Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin
Abstract
1. Introduction
2. Results
2.1. Effect of TnT1 Fragment on the Binding Affinity of Tpm Mutants to F-Actin
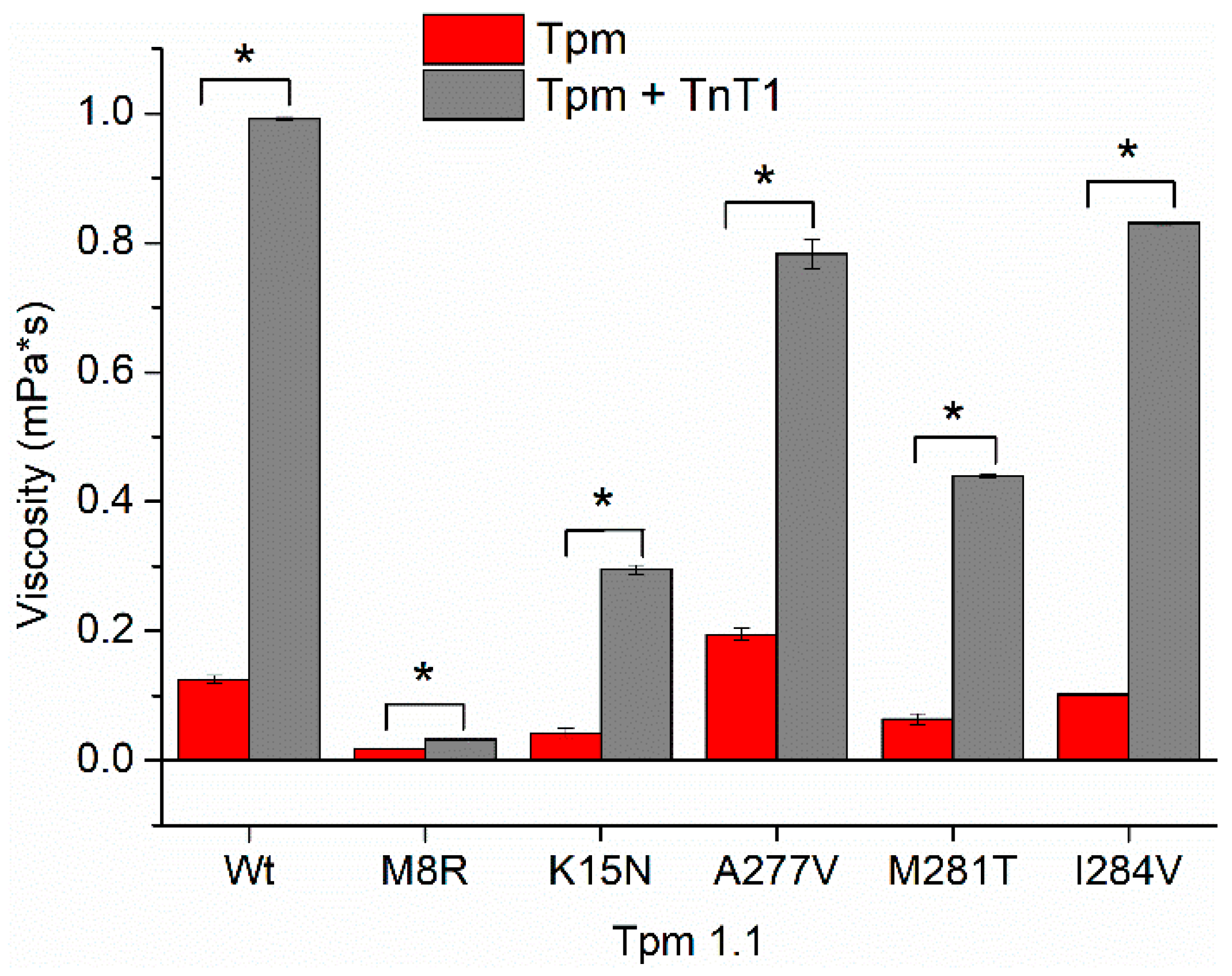
2.2. Effect of the TnT1 Fragment on the Thermally Induced Dissociation of Tpm–F-Actin Complexes
2.3. Interaction of TnT1 Fragment with Tpm Overlap Junction
2.4. Effect of TnT1 on Actin–Myosin Interaction
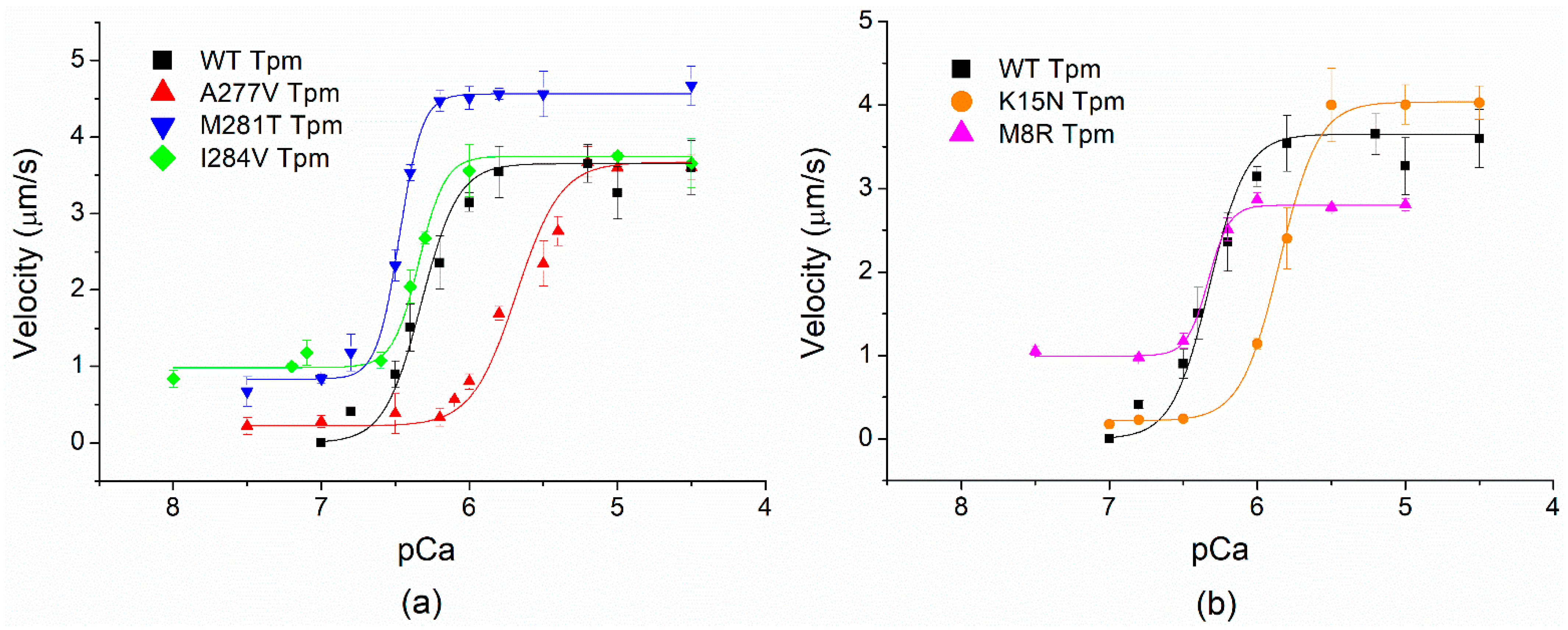
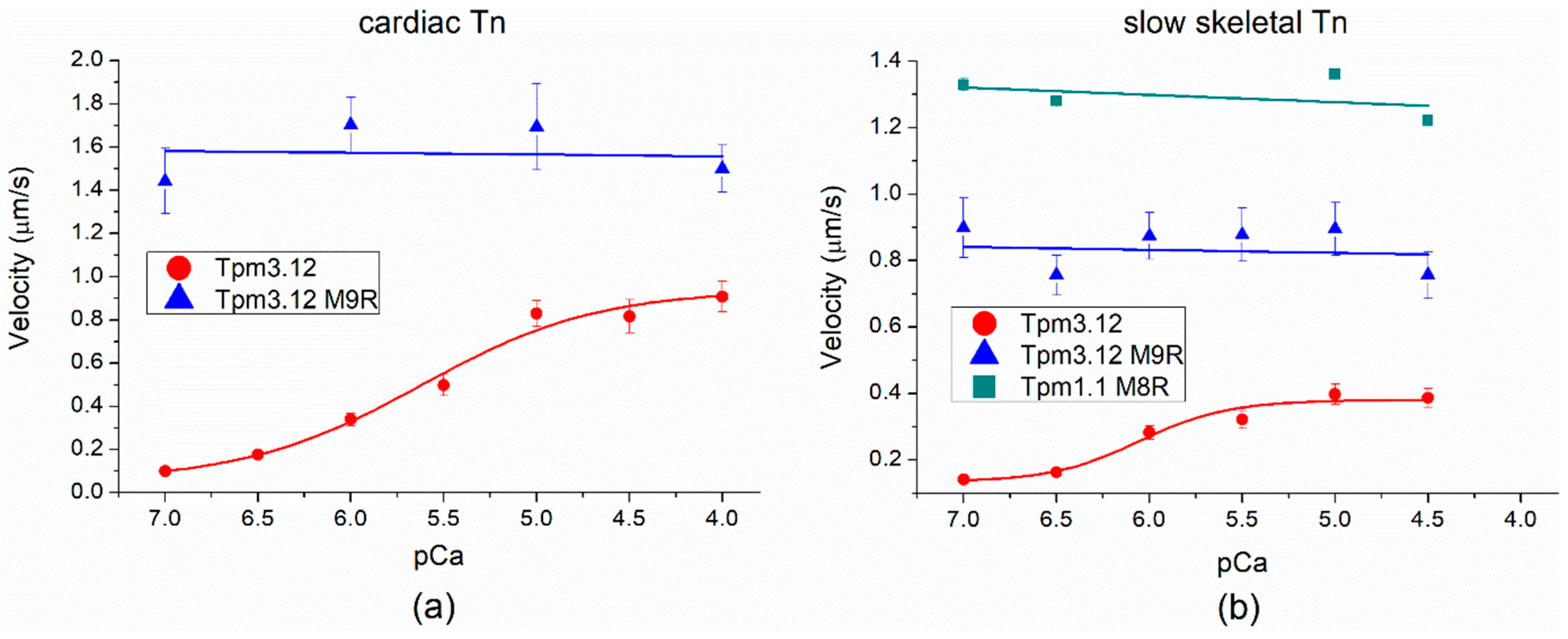
2.5. Effects of cardiomyopathy Tpm Mutations on the Ca2+-Regulation of the Actin–Myosin Interaction
2.6. Thermodynamics of Tpm Interactions with TnT1
3. Discussion
3.1. Mechanism of TnT1 and Tpm Interactions
3.2. Effects of Tpm1.1 M8R Mutation
3.3. Comparison of the Effects of Cardiomyopathy-Causing Tpm1.1 M8R and Myopathy-Causing Tpm3.12 M9R Mutations
3.4. Effects of Tpm1.1 K15N and A277V Mutations
3.5. Effects of Tpm1.1 M281T and I284V Mutations
4. Materials and Methods
4.1. Protein Preparations
4.2. Co-Sedimentation and Quantitative Electrophoresis
4.3. Light Scattering
4.4. Viscosity Measurements
4.5. Isothermal Titration Calorimetry (ITC)
4.6. In Vitro Motility Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Matyushenko, A.M.; Levitsky, D.I. Molecular mechanisms of pathologies of skeletal and cardiac muscles caused by point mutations in the tropomyosin genes. Biochemistry 2020, 85 (Suppl. 1), S20–S33. [Google Scholar] [CrossRef]
- Redwood, C.; Robinson, P. Alpha-tropomyosin mutations in inherited cardiomyopathies. J. Muscle Res. Cell Motil. 2013, 34, 285–294. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Huang, Y.J.; Swapna, G.V.; Bhattacharya, A.; Rapp, B.; Singh, A.; Montelione, G.T.; Hitchcock-DeGregori, S.E. Solution NMR structure of the junction between tropomyosin molecules: Implications for actin binding and regulation. J. Mol. Biol. 2006, 364, 80–96. [Google Scholar] [CrossRef]
- McKillop, D.F.; Geeves, M.A. Regulation of the interaction between actin and myosin subfragment-1: Evidence for three states of the thin filament. Biophys. J. 1993, 65, 693–701. [Google Scholar] [CrossRef]
- Perry, S.V. Vertebrate tropomyosin: Distribution, properties and function. J. Muscle Res. Cell Motil. 2001, 22, 5–49. [Google Scholar] [CrossRef]
- Tobacman, L.S. Thin filament-mediated regulation of cardiac contraction. Annu. Rev. Physiol. 1996, 58, 447–481. [Google Scholar] [CrossRef]
- Marston, S.; Zamora, J.E. Troponin structure and function: A view of recent progress. J. Muscle Res. Cell Motil. 2020, 41, 71–89. [Google Scholar] [CrossRef]
- Wei, B.; Jin, J.-P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene 2016, 582, 1–13. [Google Scholar] [CrossRef]
- Ohtsuki, I. Troponin: Structure, function and dysfunction. Adv. Exp. Med. Biol. 2007, 592, 21–36. [Google Scholar] [CrossRef]
- Katrukha, I.A. Human cardiac troponin complex. Structure and function. Biochemistry 2013, 78, 1447–1465. [Google Scholar] [CrossRef]
- Jin, J.-P. Evolution, regulation, and function of N-terminal variable region of troponin T: Modulation of muscle contractility and beyond international review of cell and molecular biology. Int. Rev. Cell Mol. Biol. 2016, 321, 1–28. [Google Scholar] [CrossRef]
- Tanokura, M.; Tawada, Y.; Onoyama, Y.; Nakamura, S.; Ohtsuki, I. Primary structure of chymotryptic subfragments from rabbit skeletal troponin T. J. Biochem. 1981, 90, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-P.; Chong, S.M. Localization of the two tropomyosin-binding sites of troponin T. Arch. Biochem. Biophys. 2010, 500, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Maytum, R.; Geeves, M.A.; Lehrer, S.S. A modulatory role for the troponin T tail domain in thin filament regulation. J. Biol. Chem. 2002, 277, 29774–29780. [Google Scholar] [CrossRef] [PubMed]
- Tobacman, L.S.; Nihli, M.; Butters, C.; Heller, M.; Hatch, V.; Craig, R.; Lehman, W.; Homsher, E. The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J Biol Chem. 2002, 277, 27636–27642. [Google Scholar] [CrossRef]
- Gollapudi, S.K.; Gallon, C.E.; Chandra, M. The tropomyosin binding region of cardiac troponin T modulates crossbridge recruitment dynamics in rat cardiac muscle fibers. J. Mol. Biol. 2013, 425, 1565–1581. [Google Scholar] [CrossRef]
- Wijnkera, P.J.M.; van der Velden, J. Mutation-specific pathology and treatment of hypertrophic cardiomyopathy in patients, mouse models and human engineered heart tissue. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165774. [Google Scholar] [CrossRef]
- Racca, W.; Rynkiewicz, M.J.; LaFave, N.; Ghosh, A.; Lehman, W.; Moore, J.R. M8R tropomyosin mutation disrupts actin binding and filament regulation: The beginning affects the middle and end. J. Biol. Chem. 2020, 295, 17128–17137. [Google Scholar] [CrossRef]
- Coplan, M.; Ly, T.; Grover, S.; Tolkachev, D.; Kostyukova, A.S. The cardiomyopathy-associated K15N mutation in tropomyosin alters actin filament pointed end dynamics. Arch. Biochem. Biophys. 2017, 630, 18–26. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Koubassova, N.A.; Shchepkin, D.V.; Kopylova, G.V.; Nabiev, S.R.; Nikitina, L.V.; Bershitsky, S.Y.; Levitsky, D.I.; Tsaturyan, A.K. The effects of cardiomyopathy-associated mutations in the head-to-tail overlap junction of α-tropomyosin on its properties and interaction with actin. Int. J. Biol. Macromol. 2019, 125, 1266–1274. [Google Scholar] [CrossRef]
- Sundar, S.; Rynkiewicz, M.J.; Ghosh, A.; Lehman, W.; Moore, J.R. Cardiomyopathy mutation alters end-to-end junction of tropomyosin and reduces calcium sensitivity. Biophys. J. 2020, 118, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, V.V.; Koubassova, N.A.; Borzova, V.A.; Kleymenov, S.Y.; Tsaturyan, A.K.; Matyushenko, A.M.; Levitsky, D.I. Tropomyosin pseudo-phosphorylation can rescue the effects of cardiomyopathy-associated mutations. Int. J. Biol. Macromol. 2021, 66, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Ly, T.; Pappas, C.T.; Johnson, D.; Schlecht, W.; Colpan, M.; Galkin, V.E.; Gregorio, C.C.; Dong, W.J.; Kostyukova, A.S. Effects of cardiomyopathy-linked mutations K15N and R21H in tropomyosin on thin-filament regulation and pointed-end dynamics. Mol. Biol. Cell 2019, 30, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Palm, T.; Graboski, S.; Hitchcock-DeGregori, S.E.; Greenfield, N.J. Disease-causing mutations in cardiac troponin T: Identification of a critical tropomyosin-binding Region. Biophys. J. 2001, 81, 2827–2828. [Google Scholar] [CrossRef][Green Version]
- Gangadharana, B.; Sunithaa, M.S.; Mukherjeea, S.; Chowdhuryd, R.R.; Haquea, F.; Sekara, N.; Sowdhaminic, R.; Spudich, J.A.; Mercera, J.A. Molecular mechanisms and structural features of cardiomyopathy-causing troponin T mutants in the tropomyosin overlap region. Proc. Natl. Acad. Sci. USA. 2017, 114, 11115–11120. [Google Scholar] [CrossRef]
- Geeves, M.A.; Hitchcock-DeGregori, S.E.; Gunning, P.W. A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 2015, 36, 147–153. [Google Scholar] [CrossRef]
- Ilkovski, B.; Mokbel, N.; Lewis, R.A.; Walker, K.; Nowak, K.J.; Domazetovska, A.; Laing, N.G.; Fowler, V.M.; North, K.N.; Cooper, S.T. Disease severity and thin filament regulation in M9R TPM3 nemaline myopathy. J. Neuropathol. Exp. Neurol. 2008, 67, 867–877. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Nefedova, V.V.; Shchepkin, D.V.; Kopylova, G.V.; Berg, V.Y.; Pivovarova, A.V.; Kleymenov, S.Y.; Bershitsky, S.Y.; Levitsky, D.I. Mechanisms of disturbance of the contractile function of slow skeletal muscles induced by myopathic mutations in the tropomyosin TPM3 gene. FASEB J. 2020, 34, 13507–13520. [Google Scholar] [CrossRef]
- Pavadai, E.; Rynkiewicz, M.J.; Ghosh, A.; Lehman, W. Docking troponin T onto the tropomyosin overlapping domain of thin filaments. Biophys. J. 2020, 118, 325–336. [Google Scholar] [CrossRef]
- Jackson, P.; Amphlett, G.W.; Perry, S.V. The primary structure of troponin T and the interaction with tropomyosin. Biochem. J. 1975, 151, 85–97. [Google Scholar] [CrossRef]
- Margossian, S.S.; Cohen, C. Troponin subunit interactions. J. Mol. Biol. 1973, 81, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mui, S.; Brown, J.H.; Strand, J.; Reshetnikova, L.; Tobacman, L.S.; Cohen, C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc. Natl. Acad. Sci. USA. 2002, 99, 7378–7383. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J.; Stafford, W.E.; Hitchcock-DeGregory, S.E. The effect of N-terminal acetylation on the structure of an N-terminal tropomyosin peptide and aa-tropomyosin. Protein Sci. 1994, 3, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Hammell, R.L.; Hitchcock-DeGregori, S.E. Mapping the functional domains within the carboxyl terminus of alpha-tropomyosin encoded by the alternatively spliced ninth exon. J. Biol. Chem. 1996, 271, 4236–4242. [Google Scholar] [CrossRef]
- Palm, T.; Greenfield, N.J.; Hitchcock-DeGregori, S.E. Tropomyosin ends determine the stability and functionality of overlap and troponin T complexes. Biophys. J. 2003, 84, 3181–3189. [Google Scholar] [CrossRef][Green Version]
- Moraczewska, J.; Greenfield, N.J.; Liu, Y.; Hitchcock-DeGregori, S.E. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophys J. 2000, 79, 3217–3225. [Google Scholar] [CrossRef][Green Version]
- Matyushenko, A.M.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Levitsky, D.I. Unique functional properties of slow skeletal muscle tropomyosin. Biochimie 2020, 174, 1–8. [Google Scholar] [CrossRef]
- Marston, S.B.; Redwood, C.S. Modulation of thin filament activation by breakdown or isoform switching of thin filament proteins physiological and pathological implications. Circ. Res. 2003, 93, 1170–1178. [Google Scholar] [CrossRef]
- Amarasinghe, C.; Jin, J.-P. N-terminal hypervariable region of muscle type isoforms of troponin T differentially modulates the affinity of tropomyosin-binding site 1. Biochemistry 2015, 54, 3822–3830. [Google Scholar] [CrossRef]
- Sequeira, V.; Wijnker, P.J.M.; Nijenkamp, L.L.A.M.; Kuster, D.W.D.; Najafi, A.; Witjas-Paalberends, E.R.; Regan, J.A.; Boontje, N.; ten Cate, F.J.; Germans, T.; et al. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ. Res. 2013, 112, 1491–1505. [Google Scholar] [CrossRef]
- Gupte, T.M.; Haque, F.; Gangadharan, B.; Sunitha, M.S.; Mukherjee, S.; Anandhan, S.; Rani, D.S.; Mukundan, N.; Jambekar, A.; Thangaraj, K.; et al. Mechanistic Heterogeneity in Contractile Properties of α-Tropomyosin (TPM1) Mutants Associated with Inherited Cardiomyopathies. J. Biol. Chem. 2015, 290, 7003–7015. [Google Scholar] [CrossRef]
- Doran, M.H.; Pavadai, E.; Rynkiewicz, M.J.; Walklate, J.; Bullitt, E.; Moore, J.R.; Regnier, M.; Geeves, M.A.; Lehman, W. Cryo-EM and molecular docking shows myosin loop 4 contacts actin and tropomyosin on thin filaments. Biophys J. 2020, 119, 821–830. [Google Scholar] [CrossRef]
- Monteiro, P.B.; Lataro, R.C.; Ferro, J.A.; Reinach, F.d.C. Functional α-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J. Biol. Chem. 1994, 269, 10461–10466. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Palm, T.; Hitchcock-DeGregori, S.E. Structure and interactions of the carboxyl terminus of striated muscle alpha-tropomyosin: It is important to be flexible. Biophys. J. 2002, 83, 2754–2766. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Artemova, N.V.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Tsaturyan, A.K.; Sluchanko, N.N.; Levitsky, D.I. Structural and functional effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin molecule. FEBS J. 2014, 281, 2004–2016. [Google Scholar] [CrossRef]
- Mariasina, S.S.; Chang, C.-F.; Petrova, O.A.; Efimov, S.V.; Klochkov, V.V.; Kechko, O.I.; Mitkevich, V.A.; Sergiev, P.V.; Dontsova, O.A.; Polshakov, V.I. Williams-Beuren syndrome-related methyltransferase WBSCR27: Cofactor binding and cleavage. FEBS J. 2020, 287, 5375–5393. [Google Scholar] [CrossRef]
- Margossian, S.S.; Lowey, S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982, 85, 55–71. [Google Scholar] [CrossRef]
- Potter, J.D. Preparation of troponin and its subunits. Methods Enzymol. 1982, 85, 241–263. [Google Scholar] [CrossRef]
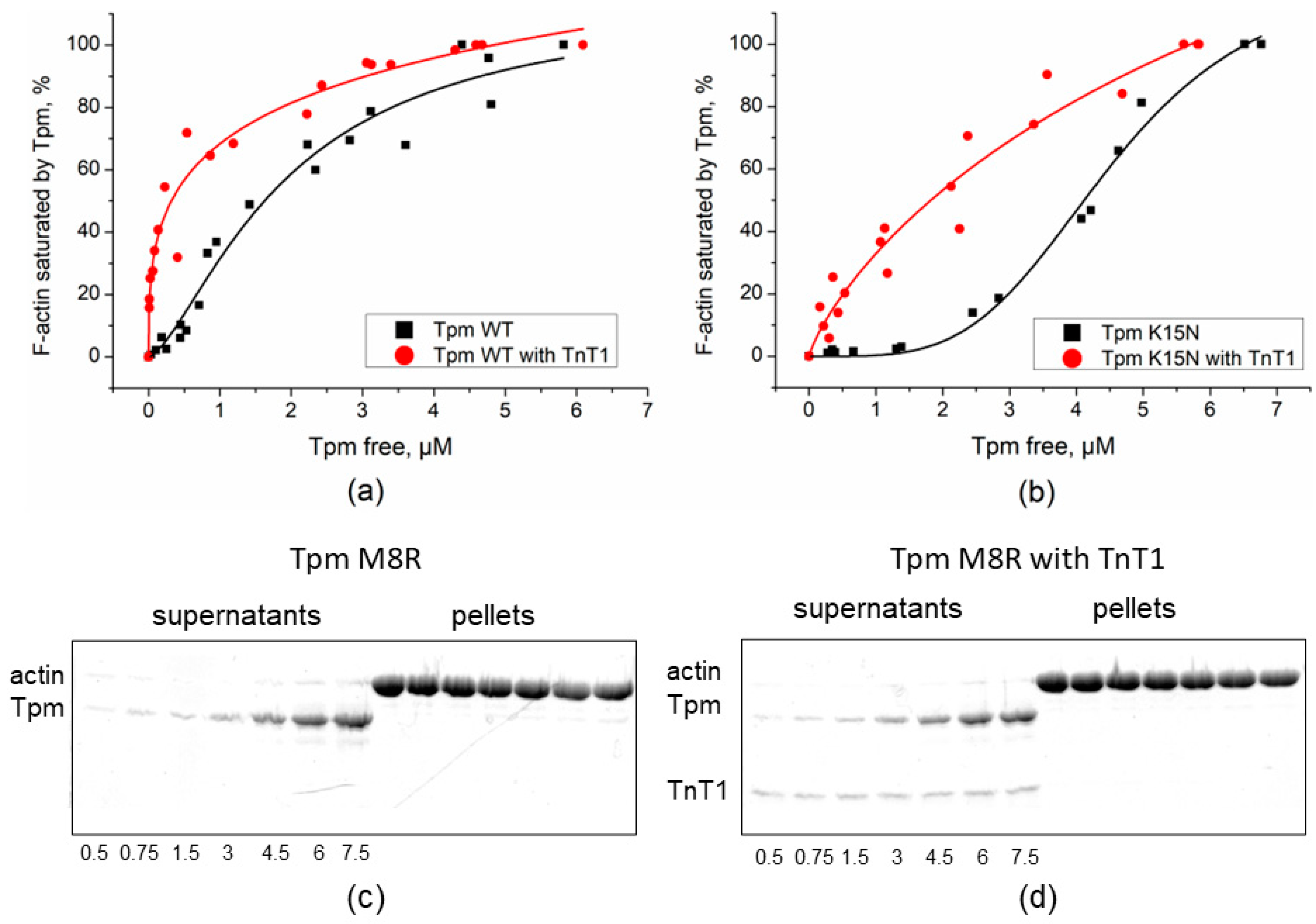
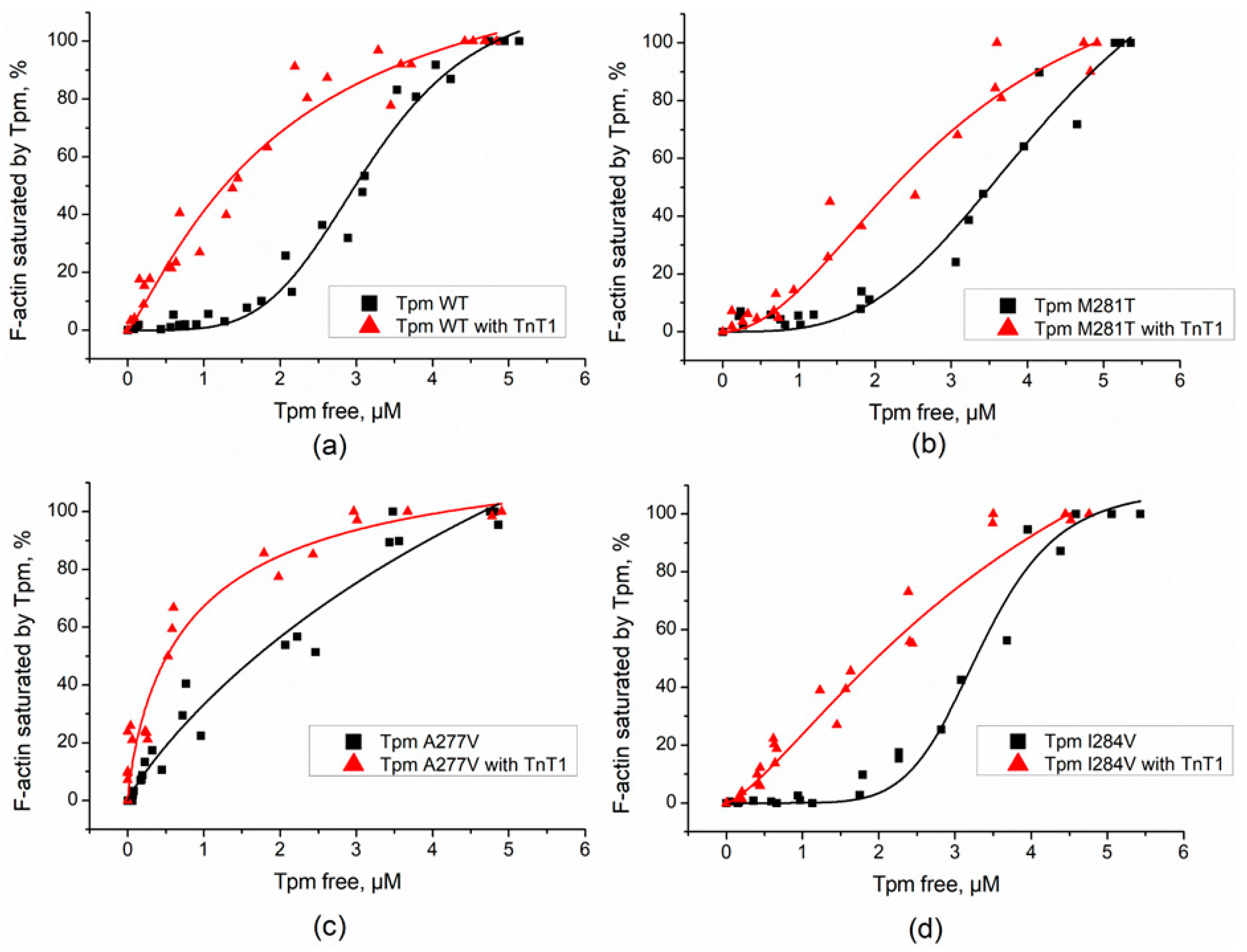



| Tpm | Tdiss, °C without TnT1 | Tdiss, °C with TnT1 |
|---|---|---|
| WT | 46.5 ± 0.03 | 48.1 ± 0.02 |
| M8R | ND | ND |
| K15N | 39.4 ± 0.06 | 44.1 ± 0.04 |
| A277V | 48.7 ± 0.06 | 50.3 ± 0.04 |
| M281T | 45.4 ± 0.03 | 46.8 ± 0.02 |
| I284V | 45.4 ± 0.03 | 47.7 ± 0.04 |
| Tpm | Viscosity (mPa·s) without TnT1 | Viscosity (mPa·s) with TnT1 |
|---|---|---|
| WT | 0.125 ± 0.001 | 0.992 ± 0.001 # |
| M8R | 0.018 ± 0.001 § | 0.033 ± 0.001 §,# |
| K15N | 0.043 ± 0.001 § | 0.295 ± 0.007 §,# |
| A277V | 0.195 ± 0.001 § | 0.783 ± 0.023 §,# |
| M281T | 0.064 ± 0.001 § | 0.439 ± 0.002 §,# |
| I284V | 0.102 ± 0.001 § | 0.830 ± 0.003 §,# |
| Tpm | Vmax (µm/s) | V0 (µm/s) | h | pCa50 |
|---|---|---|---|---|
| WT | 3.7 ± 0.1 | 0 | 2 ± 0.2 | 6.33 ± 0.01 |
| M8R | 2.8 ± 0.1 * | 1.0 ± 0.1 * | 2.2 ± 0.2 | 6.33 ± 0.03 |
| K15N | 4.0 ± 0.1 * | 0.2 ± 0.1 | 1.8 ± 0.3 | 5.85 ± 0.01 * |
| A277V | 3.7 ± 0.1 | 0.2 ± 0.1 | 1.6 ± 0.2 | 5.68 ± 0.01 * |
| M281T | 4.6 ± 0.1 * | 0.8 ± 0.1 * | 2.7 ± 0.3 | 6.47 ± 0.01 * |
| I284V | 3.7 ± 0.1 | 1.0 ± 0.1 * | 2.1 ± 0.2 | 6.34 ± 0.01 |
| Myosin and Tn | Tpm | Vmax (µm/s) | pCa50 |
|---|---|---|---|
| cardiac myosin + cardiac Tn | γ-Tpm | 0.94 ± 0.08 | 5.62 ± 0.12 |
| M9R γ-Tpm | 1.55 ± 0.17 | ||
| cardiac myosin + slow skeletal Tn | γ-Tpm | 0.38 ± 0.02 | 6.05 ± 0.11 |
| M9R γ-Tpm | 0.84 ± 0.06 | ||
| M8R α-Tpm | 1.30 ± 0.06 |
| Sample | N | KD, µM | ΔH, kcal/mol | ΔG, kcal/mol | TΔS, kcal/mol |
|---|---|---|---|---|---|
| Tpm WT | 0.629 ± 0.009 | 0.91 ± 0.14 | −9.69 ± 0.26 | −8.25 | −1.5 |
| Tpm M8R | 1.15 ± 0.03 | 1.9 ± 0.4 | 2.51 ± 0.13 | −7.8 | 10.3 |
| Tpm K15N | 1.05 ± 0.02 | 1.3 ± 0.3 | −12.1 ± 0.5 | −8.0 | −4.1 |
| Tpm A277V | 1.03 ± 0.01 | 2.51 ± 0.15 | −10.60 ± 0.02 | −7.6 | −3.0 |
| Tpm M281T | 0.857 ± 0.014 | 1.52 ± 0.21 | −8.83 ± 0.28 | −7.9 | −0.9 |
| Tpm I284V | 0.775 ± 0.008 | 1.89 ± 0.16 | −14.40 ± 0.29 | −7.9 | −6.6 |
| Sample | Ligand | N | KD, µM | ΔH, kcal/mol | ΔG, kcal/mol | TΔS, kcal/mol |
|---|---|---|---|---|---|---|
| Tpm WT1–133/ Tpm134–284 | TnT1 | 0.69 ± 0.012 | 0.90 ± 0.15 | −7.7 ± 0.3 | −8.25 | 0.6 |
| Tpm WT1–133 | TnT1 | no binding | - | - | - | - |
| Tpm WT134–284 | TnT1 | 1.06 ± 0.03 | 1.9 ± 0.5 | 2.10 ± 0.11 | −7.8 | 9.9 |
| Tpm WT134–284 | Tpm WT1–133 | 1.17 ± 0.05 | 10.2 ± 0.3 | 1.95 ± 0.15 | −6.8 | 8.8 |
| TnT1/ TpmWT134–284 | Tpm WT1–133 | 1.07 ± 0.03 | 1.7 ± 0.4 | −6.5 ± 0.4 | −7.9 | 1.4 |
| TnT1/ TpmWT134–284 | Tpm M8R1–133 | no binding | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nefedova, V.V.; Kopylova, G.V.; Shchepkin, D.V.; Kochurova, A.M.; Kechko, O.I.; Borzova, V.A.; Ryabkova, N.S.; Katrukha, I.A.; Mitkevich, V.A.; Bershitsky, S.Y.; et al. Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin. Int. J. Mol. Sci. 2022, 23, 15723. https://doi.org/10.3390/ijms232415723
Nefedova VV, Kopylova GV, Shchepkin DV, Kochurova AM, Kechko OI, Borzova VA, Ryabkova NS, Katrukha IA, Mitkevich VA, Bershitsky SY, et al. Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin. International Journal of Molecular Sciences. 2022; 23(24):15723. https://doi.org/10.3390/ijms232415723
Chicago/Turabian StyleNefedova, Victoria V., Galina V. Kopylova, Daniil V. Shchepkin, Anastasia M. Kochurova, Olga I. Kechko, Vera A. Borzova, Natalia S. Ryabkova, Ivan A. Katrukha, Vladimir A. Mitkevich, Sergey Y. Bershitsky, and et al. 2022. "Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin" International Journal of Molecular Sciences 23, no. 24: 15723. https://doi.org/10.3390/ijms232415723
APA StyleNefedova, V. V., Kopylova, G. V., Shchepkin, D. V., Kochurova, A. M., Kechko, O. I., Borzova, V. A., Ryabkova, N. S., Katrukha, I. A., Mitkevich, V. A., Bershitsky, S. Y., Levitsky, D. I., & Matyushenko, A. M. (2022). Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin. International Journal of Molecular Sciences, 23(24), 15723. https://doi.org/10.3390/ijms232415723






