y.; Wu, M.; Yu, Y. Bisphenol AF Exerts Estrogenic Activity in MCF-7 cells through Activation of Erk and PI3K/Akt Signals via GPER Signaling Pathway. Chemosphere 2019, 220, 362–370. [Google Scholar] [CrossRef]
- Hsu, L.-H.; Chu, N.-M.; Lin, Y.-F.; Kao, S.-H. G-Protein Coupled Estrogen Receptor in Breast Cancer. Int. J. Mol. Sci. 2019, 20, E306. [Google Scholar] [CrossRef]
- Yu, T.; Liu, M.; Luo, H.; Wu, C.; Tang, X.; Tang, S.; Hu, P.; Yan, Y.; Wang, Z.; Tu, G. GPER Mediates Enhanced Cell Viability and Motility via Non-Genomic Signaling Induced by 17β-Estradiol in Triple-Negative Breast Cancer Cells. J. Steroid Biochem. Mol. Biol. 2014, 143, 392–403. [Google Scholar] [CrossRef]
- Filardo, E.; Quinn, J.; Pang, Y.; Graeber, C.; Shaw, S.; Dong, J.; Thomas, P. Activation of the Novel Estrogen Receptor G Protein-Coupled Receptor 30 (GPR30) at the Plasma Membrane. Endocrinology 2007, 148, 3236–3245. [Google Scholar] [CrossRef]
- Lappano, R.; Rigiracciolo, D.C.; Belfiore, A.; Maggiolini, M.; De Francesco, E.M. Cancer Associated Fibroblasts: Role in Breast Cancer and Potential as Therapeutic Targets. Expert Opin. Ther. Targets 2020, 24, 559–572. [Google Scholar] [CrossRef]
- Madeo, A.; Maggiolini, M. Nuclear Alternate Estrogen Receptor GPR30 Mediates 17beta-Estradiol-Induced Gene Expression and Migration in Breast Cancer-Associated Fibroblasts. Cancer Res. 2010, 70, 6036–6046. [Google Scholar] [CrossRef]
- Molina, L.; Figueroa, C.D.; Bhoola, K.D.; Ehrenfeld, P. GPER-1/GPR30 a Novel Estrogen Receptor Sited in the Cell Membrane: Therapeutic Coupling to Breast Cancer. Expert Opin. Ther. Targets 2017, 21, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Rigiracciolo, D.; De Marco, P.; Avino, S.; Cappello, A.R.; Rosano, C.; Maggiolini, M.; De Francesco, E.M. Recent Advances on the Role of G Protein-Coupled Receptors in Hypoxia-Mediated Signaling. AAPS J. 2016, 18, 305–310. [Google Scholar] [CrossRef] [PubMed]
- De Marco, P.; Lappano, R.; Francesco, E.M.D.; Cirillo, F.; Pupo, M.; Avino, S.; Vivacqua, A.; Abonante, S.; Picard, D.; Maggiolini, M. GPER Signalling in Both Cancer-Associated Fibroblasts and Breast Cancer Cells Mediates a Feedforward IL1β/IL1R1 Response. Sci. Rep. 2016, 6, 24354. [Google Scholar] [CrossRef] [PubMed]
- Talia, M.; De Francesco, E.M.; Rigiracciolo, D.C.; Muoio, M.G.; Muglia, L.; Belfiore, A.; Maggiolini, M.; Sims, A.H.; Lappano, R. The G Protein-Coupled Estrogen Receptor (GPER) Expression Correlates with Pro-Metastatic Pathways in ER-Negative Breast Cancer: A Bioinformatics Analysis. Cells 2020, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J.; Thomas, P. Minireview: G Protein-Coupled Estrogen Receptor-1, GPER-1: Its Mechanism of Action and Role in Female Reproductive Cancer, Renal and Vascular Physiology. Endocrinology 2012, 153, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Vivacqua, A.; De Marco, P.; Santolla, M.F.; Cirillo, F.; Pellegrino, M.; Panno, M.L.; Abonante, S.; Maggiolini, M. Estrogenic Gper Signaling Regulates Mir144 Expression in Cancer Cells and Cancer-Associated Fibroblasts (Cafs). Oncotarget 2015, 6, 16573–16587. [Google Scholar] [CrossRef]
- Treeck, O.; Schüler-Toprak, S.; Ortmann, O. Estrogen Actions in Triple-Negative Breast Cancer. Cells 2020, 9, E2358. [Google Scholar] [CrossRef]
- Huang, B.; Omoto, Y.; Iwase, H.; Yamashita, H.; Toyama, T.; Coombes, R.C.; Filipovic, A.; Warner, M.; Gustafsson, J.-Å. Differential Expression of Estrogen Receptor α, Β1, and Β2 in Lobular and Ductal Breast Cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 1933–1938. [Google Scholar] [CrossRef]
- Hawse, J.R.; Carter, J.M.; Aspros, K.G.M.; Bruinsma, E.S.; Koepplin, J.W.; Negron, V.; Subramaniam, M.; Ingle, J.N.; Rech, K.L.; Goetz, M.P. Optimized Immunohistochemical Detection of Estrogen Receptor Beta Using Two Validated Monoclonal Antibodies Confirms Its Expression in Normal and Malignant Breast Tissues. Breast Cancer Res. Treat. 2020, 179, 241–249. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, Z. MiR-191/DAB2 Axis Regulates the Tumorigenicity of Estrogen Receptor-Positive Breast Cancer: MIR-191/AXIS REGULATES ER+ BREAST CANCER. IUBMB Life 2018, 70, 71–80. [Google Scholar] [CrossRef]
- Liu, B.-W.; Yu, Z.-H.; Chen, A.-X.; Chi, J.-R.; Ge, J.; Yu, Y.; Cao, X.-C. Estrogen Receptor-α-MiR-1271-SNAI2 Feedback Loop Regulates Transforming Growth Factor-β-Induced Breast Cancer Progression. J. Exp. Clin. Cancer Res. 2019, 38, 109. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Gjerstorff, M.F.; Shirjang, S.; Asadzadeh, Z.; Khaze, V.; Holmskov, U.; Kazemi, T.; Duijf, P.H.G.; Baradaran, B. MiR-142-3p Is a Tumor Suppressor That Inhibits Estrogen Receptor Expression in ER-Positive Breast Cancer. J. Cell. Physiol. 2019, 234, 16043–16053. [Google Scholar] [CrossRef]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke Off. J. Int. Stroke Soc. 2018, 13, 612–632. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chao, L.; Wang, J.; Sun, Y. MiRNA-148a Regulates the Expression of the Estrogen Receptor through DNMT1-Mediated DNA Methylation in Breast Cancer Cells. Oncol. Lett. 2017, 14, 4736–4740. [Google Scholar] [CrossRef]
- Baravalle, R.; Di Nardo, G.; Bandino, A.; Barone, I.; Catalano, S.; Andò, S.; Gilardi, G. Impact of R264C and R264H Polymorphisms in Human Aromatase Function. J. Steroid Biochem. Mol. Biol. 2017, 167, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Liu, H.; Hu, H.; Li, L.-W.; Zong, Q.-B.; Wu, T.-W.; Li, X.-Y.; Fang, S.-Q.; Liu, Y.-W.; Zhan, Y.; et al. LINC00094/MiR-19a-3p/CYP19A1 Axis Affects the Sensitivity of ER Positive Breast Cancer Cells to Letrozole through EMT Pathway. Aging 2022, 14, 4755–4768. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-F.; Shi, Z.-M.; Li, D.-M.; Qian, Y.-C.; Ren, Y.; Bai, X.-M.; Xie, Y.-X.; Wang, L.; Ge, X.; Liu, W.-T.; et al. Estrogen-Induced MiR-196a Elevation Promotes Tumor Growth and Metastasis via Targeting SPRED1 in Breast Cancer. Mol. Cancer 2018, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-Regulated MicroRNAs Control Estradiol Response in Breast Cancer Cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef] [PubMed]
- Jacenik, D.; Cygankiewicz, A.I.; Krajewska, W.M. The G Protein-Coupled Estrogen Receptor as a Modulator of Neoplastic Transformation. Mol. Cell. Endocrinol. 2016, 429, 10–18. [Google Scholar] [CrossRef]
- Yu, T.; Cheng, H.; Ding, Z.; Wang, Z.; Zhou, L.; Zhao, P.; Tan, S.; Xu, X.; Huang, X.; Liu, M.; et al. GPER Mediates Decreased Chemosensitivity via Regulation of ABCG2 Expression and Localization in Tamoxifen-Resistant Breast Cancer Cells. Mol. Cell. Endocrinol. 2020, 506, 110762. [Google Scholar] [CrossRef]
- Ignatov, A.; Ignatov, T.; Weißenborn, C.; Eggemann, H.; Bischoff, J.; Semczuk, A.; Roessner, A.; Costa, S.D.; Kalinski, T. G-Protein-Coupled Estrogen Receptor GPR30 and Tamoxifen Resistance in Breast Cancer. Breast Cancer Res. Treat. 2011, 128, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Liu, M.; Yang, F.; Luo, H.; Li, Z.; Tu, G.; Yang, G. GPR30 as an Initiator of Tamoxifen Resistance in Hormone-Dependent Breast Cancer. Breast Cancer Res. 2013, 15, R114. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Bustamante, F.; Ortloff, A.; Ramos, I.; Ehrenfeld, P.; Figueroa, C.D. Continuous Exposure of Breast Cancer Cells to Tamoxifen Upregulates GPER-1 and Increases Cell Proliferation. Front. Endocrinol. 2020, 11, 563165. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, T.; Claus, M.; Nass, N.; Haybaeck, J.; Seifert, B.; Kalinski, T.; Ortmann, O.; Ignatov, A. G-Protein-Coupled Estrogen Receptor GPER-1 Expression in Hormone Receptor-Positive Breast Cancer Is Associated with Poor Benefit of Tamoxifen. Breast Cancer Res. Treat. 2019, 174, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Howard, E.W.; Yang, X. MicroRNA Regulation in Estrogen Receptor-Positive Breast Cancer and Endocrine Therapy. Biol. Proced. Online 2018, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Chen, B.; Song, X.; Li, Y.; Liang, Y.; Han, D.; Zhang, N.; Zhang, H.; Liu, Y.; Chen, T.; et al. CircRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the MiR-182-5p/FOXO3a Axis in Breast Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2019, 27, 1638–1652. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-S.; Li, Y.; Zhang, H.; Zhang, D.; Zhang, X.-B.; Wang, X.; Yu, Y. The ERα-MiR-575-P27 Feedback Loop Regulates Tamoxifen Sensitivity in ER-Positive Breast Cancer. Theranostics 2020, 10, 10729–10742. [Google Scholar] [CrossRef]
- Zhou, Q.; Zeng, H.; Ye, P.; Shi, Y.; Guo, J.; Long, X. Differential MicroRNA Profiles between Fulvestrant-Resistant and Tamoxifen-Resistant Human Breast Cancer Cells. Anticancer Drugs 2018, 29, 539–548. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, Y.S.; Kang, K.N.; Kim, K.H.; Park, Y.J.; Kim, C.W. Multiple MicroRNAs as Biomarkers for Early Breast Cancer Diagnosis. Mol. Clin. Oncol. 2021, 14, 31. [Google Scholar] [CrossRef]
- Min, W.; Wang, B.; Li, J.; Han, J.; Zhao, Y.; Su, W.; Dai, Z.; Wang, X.; Ma, Q. The Expression and Significance of Five Types of MiRNAs in Breast Cancer. Med. Sci. Monit. Basic Res. 2014, 20, 97–104. [Google Scholar] [CrossRef]
- Mansoori, B.; Duijf, P.H.G.; Mohammadi, A.; Safarzadeh, E.; Ditzel, H.J.; Gjerstorff, M.F.; Cho, W.C.-S.; Baradaran, B. MiR-142-3p Targets HMGA2 and Suppresses Breast Cancer Malignancy. Life Sci. 2021, 276, 119431. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S.; Zhou, J.; Qian, Q. MiR-424-5p Regulates Cell Proliferation, Migration and Invasion by Targeting Doublecortin-like Kinase 1 in Basal-like Breast Cancer. Biomed. Pharmacother. 2018, 102, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Radojicic, J.; Zaravinos, A.; Vrekoussis, T.; Kafousi, M.; Spandidos, D.A.; Stathopoulos, E.N. MicroRNA Expression Analysis in Triple-Negative (ER, PR and Her2/Neu) Breast Cancer. Cell Cycle Georget. Tex 2011, 10, 507–517. [Google Scholar] [CrossRef]
- Kahraman, M.; Röske, A.; Laufer, T.; Fehlmann, T.; Backes, C.; Kern, F.; Kohlhaas, J.; Schrörs, H.; Saiz, A.; Zabler, C.; et al. MicroRNA in Diagnosis and Therapy Monitoring of Early-Stage Triple-Negative Breast Cancer. Sci. Rep. 2018, 8, 11584. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, L.; Sun, S.; Wu, J.; Wang, Q. Quantitative Measurement of Serum MicroRNA-21 Expression in Relation to Breast Cancer Metastasis in Chinese Females. Ann. Lab. Med. 2015, 35, 226–232. [Google Scholar] [CrossRef]
- Krukovets, I.; Legerski, M.; Sul, P.; Stenina-Adognravi, O. Inhibition of Hyperglycemia-Induced Angiogenesis and Breast Cancer Tumor Growth by Systemic Injection of MicroRNA-467 Antagonist. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 3726–3736. [Google Scholar] [CrossRef]
- Gajeton, J.; Krukovets, I.; Yendamuri, R.; Verbovetskiy, D.; Vasanji, A.; Sul, L.; Stenina-Adognravi, O. MiR-467 Regulates Inflammation and Blood Insulin and Glucose. J. Cell. Mol. Med. 2021, 25, 2549–2562. [Google Scholar] [CrossRef]
- Hodge, J.; Wang, F.; Wang, J.; Liu, Q.; Saaoud, F.; Wang, Y.; Singh, U.P.; Chen, H.; Luo, M.; Ai, W.; et al. Overexpression of MicroRNA-155 Enhances the Efficacy of Dendritic Cell Vaccine against Breast Cancer. Oncoimmunology 2020, 9, 1724761. [Google Scholar] [CrossRef]
- Pasculli, B.; Barbano, R.; Fontana, A.; Biagini, T.; Di Viesti, M.P.; Rendina, M.; Valori, V.M.; Morritti, M.; Bravaccini, S.; Ravaioli, S.; et al. Hsa-MiR-155-5p Up-Regulation in Breast Cancer and Its Relevance for Treatment With Poly[ADP-Ribose] Polymerase 1 (PARP-1) Inhibitors. Front. Oncol. 2020, 10, 1415. [Google Scholar] [CrossRef]
- Kim, S.; Lee, E.S.; Lee, E.J.; Jung, J.Y.; Lee, S.B.; Lee, H.J.; Kim, J.; Kim, H.J.; Lee, J.W.; Son, B.H.; et al. Targeted Eicosanoids Profiling Reveals a Prostaglandin Reprogramming in Breast Cancer by MicroRNA-155. J. Exp. Clin. Cancer Res. 2021, 40, 43. [Google Scholar] [CrossRef]
- Bahiraee, A.; Ebrahimi, R.; Halabian, R.; Aghabozorgi, A.S.; Amani, J. The Role of Inflammation and Its Related MicroRNAs in Breast Cancer: A Narrative Review. J. Cell. Physiol. 2019, 234, 19480–19493. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.M.; Salvatore, M.; Incoronato, M. MiRNA-Based Therapeutics in Breast Cancer: A Systematic Review. Front. Oncol. 2021, 11, 668464. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, N.; Safaralizadeh, R.; Khojasteh, S.M.B.; Shadbad, M.A.; Hosseinpourfeizi, M.A.; Azarbarzin, S.; Rajabi, A.; Baradaran, B. The Combined Restoration of MiR-424-5p and MiR-142-3p Effectively Inhibits MCF-7 Breast Cancer Cell Line via Modulating Apoptosis, Proliferation, Colony Formation, Cell Cycle and Autophagy. Mol. Biol. Rep. 2022, 49, 8325–8335. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; Wang, Y.C.; et al. Relevance of Breast Cancer Hormone Receptors and Other Factors to the Efficacy of Adjuvant Tamoxifen: Patient-Level Meta-Analysis of Randomised Trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Munzone, E.; Colleoni, M. Optimal Management of Luminal Breast Cancer: How Much Endocrine Therapy Is Long Enough? Ther. Adv. Med. Oncol. 2018, 10, 175883591877743. [Google Scholar] [CrossRef]
- Amiruddin, A.; Massi, M.N.; Islam, A.A.; Patellongi, I.; Pratama, M.Y.; Sutandyo, N.; Natzir, R.; Hatta, M.; Md Latar, N.H.; Wahid, S. MicroRNA-221 and Tamoxifen Resistance in Luminal-Subtype Breast Cancer Patients: A Case-Control Study. Ann. Med. Surg. 2022, 73, 103092. [Google Scholar] [CrossRef]
- Ouyang, Y.X.; Feng, J.; Wang, Z.; Zhang, G.J.; Chen, M. MiR-221/222 Sponge Abrogates Tamoxifen Resistance in ER-Positive Breast Cancer Cells through Restoring the Expression of ERα. Mol. Biomed. 2021, 2, 20. [Google Scholar] [CrossRef]
- Cheng, C.-W.; Yu, J.-C.; Hsieh, Y.-H.; Liao, W.-L.; Shieh, J.-C.; Yao, C.-C.; Lee, H.-J.; Chen, P.-M.; Wu, P.-E.; Shen, C.-Y. Increased Cellular Levels of MicroRNA-9 and MicroRNA-221 Correlate with Cancer Stemness and Predict Poor Outcome in Human Breast Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 48, 2205–2218. [Google Scholar] [CrossRef]
- Li, X.; Tang, X.; Li, K.; Lu, L. Evaluation of Serum MicroRNAs (MiR-9-5p, MiR-17-5p, and MiR-148a-3p) as Potential Biomarkers of Breast Cancer. BioMed Res. Int. 2022, 2022, 9961412. [Google Scholar] [CrossRef]
- Xing, A.-Y.; Wang, B.; Li, Y.-H.; Chen, X.; Wang, Y.-W.; Liu, H.-T.; Gao, P. Identification of MiRNA Signature in Breast Cancer to Predict Neoadjuvant Chemotherapy Response. Pathol. Oncol. Res. 2021, 27, 1609753. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanusek, K.; Karczmarski, J.; Litwiniuk, A.; Urbańska, K.; Ambrozkiewicz, F.; Kwiatkowski, A.; Martyńska, L.; Domańska, A.; Bik, W.; Paziewska, A. Obesity as a Risk Factor for Breast Cancer—The Role of miRNA. Int. J. Mol. Sci. 2022, 23, 15683. https://doi.org/10.3390/ijms232415683
Hanusek K, Karczmarski J, Litwiniuk A, Urbańska K, Ambrozkiewicz F, Kwiatkowski A, Martyńska L, Domańska A, Bik W, Paziewska A. Obesity as a Risk Factor for Breast Cancer—The Role of miRNA. International Journal of Molecular Sciences. 2022; 23(24):15683. https://doi.org/10.3390/ijms232415683
Chicago/Turabian StyleHanusek, Karolina, Jakub Karczmarski, Anna Litwiniuk, Katarzyna Urbańska, Filip Ambrozkiewicz, Andrzej Kwiatkowski, Lidia Martyńska, Anita Domańska, Wojciech Bik, and Agnieszka Paziewska. 2022. "Obesity as a Risk Factor for Breast Cancer—The Role of miRNA" International Journal of Molecular Sciences 23, no. 24: 15683. https://doi.org/10.3390/ijms232415683
APA StyleHanusek, K., Karczmarski, J., Litwiniuk, A., Urbańska, K., Ambrozkiewicz, F., Kwiatkowski, A., Martyńska, L., Domańska, A., Bik, W., & Paziewska, A. (2022). Obesity as a Risk Factor for Breast Cancer—The Role of miRNA. International Journal of Molecular Sciences, 23(24), 15683. https://doi.org/10.3390/ijms232415683





