Transcriptional Regulation of zma-MIR528a by Action of Nitrate and Auxin in Maize
Abstract
1. Introduction
2. Results
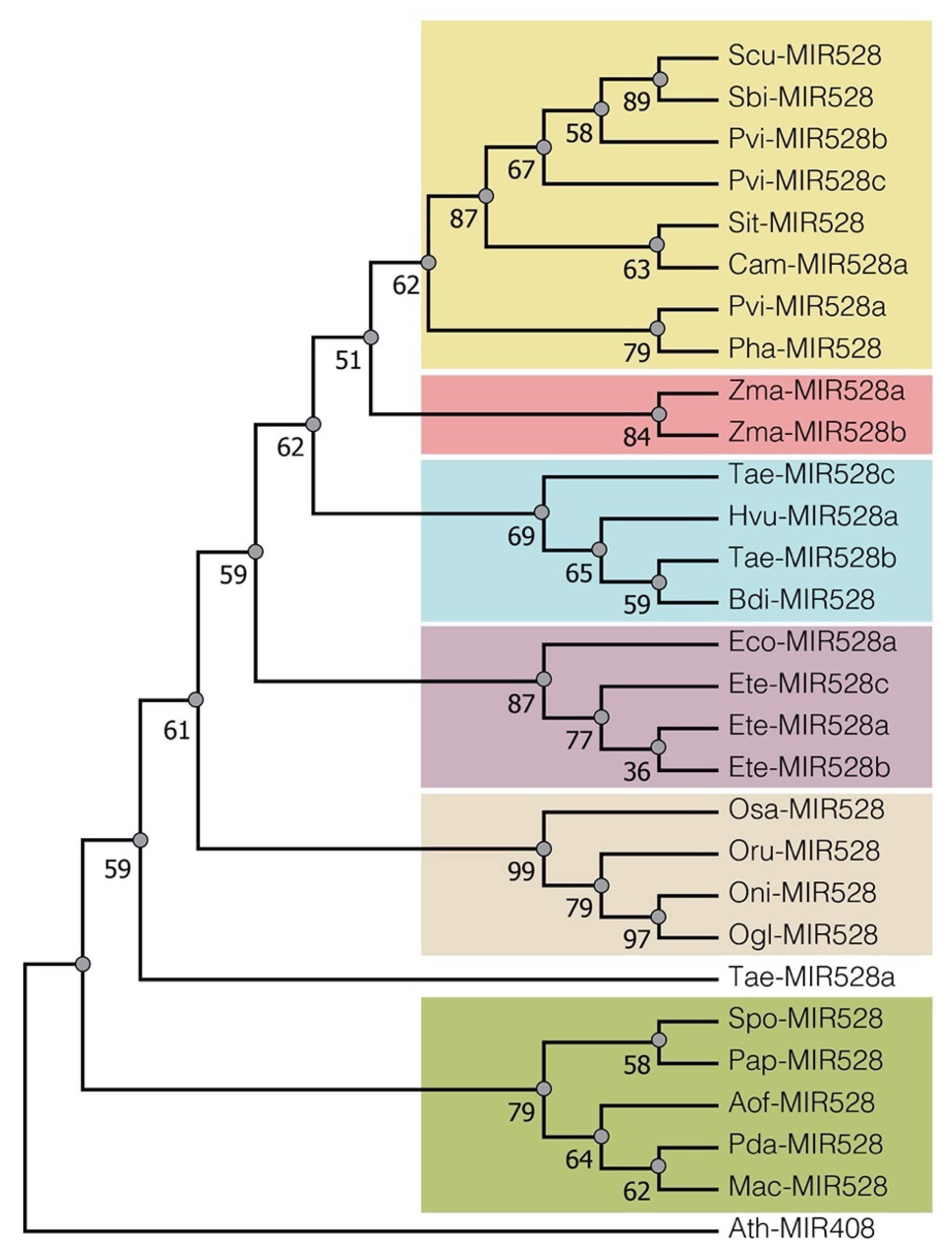
2.1. Conservation of pre-miR528 in Monocots
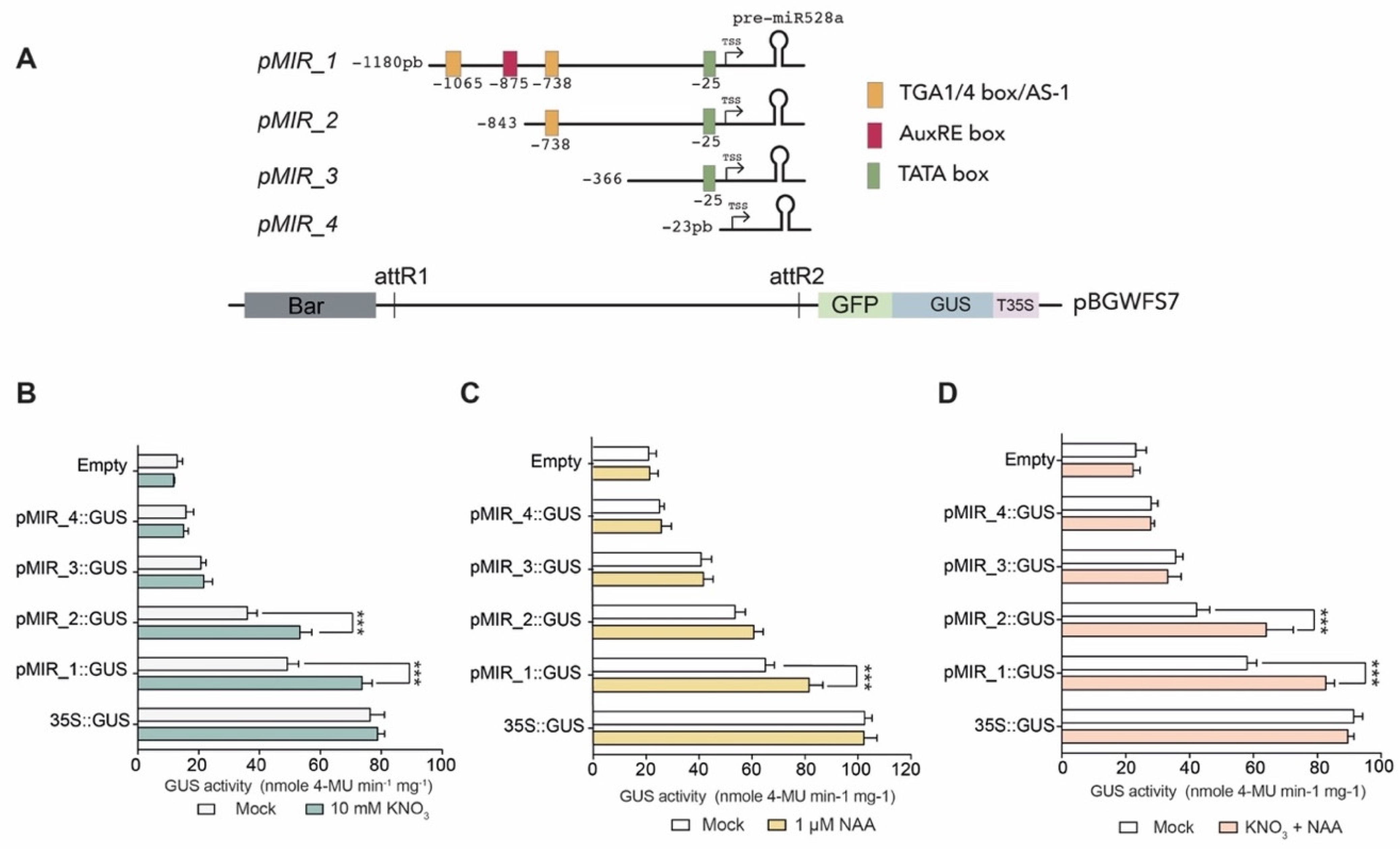
2.2. Identification of Promoter cis-Acting Elements in zma-MIR528a
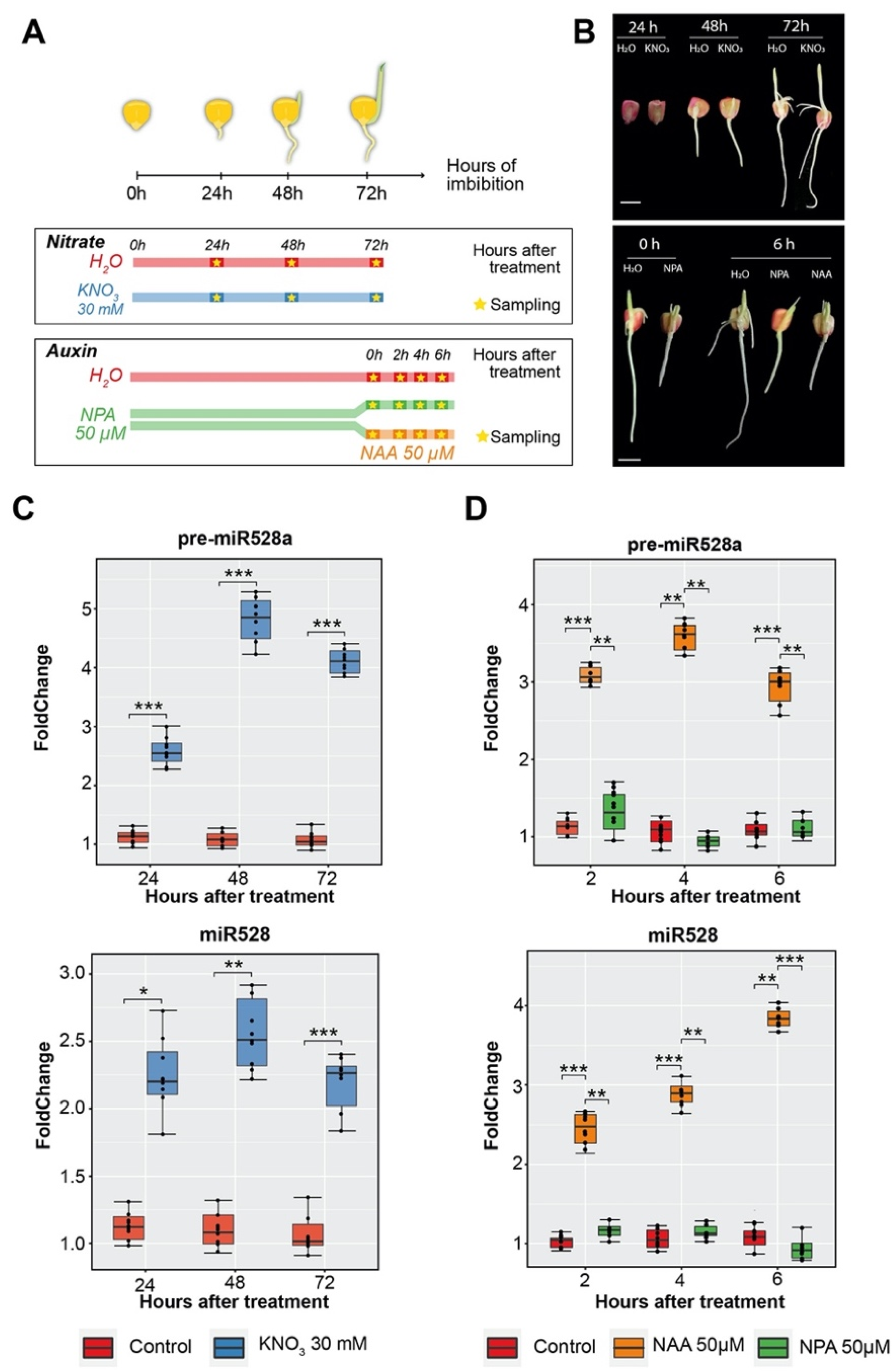
2.3. Accumulation of pre-miR528a Decreases as Seedling Establishment Takes Place
2.4. Exogenous Nitrate and Auxin Treatments during Maize Seed Imbibition Trigger Increases in pre-miR528a and Mature miR528 Levels
2.5. zma-MIR528a Promoter Harbors Regulatory Elements Responsible for Transcription Enhancement in Response to Nitrate and Auxins
2.6. TFBS within the zma-MIR528a Promoter Contribute Differentially to Nitrate and Auxin Induction
2.7. ARF34 Contributes to Transcriptional Activation of the zma-MIR528a Promoter
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Isolation, Size Fractionation, and Purification
4.3. Precursor, Mature microRNA, and Target Quantification by RT-qPCR
4.4. Experimental Validation of the Transcription Start Site (TSS) by 5′ RLM-RACE
4.5. Phylogenetic Analysis of miR528 Precursors and Promoters in Monocots
4.6. Promoter cis-Element Analysis
4.7. Generation of Plasmid Constructions
4.8. Maize Leaf Protoplast Isolation and Transfection
4.9. Promoter Activity Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lucero, L.; Fonouni-Farde, C.; Crespi, M.; Ariel, F. Long noncoding RNAs shape transcription in plants. Transcription 2020, 11, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Waititu, J.K.; Zhang, C.; Liu, J.; Wang, H. Plant Non-Coding RNAs: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 8401. [Google Scholar] [CrossRef]
- Li, M.; Yu, B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021, 18, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Nie, J.; Wang, H. microRNA biogenesis in plant. Plant Growth Regul. 2021, 93, 1–12. [Google Scholar] [CrossRef]
- Pandey, P.; Srivastava, P.K.; Pandey, S.P. Prediction of Plant miRNA Targets. Methods Mol. Biol. 2019, 1932, 99–107. [Google Scholar] [CrossRef]
- Pegler, J.L.; Grof, C.P.L.; Eamens, A.L. The Plant microRNA Pathway: The Production and Action Stages. Methods Mol. Biol. 2019, 1932, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Barman, A.; Phukan, T.; Ray, S.K. Harnessing Perks of MiRNA Principles for Betterment of Agriculture and Food Security. In Omics Technologies for Sustainable Agriculture and Global Food Security; Kumar, A., Kumar, R., Shukla, P., Patel, H.K., Eds.; Springer: Singapore, 2021; Volume 2, pp. 123–191. [Google Scholar]
- Yan, J.; Zhang, H.; Zheng, Y.; Ding, Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta 2015, 241, 109–123. [Google Scholar] [CrossRef]
- Njaci, I.; Williams, B.; Castillo-Gonzalez, C.; Dickman, M.B.; Zhang, X.; Mundree, S. Genome-Wide Investigation of the Role of MicroRNAs in Desiccation Tolerance in the Resurrection Grass Tripogon loliiformis. Plants 2018, 7, 68. [Google Scholar] [CrossRef]
- Kravchik, M.; Stav, R.; Belausov, E.; Arazi, T. Functional Characterization of microRNA171 Family in Tomato. Plants 2019, 8, 10. [Google Scholar] [CrossRef]
- Pegler, J.L.; Grof, C.P.L.; Eamens, A.L. Profiling of the Differential Abundance of Drought and Salt Stress-Responsive MicroRNAs Across Grass Crop and Genetic Model Plant Species. Agronomy 2018, 8, 118. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Profiling the Abiotic Stress Responsive microRNA Landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Allen, E.; Fahlgren, N.; Calamar, A.; Givan, S.A.; Carrington, J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005, 138, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, M.G.; Scarpa, A.; Magele, I.; Ruiz-Duarte, P.; von Born, P.; Bald, L.; Jouannet, V.; Maizel, A. ARF5/MONOPTEROS directly regulates miR390 expression in the Arabidopsis thaliana primary root meristem. Plant Direct 2019, 3, e00116. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional Regulation of miR528 by OsSPL9 Orchestrates Antiviral Response in Rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, P.; Mei, H.; Wang, D.; Sun, J.; Yang, C.; Hao, L.; Cao, S.; Chu, C.; Hu, S.; et al. Fine-Tuning of MiR528 Accumulation Modulates Flowering Time in Rice. Mol. Plant 2019, 12, 1103–1113. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive Expression of Rice MicroRNA528 Alters Plant Development and Enhances Tolerance to Salinity Stress and Nitrogen Starvation in Creeping Bentgrass. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W.X. MicroRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.; Zeng, J.; Yun, Z.; Liu, Y.; Qu, H.; Jiang, Y.; Duan, X.; Xia, R. MicroRNA528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol. 2020, 225, 385–399. [Google Scholar] [CrossRef]
- Lujan-Soto, E.; Juarez-Gonzalez, V.T.; Reyes, J.L.; Dinkova, T.D. MicroRNA Zma-miR528 Versatile Regulation on Target mRNAs during Maize Somatic Embryogenesis. Int. J. Mol. Sci. 2021, 22, 5310. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Liu, X.; Cui, D.; Chen, T.; Zhang, H.; Jiang, C.; Xu, C.; Li, P.; Li, S.; et al. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS ONE 2013, 8, e55107. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, Z.; Gao, L.; Wang, L.; Gao, M.; Jiao, Z.; Qiao, H.; Yang, J.; Chen, M.; Yao, L.; et al. Genome-Wide Identification and Characterization of microRNAs in Developing Grains of Zea mays L. PLoS ONE 2016, 11, e0153168. [Google Scholar] [CrossRef]
- Shen, Y.; Jiang, Z.; Lu, S.; Lin, H.; Gao, S.; Peng, H.; Yuan, G.; Liu, L.; Zhang, Z.; Zhao, M.; et al. Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem. Biophys. Res. Commun. 2013, 441, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Hernandez, E.C.; Alejandri-Ramirez, N.D.; Juarez-Gonzalez, V.T.; Dinkova, T.D. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front. Plant Sci. 2015, 6, 555. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, Y.; Xia, R. Jack of Many Trades: The Multifaceted Role of miR528 in Monocots. Mol. Plant 2019, 12, 1044–1046. [Google Scholar] [CrossRef]
- Mejia-Guerra, M.K.; Li, W.; Galeano, N.F.; Vidal, M.; Gray, J.; Doseff, A.I.; Grotewold, E. Core Promoter Plasticity Between Maize Tissues and Genotypes Contrasts with Predominance of Sharp Transcription Initiation Sites. Plant Cell 2015, 27, 3309–3320. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Printz, B.; Lutts, S.; Hausman, J.F.; Sergeant, K. Copper Trafficking in Plants and Its Implication on Cell Wall Dynamics. Front. Plant Sci. 2016, 7, 601. [Google Scholar] [CrossRef]
- Le Hir, R.; Bellini, C. The plant-specific dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-Lopez, O.; Tamayo, K.P.; Aceituno, F.; Gomez, I.; Ruffel, S.; Lejay, L.; et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Boer, D.R.; et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef] [PubMed]
- Alejandri-Ramirez, N.D.; Chavez-Hernandez, E.C.; Contreras-Guerra, J.L.; Reyes, J.L.; Dinkova, T.D. Small RNA differential expression and regulation in Tuxpeno maize embryogenic callus induction and establishment. Plant Physiol. Biochem. 2018, 122, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Osuna, D.; Prieto, P.; Aguilar, M. Control of Seed Germination and Plant Development by Carbon and Nitrogen Availability. Front. Plant Sci. 2015, 6, 1023. [Google Scholar] [CrossRef] [PubMed]
- Crombez, H.; Roberts, I.; Vangheluwe, N.; Motte, H.; Jansen, L.; Beeckman, T.; Parizot, B. Lateral Root Inducible System in Arabidopsis and Maize. J. Vis. Exp. 2016, 107, e53481. [Google Scholar] [CrossRef]
- Galli, M.; Khakhar, A.; Lu, Z.; Chen, Z.; Sen, S.; Joshi, T.; Nemhauser, J.L.; Schmitz, R.J.; Gallavotti, A. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 2018, 9, 4526. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L. Comparative analysis of microRNA promoters in Arabidopsis and rice. Genom. Proteom. Bioinform. 2013, 11, 56–60. [Google Scholar] [CrossRef]
- Trevisan, S.; Nonis, A.; Begheldo, M.; Manoli, A.; Palme, K.; Caporale, G.; Ruperti, B.; Quaggiotti, S. Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 2012, 35, 1137–1155. [Google Scholar] [CrossRef]
- Meng, Y.; Huang, F.; Shi, Q.; Cao, J.; Chen, D.; Zhang, J.; Ni, J.; Wu, P.; Chen, M. Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta 2009, 230, 883–898. [Google Scholar] [CrossRef]
- Megraw, M.; Baev, V.; Rusinov, V.; Jensen, S.T.; Kalantidis, K.; Hatzigeorgiou, A.G. MicroRNA promoter element discovery in Arabidopsis. RNA 2006, 12, 1612–1619. [Google Scholar] [CrossRef]
- Jores, T.; Tonnies, J.; Wrightsman, T.; Buckler, E.S.; Cuperus, J.T.; Fields, S.; Queitsch, C. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat. Plants 2021, 7, 842–855. [Google Scholar] [CrossRef]
- Megraw, M.; Cumbie, J.S.; Ivanchenko, M.G.; Filichkin, S.A. Small Genetic Circuits and MicroRNAs: Big Players in Polymerase II Transcriptional Control in Plants. Plant Cell 2016, 28, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R.; et al. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Sen, S.; Schott, D.; Portwood, J.L.; Freeling, M.; Walley, J.W.; Andorf, C.M.; Schnable, J.C. qTeller: A tool for comparative multi-genomic gene expression analysis. Bioinformatics 2021, 38, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Porto, M.S.; Pinheiro, M.P.; Batista, V.G.; dos Santos, R.C.; Filho Pde, A.; de Lima, L.M. Plant promoters: An approach of structure and function. Mol. Biotechnol. 2014, 56, 38–49. [Google Scholar] [CrossRef]
- Lu, S. De novo origination of MIRNAs through generation of short inverted repeats in target genes. RNA Biol. 2019, 16, 846–859. [Google Scholar] [CrossRef]
- Chorostecki, U.; Moro, B.; Rojas, A.M.L.; Debernardi, J.M.; Schapire, A.L.; Notredame, C.; Palatnik, J.F. Evolutionary Footprints Reveal Insights into Plant MicroRNA Biogenesis. Plant Cell 2017, 29, 1248–1261. [Google Scholar] [CrossRef]
- Narjala, A.; Nair, A.; Tirumalai, V.; Hari Sundar, G.V.; Shivaprasad, P.V. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res. 2020, 48, 3103–3118. [Google Scholar] [CrossRef]
- Li, N.; Muthreich, M.; Huang, L.J.; Thurow, C.; Sun, T.; Zhang, Y.; Gatz, C. TGACG-BINDING FACTORs (TGAs) and TGA-interacting CC-type glutaredoxins modulate hyponastic growth in Arabidopsis thaliana. New Phytol. 2019, 221, 1906–1918. [Google Scholar] [CrossRef]
- Ullah, I.; Magdy, M.; Wang, L.; Liu, M.; Li, X. Genome-wide identification and evolutionary analysis of TGA transcription factors in soybean. Sci. Rep. 2019, 9, 11186. [Google Scholar] [CrossRef]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Walcher, C.L.; Nemhauser, J.L. Bipartite promoter element required for auxin response. Plant Physiol. 2012, 158, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.Q.; Shu, J.Q.; Li, W.M.; Wang, G.Z. Role of Auxin and Nitrate Signaling in the Development of Root System Architecture. Front. Plant Sci. 2021, 12, 690363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Bao, M.L.; Sun, Y.Z.; Yang, Y.J.; Xu, X.H.; Wang, J.H.; Han, N.; Bian, H.W.; Zhu, M.Y. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutierrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef]
- Luo, P.; Di, D.; Wu, L.; Yang, J.; Lu, Y.; Shi, W. MicroRNAs Are Involved in Regulating Plant Development and Stress Response through Fine-Tuning of TIR1/AFB-Dependent Auxin Signaling. Int. J. Mol. Sci. 2022, 23, 510. [Google Scholar] [CrossRef]
- Wojcik, A.M.; Gaj, M.D. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta 2016, 244, 231–243. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Moyano, T.C.; Zhang, T.; Gras, D.E.; Herrera, F.J.; Araus, V.; O’Brien, J.A.; Carrillo, L.; Medina, J.; Vicente-Carbajosa, J.; et al. Local Changes in Chromatin Accessibility and Transcriptional Networks Underlying the Nitrate Response in Arabidopsis Roots. Mol. Plant 2019, 12, 1545–1560. [Google Scholar] [CrossRef]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.L.; Varala, K.; Gutierrez, R.A.; Krouk, G.; et al. Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 2019, 10, 1569. [Google Scholar] [CrossRef]
- Gatz, C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 2013, 26, 151–159. [Google Scholar] [CrossRef]
- Weiste, C.; Droge-Laser, W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 2014, 5, 3883. [Google Scholar] [CrossRef] [PubMed]
- Noshi, M.; Mori, D.; Tanabe, N.; Maruta, T.; Shigeoka, S. Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flg22. Plant Sci. 2016, 252, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Boer, D.R.; Freire-Rios, A.; van den Berg, W.A.; Saaki, T.; Manfield, I.W.; Kepinski, S.; Lopez-Vidrieo, I.; Franco-Zorrilla, J.M.; de Vries, S.C.; Solano, R.; et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef]
- Pierre-Jerome, E.; Moss, B.L.; Lanctot, A.; Hageman, A.; Nemhauser, J.L. Functional analysis of molecular interactions in synthetic auxin response circuits. Proc. Natl. Acad. Sci. USA 2016, 113, 11354–11359. [Google Scholar] [CrossRef]
- Lieberman-Lazarovich, M.; Yahav, C.; Israeli, A.; Efroni, I. Deep Conservation of cis-Element Variants Regulating Plant Hormonal Responses. Plant Cell 2019, 31, 2559–2572. [Google Scholar] [CrossRef]
- Guan, P. Dancing with Hormones: A Current Perspective of Nitrate Signaling and Regulation in Arabidopsis. Front. Plant Sci. 2017, 8, 1697. [Google Scholar] [CrossRef]
- Asim, M.; Ullah, Z.; Oluwaseun, A.; Wang, Q.; Liu, H. Signalling Overlaps between Nitrate and Auxin in Regulation of The Root System Architecture: Insights from the Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2880. [Google Scholar] [CrossRef]
- Matilla, A.J. Auxin: Hormonal Signal Required for Seed Development and Dormancy. Plants 2020, 9, 705. [Google Scholar] [CrossRef]
- Wu, M.; Wu, J.; Gan, Y. The new insight of auxin functions: Transition from seed dormancy to germination and floral opening in plants. Plant Growth Regul. 2020, 91, 169–174. [Google Scholar] [CrossRef]
- Wang, L.; Liu, H.; Li, D.; Chen, H. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genom. 2011, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ruiz, B.A.; Juarez-Gonzalez, V.T.; Chavez-Hernandez, E.C.; Dinkova, T.D. MicroRNA Expression and Regulation During Maize Somatic Embryogenesis. Methods Mol. Biol. 2018, 1815, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Liu, F.; Zheng, K.; Chen, H.C.; Liu, Z.F. Capping-RACE: A simple, accurate, and sensitive 5’ RACE method for use in prokaryotes. Nucleic Acids Res. 2018, 46, e129. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Wang, Y.; Zhao, Y.; Tao, Y.; Cheng, C.; Yang, J.; Lu, X.; Hao, C.; Wang, T.; et al. PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020, 48, D1114–D1121. [Google Scholar] [CrossRef]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods Mol. Biol. 2016, 1374, 115–140. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Ting, Z.T.; Yang, W.; Chunyu, Z.; Qing, M.; Lijun, Z.; Feng, L. Modification of CTAB protocol for maize genomic DNA extraction. Res. J. Biotechnol. 2013, 8, 41–45. [Google Scholar]
- Hussain, H.; Chong, N.F. Combined Overlap Extension PCR Method for Improved Site Directed Mutagenesis. Biomed. Res. Int. 2016, 2016, 8041532. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136. [Google Scholar] [CrossRef]
- Hanifiah, F.H.A.; Abdullah, S.N.A.; Othman, A.; Shaharuddin, N.A.; Saud, H.M.; Hasnulhadi, H.A.H.; Munusamy, U. GCTTCA as a novel motif for regulating mesocarp-specific expression of the oil palm (Elaeis guineensis Jacq.) stearoyl-ACP desaturase gene. Plant Cell Rep. 2018, 37, 1127–1143. [Google Scholar] [CrossRef]
- Cervera, M. Histochemical and fluorometric assays for uidA (GUS) gene detection. Methods Mol. Biol. 2005, 286, 203–214. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luján-Soto, E.; Aguirre de la Cruz, P.I.; Juárez-González, V.T.; Reyes, J.L.; Sanchez, M.d.l.P.; Dinkova, T.D. Transcriptional Regulation of zma-MIR528a by Action of Nitrate and Auxin in Maize. Int. J. Mol. Sci. 2022, 23, 15718. https://doi.org/10.3390/ijms232415718
Luján-Soto E, Aguirre de la Cruz PI, Juárez-González VT, Reyes JL, Sanchez MdlP, Dinkova TD. Transcriptional Regulation of zma-MIR528a by Action of Nitrate and Auxin in Maize. International Journal of Molecular Sciences. 2022; 23(24):15718. https://doi.org/10.3390/ijms232415718
Chicago/Turabian StyleLuján-Soto, Eduardo, Paola I. Aguirre de la Cruz, Vasti T. Juárez-González, José L. Reyes, María de la Paz Sanchez, and Tzvetanka D. Dinkova. 2022. "Transcriptional Regulation of zma-MIR528a by Action of Nitrate and Auxin in Maize" International Journal of Molecular Sciences 23, no. 24: 15718. https://doi.org/10.3390/ijms232415718
APA StyleLuján-Soto, E., Aguirre de la Cruz, P. I., Juárez-González, V. T., Reyes, J. L., Sanchez, M. d. l. P., & Dinkova, T. D. (2022). Transcriptional Regulation of zma-MIR528a by Action of Nitrate and Auxin in Maize. International Journal of Molecular Sciences, 23(24), 15718. https://doi.org/10.3390/ijms232415718








