The N6-Methyladenosine Regulator ALKBH5 Mediated Stromal Cell–Macrophage Interaction via VEGF Signaling to Promote Recurrent Spontaneous Abortion: A Bioinformatic and In Vitro Study
Abstract
1. Introduction
2. Results
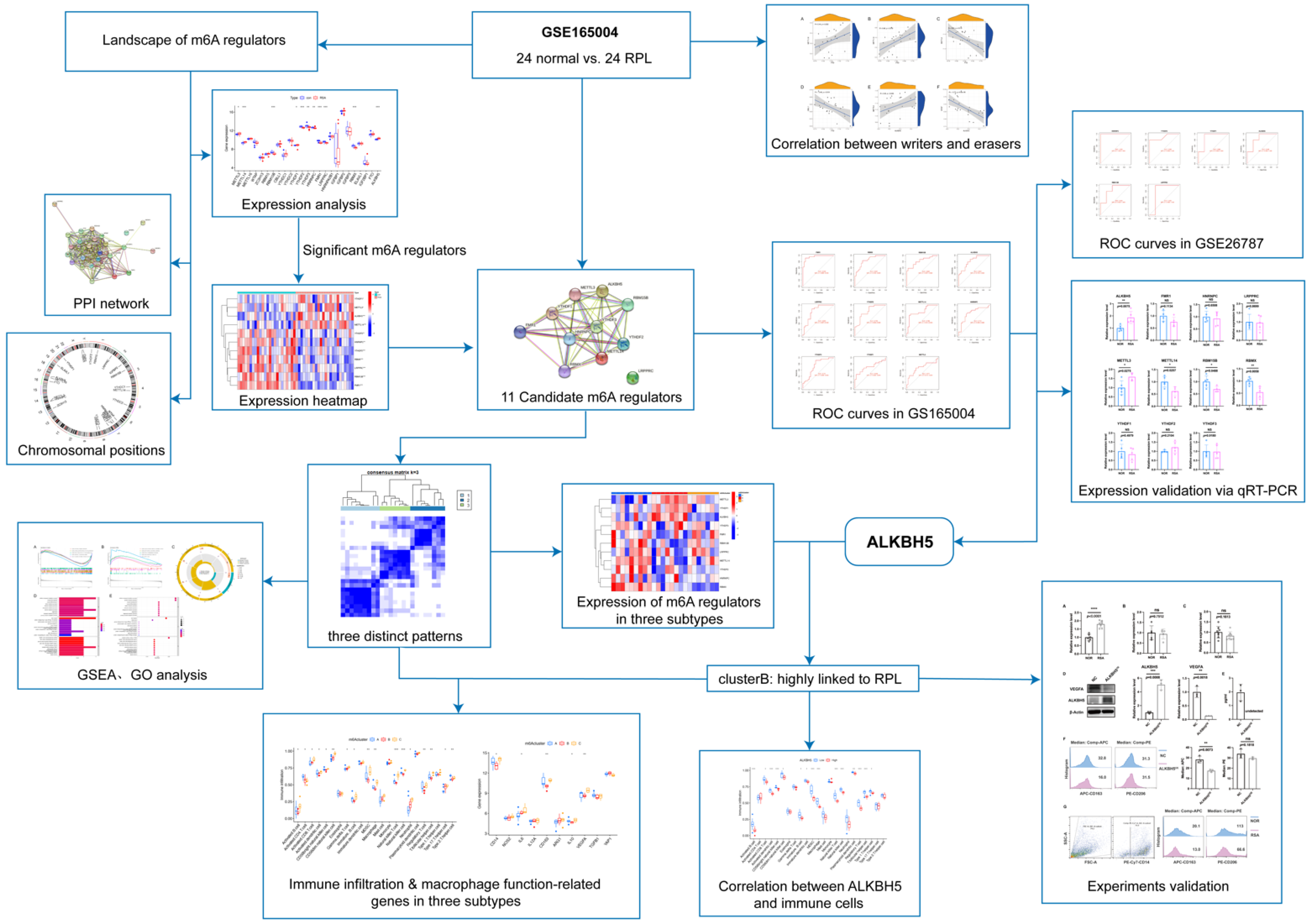
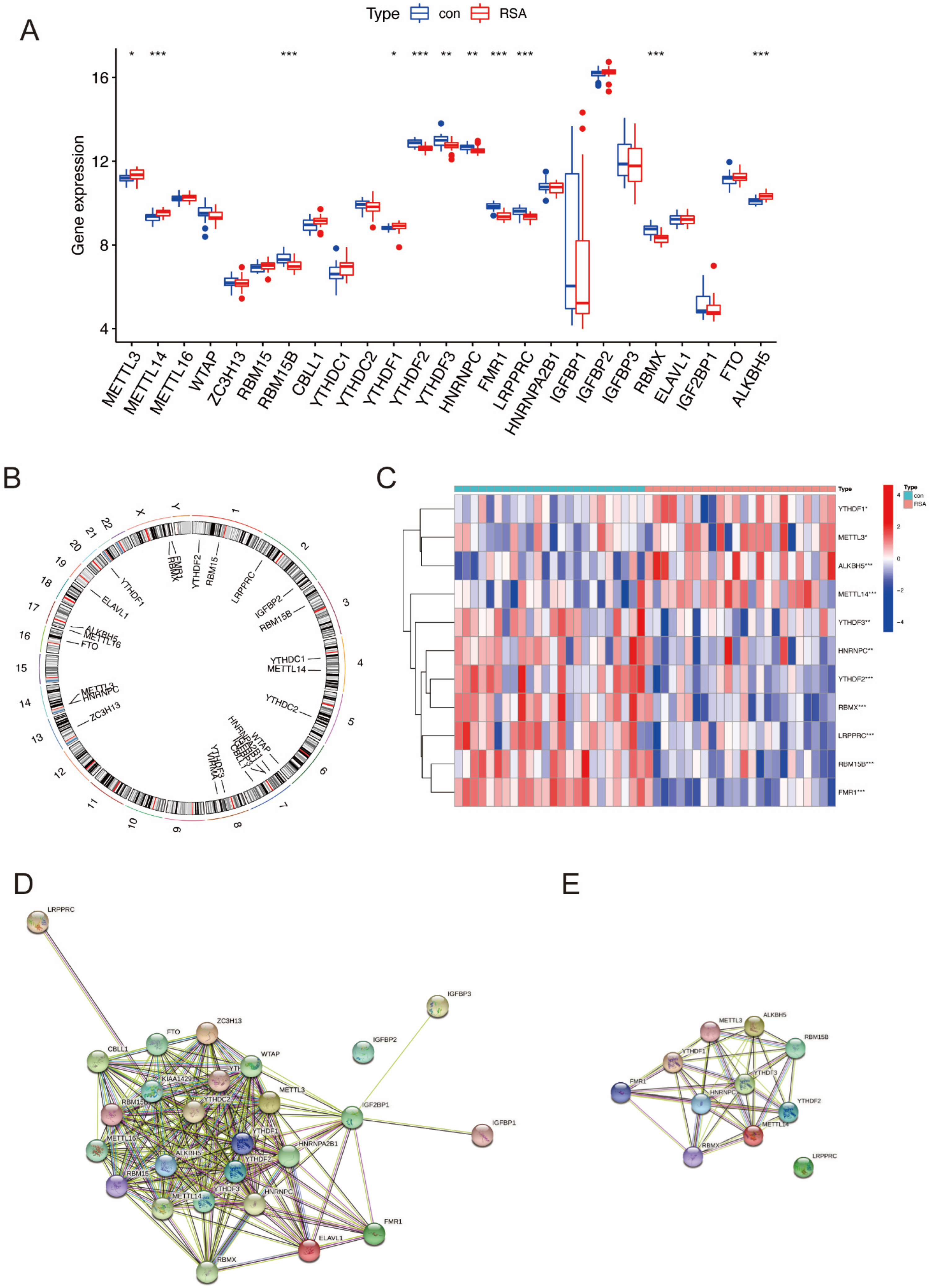
2.1. Landscape of the m6A Regulators in RSA
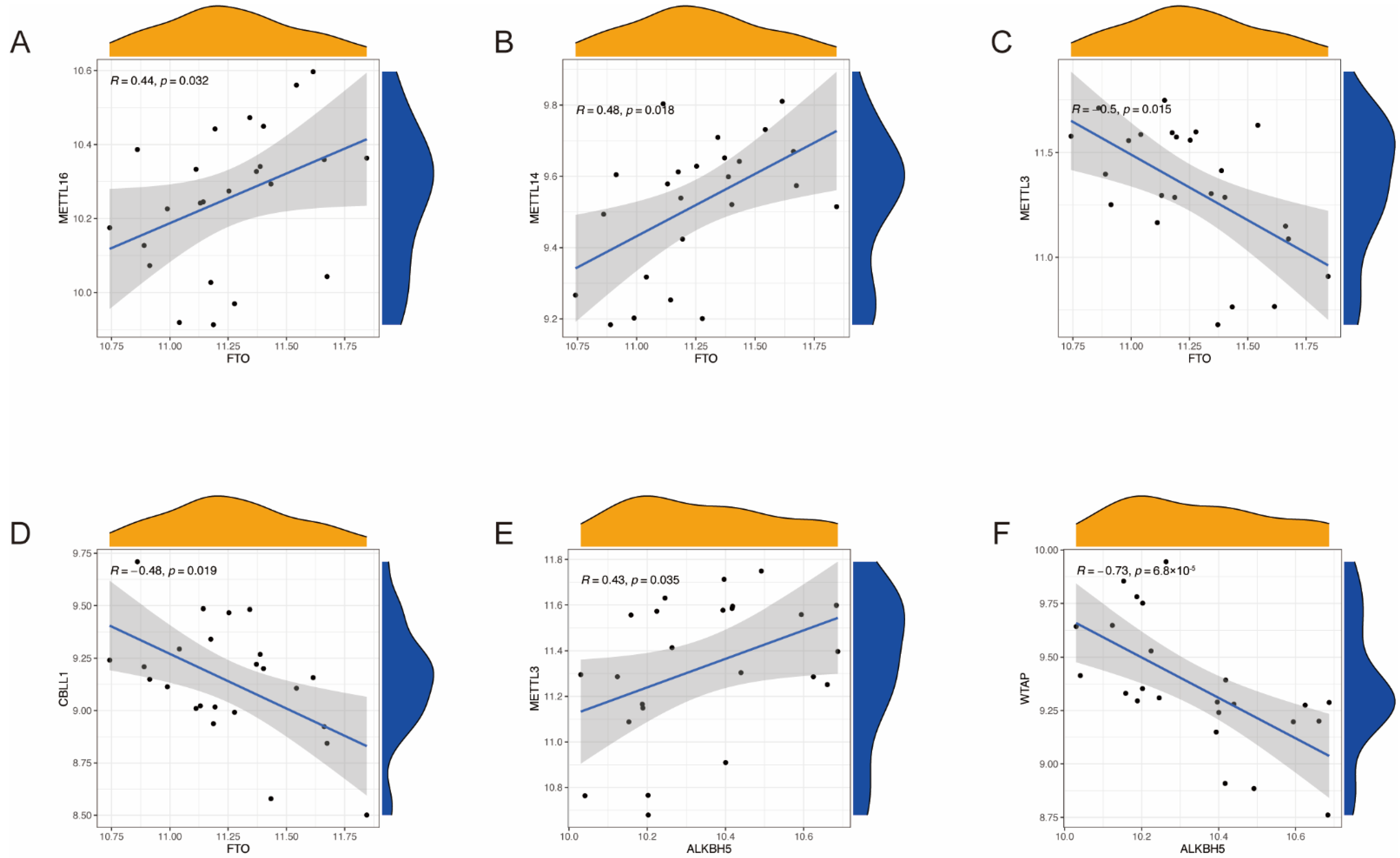
2.2. Identification of the Correlation between Writers and Erasers in RSA
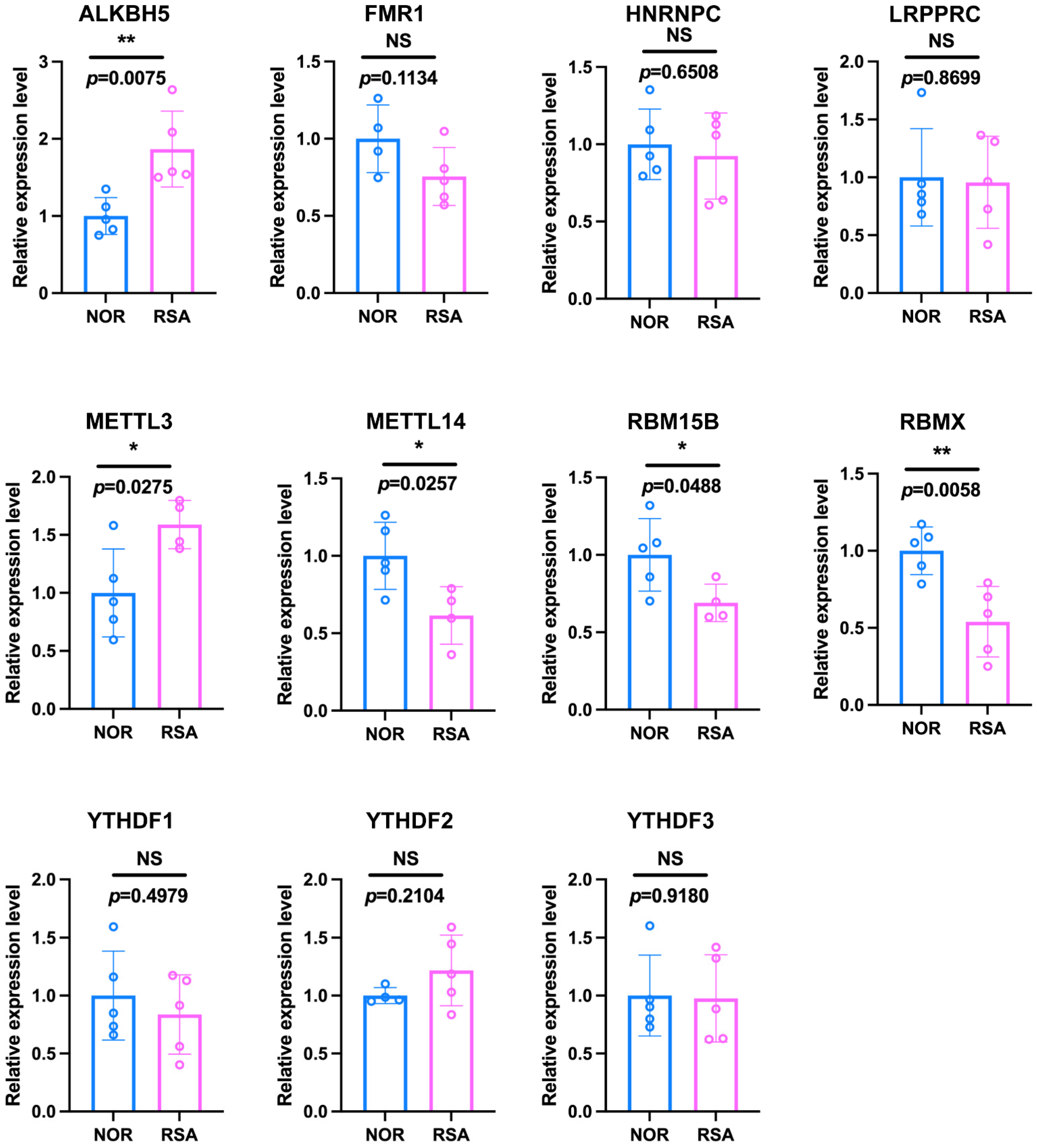
2.3. Identification and Validation of the Predictive Ability of 11 m6A Regulators
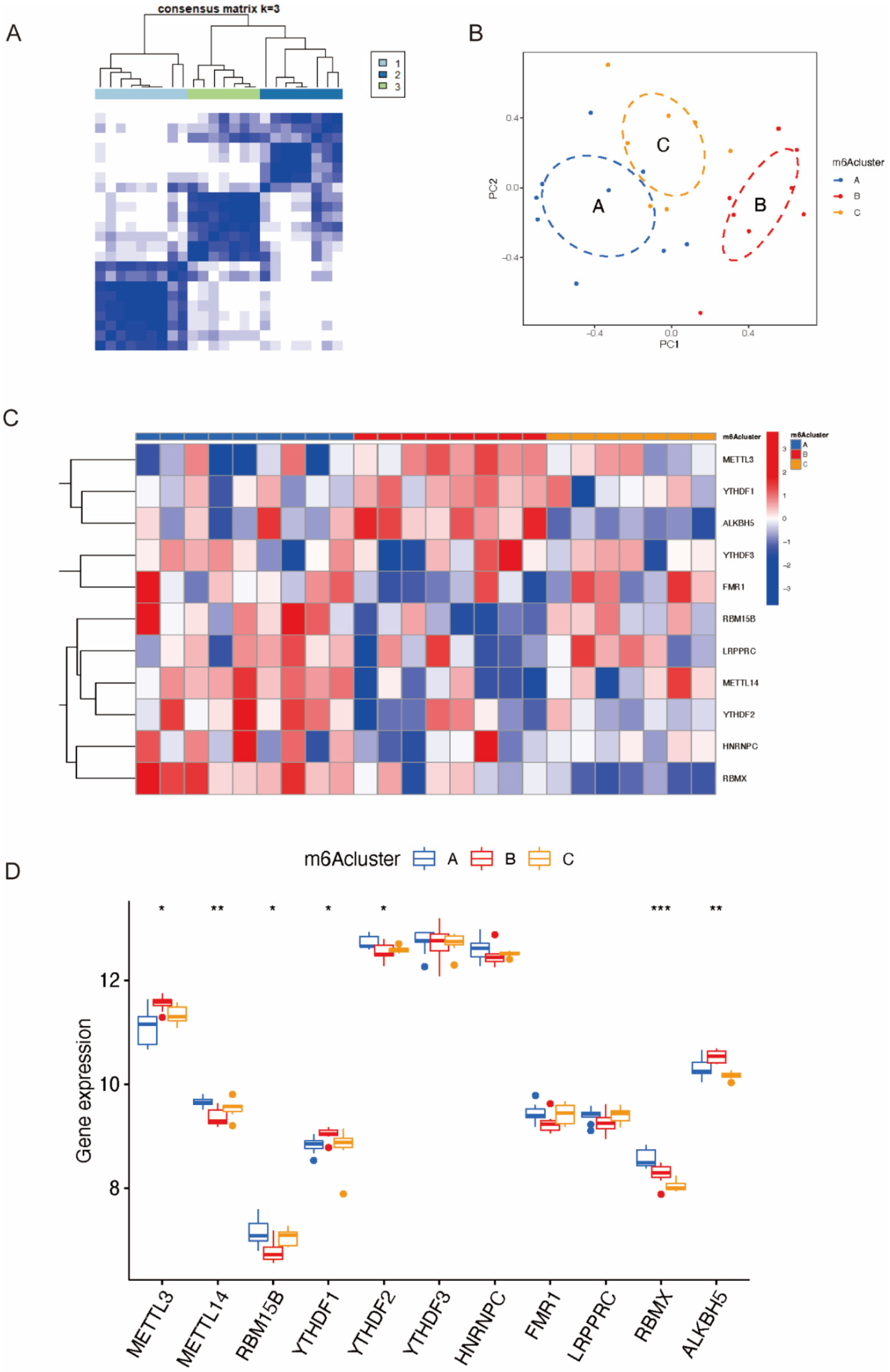
2.4. Identification of Three Distinct Patterns in RSA and Functional Enrichment Analysis
2.5. Genes Related to Immune Infiltration and Macrophage Function Related in Different m6A Patterns
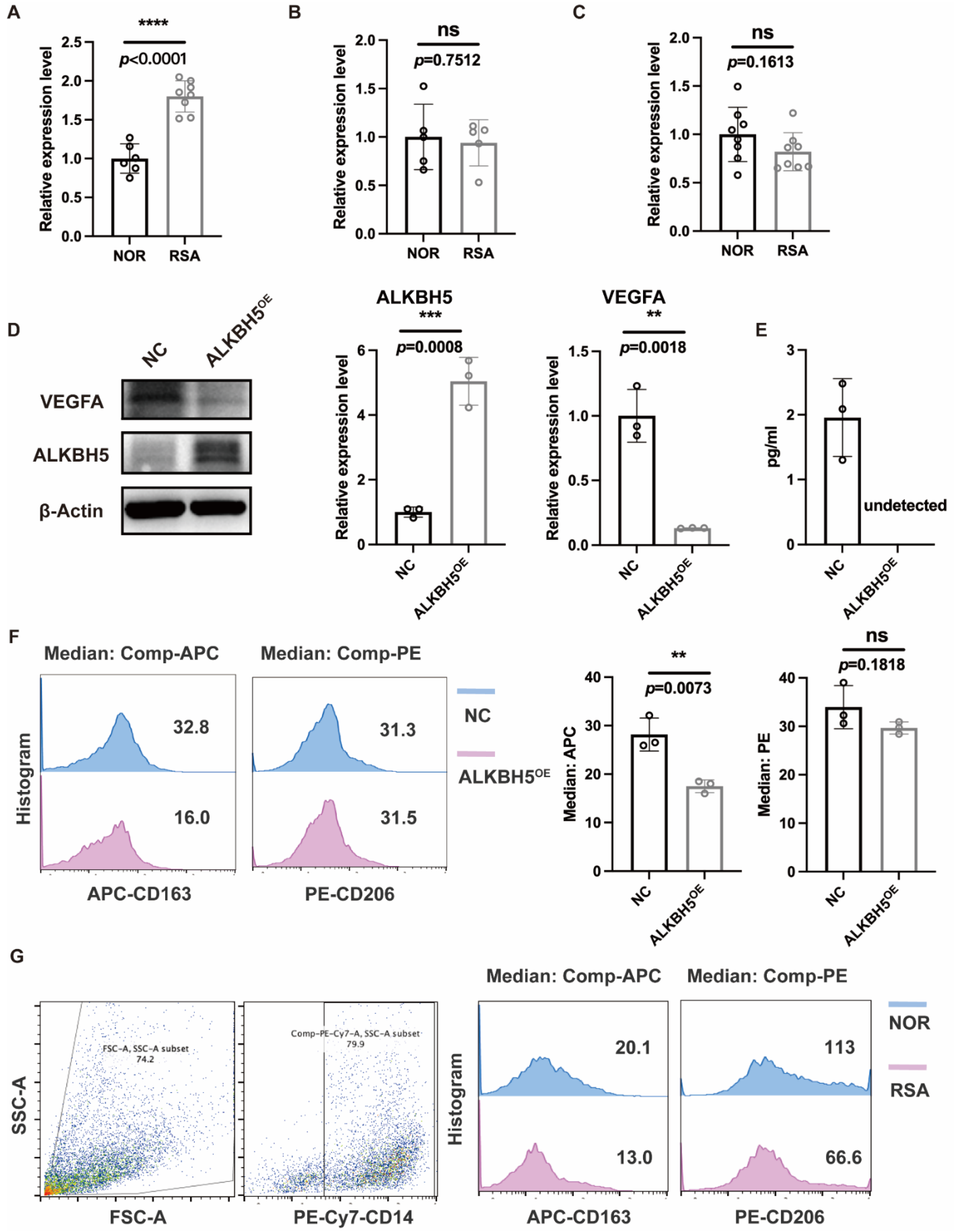
2.6. Overexpression of ALKBH5 in ESCs Affected Macrophage Differentiation via VEGF Secretion
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and PPI Network Construction
4.2. Construction of Receiver Operating Characteristic (ROC) Curves
4.3. Classification of Three Distinct m6A Subtypes and Extraction of DEGs
4.4. Functional Enrichment Analysis of DEGs
4.5. Single-Sample Gene Set Enrichment Analysis to Identify Immune-Cell Infiltration
4.6. Cell Culture
4.7. Antibodies for Flow Cytometry (FCM)
4.8. Construction of an ALKBH5-Overexpressing ESC Cell Line
4.9. Enzyme-Linked Immunosorbent Assay
4.10. Quantitative Real-Time PCR (qRT-PCR)
4.11. Western Blot
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lédée, N.; Petitbarat, M.; Prat-Ellenberg, L.; Dray, G.; Cassuto, G.N.; Chevrier, L.; Kazhalawi, A.; Vezmar, K.; Chaouat, G. Endometrial Immune Profiling: A Method to Design Personalized Care in Assisted Reproductive Medicine. Front. Immunol. 2020, 11, 1032. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.P.; Hamilton, A.E.; Talbi, S.; Dosiou, C.; Nyegaard, M.; Nayak, N.; Genbecev-Krtolica, O.; Mavrogianis, P.; Ferrer, K.; Kruessel, J.; et al. Decidual stromal cell response to paracrine signals from the trophoblast: Amplification of immune and angiogenic modulators. Biol. Reprod. 2007, 76, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Tanaka, S.; Okada, H. Immune Tolerance of the Human Decidua. J. Clin. Med. 2021, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Nallasamy, S.; Li, Q.; Bagchi, M.K.; Bagchi, I.C. Msx Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium. PLoS Genet. 2012, 8, e1002500. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Cardenas, I.; Abrahams, V.; Guller, S. Inflammation and pregnancy: The role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 2011, 1221, 80–87. [Google Scholar] [CrossRef]
- Du, L.; Deng, W.; Zeng, S.; Xu, P.; Huang, L.; Liang, Y.; Wang, Y.; Xu, H.; Tang, J.; Bi, S.; et al. Single-cell transcriptome analysis reveals defective decidua stromal niche attributes to recurrent spontaneous abortion. Cell Prolif. 2021, 54, e13125. [Google Scholar] [CrossRef]
- Dai, D.; Wang, H.; Zhu, L.; Jin, H.; Wang, X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018, 9, 124. [Google Scholar] [CrossRef]
- Li, X.-C.; Jin, F.; Wang, B.-Y.; Yin, X.-J.; Hong, W.; Tian, F.-J. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics 2019, 9, 3853–3865. [Google Scholar] [CrossRef]
- Qiu, W.; Zhou, Y.; Wu, H.; Lv, X.; Yang, L.; Ren, Z.; Tian, H.; Yu, Q.; Li, J.; Lin, W.; et al. RNA Demethylase FTO Mediated RNA m6A Modification Is Involved in Maintaining Maternal-Fetal Interface in Spontaneous Abortion. Front. Cell Dev. Biol. 2021, 9, 617172. [Google Scholar] [CrossRef]
- Morita, K.; Ono, Y.; Takeshita, T.; Sugi, T.; Fujii, T.; Yamada, H.; Nakatsuka, M.; Fukui, A.; Saito, S. Risk Factors and Outcomes of Recurrent Pregnancy Loss in Japan. J. Obstet. Gynaecol. Res. 2019, 45, 1997–2006. [Google Scholar] [CrossRef]
- Zhao, S.; Lu, J.; Chen, Y.; Wang, Z.; Cao, J.; Dong, Y. Exploration of the potential roles of m6A regulators in the uterus in pregnancy and infertility. J. Reprod. Immunol. 2021, 146, 103341. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Gu, Q.; Ma, Y.; Yang, Y.; Zhu, J.; Zhang, Q. The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell Int. 2020, 20, 347. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Zhang, J.; Zhu, J.-S. The role of m6A RNA methylation in human cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, F.; Gan, H.; Jin, L. Hypoxia induced ALKBH5 prevents spontaneous abortion by mediating m6A-demethylation of SMAD1/5 mRNAs. Biochim. Biophys. Acta 2022, 1869, 119316. [Google Scholar] [CrossRef]
- Care, A.S.; Diener, K.; Jasper, M.J.; Brown, H.M.; Ingman, W.V.; Robertson, S.A. Macrophages regulate corpus luteum development during embryo implantation in mice. J. Clin. Investig. 2013, 123, 3472–3487. [Google Scholar] [CrossRef]
- Co, E.C.; Gormley, M.; Kapidzic, M.; Rosen, D.B.; Scott, J.; Stolp, H.A.; McMaster, M.; Lanier, L.L.; Bárcena, A.; Fisher, S.J. Maternal Decidual Macrophages Inhibit NK Cell Killing of Invasive Cytotrophoblasts During Human Pregnancy. Biol. Reprod. 2013, 88, 155. [Google Scholar] [CrossRef]
- Erlebacher, A. Immunology of the Maternal-Fetal Interface. Annu. Rev. Immunol. 2013, 31, 387–411. [Google Scholar] [CrossRef]
- Nagamatsu, T.; Schust, D.J. The Immunomodulatory Roles of Macrophages at the Maternal—Fetal Interface. Reprod. Sci. 2010, 17, 209–218. [Google Scholar] [CrossRef]
- Yao, Y.; Hao, F.; Tang, L.-C.; Xu, X.-H.; Jin, L. Downregulation of HDAC8 expression decreases CD163 levels and promotes the apoptosis of macrophages by activating the ERK signaling pathway in recurrent spontaneous miscarriage. Mol. Hum. Reprod. 2020, 26, 521–531. [Google Scholar] [CrossRef]
- Kim, S.Y.; Romero, R.; Tarca, A.L.; Bhatti, G.; Kim, C.J.; Lee, J.; Elsey, A.; Than, N.G.; Chaiworapongsa, T.; Hassan, S.S.; et al. Methylome of Fetal and Maternal Monocytes and Macrophages at the Feto-Maternal Interface. Am. J. Reprod. Immunol. 2012, 68, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Heikkinen, J.; Möttönen, M.; Komi, J.; Alanen, A.; Lassila, O. Phenotypic characterization of human decidual macrophages. Clin. Exp. Immunol. 2003, 131, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Joerink, M.; Rindsjö, E.; van Riel, B.; Alm, J.; Papadogiannakis, N. Placental macrophage (Hofbauer cell) polarization is independent of maternal allergen-sensitization and presence of chorioamnionitis. Placenta 2011, 32, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, K.C.; Jena, M.; Pradhan, B.; Nayak, N.; Das, S.; Hsu, C.-D.; Wheeler, D.S.; Chen, K.; Nayak, N.R. VEGF may contribute to macrophage recruitment and M2 polarization in the decidua. PLoS ONE 2018, 13, e0191040. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, Z.; Yuan, M.; Zhao, Y.; Zhou, Y.; Pei, H.; Bai, L. M6A Regulatory Genes Play an Important Role in the Prognosis, Progression and Immune Microenvironment of Pancreatic Adenocarcinoma. Cancer Investig. 2020, 39, 39–54. [Google Scholar] [CrossRef]
- Liu, X.; Guo, A.; Tu, Y.; Li, W.; Li, L.; Liu, W.; Zhou, Y.; Sang, A.; Zhu, M. Fruquintinib inhibits VEGF/VEGFR2 axis of choroidal endothelial cells and M1-type macrophages to protect against mouse laser-induced choroidal neovascularization. Cell Death Dis. 2020, 11, 1016. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Lédée, N.; Munaut, C.; Aubert, J.; Sérazin, V.; Rahmati, M.; Chaouat, G.; Sandra, O.; Foidart, J.M. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J. Pathol. 2011, 225, 554–564. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Denny, P.; Feuermann, M.; Hill, D.P.; Lovering, R.C.; Plun-Favreau, H.; Roncaglia, P. Exploring autophagy with Gene Ontology. Autophagy 2018, 14, 419–436. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Chang, K.-K.; Liu, L.-B.; Jin, L.-P.; Zhang, B.; Mei, J.; Li, H.; Wei, C.-Y.; Zhou, W.-J.; Zhu, X.-Y.; Shao, J.; et al. IL-27 triggers IL-10 production in Th17 cells via a c-Maf/RORγt/Blimp-1 signal to promote the progression of endometriosis. Cell Death Dis. 2017, 8, e2666. [Google Scholar] [CrossRef]
- Hao, F.; Tang, L.-C.; Sun, J.-X.; Li, W.-X.; Zhao, Y.; Xu, X.-H.; Jin, L.-P. Decreased nitric oxide content mediated by asymmetrical dimethylarginine and protein l-arginine methyltransferase 3 in macrophages induces trophoblast apoptosis: A potential cause of recurrent miscarriage. Hum. Reprod. 2021, 36, 3049–3061. [Google Scholar] [CrossRef]



| ID | Category | GO Name | p. Value | Count | Gene ID |
|---|---|---|---|---|---|
| GO:0120254 | BP | olefinic compound metabolic process | 0.00014411 | 3 | DHRS3/RDH10/CYP2J2 |
| GO:0048599 | BP | oocyte development | 0.00043323 | 2 | PLD6/ANG |
| GO:0009994 | BP | oocyte differentiation | 0.00051382 | 2 | PLD6/ANG |
| GO:0042572 | BP | retinol metabolic process | 0.00053502 | 2 | DHRS3/RDH10 |
| GO:0005811 | CC | lipid droplet | 6.13 × 105 | 3 | DHRS3/RDH10/CES1 |
| GO:0052650 | MF | NADP-retinol dehydrogenase activity | 4.83 × 105 | 2 | DHRS3/RDH10 |
| GO:0004745 | MF | NAD-retinol dehydrogenase activity | 0.00010559 | 2 | DHRS3/RDH10 |
| GO:0008106 | MF | alcohol dehydrogenase (NADP+) activity | 0.00014244 | 2 | DHRS3/RDH10 |
| GO:0004033 | MF | aldo-keto reductase (NADP) activity | 0.00024952 | 2 | DHRS3/RDH10 |
| GO:0004521 | MF | endoribonuclease activity | 0.00141489 | 2 | PLD6/ANG |
| GO:0140098 | MF | catalytic activity, acting on RNA | 0.00347205 | 3 | PLD6/RAD54B/ANG |
| GO:0004540 | MF | ribonuclease activity | 0.00393461 | 2 | PLD6/ANG |
| GO:0016616 | MF | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 0.00448163 | 2 | DHRS3/RDH10 |
| GO:0004519 | MF | endonuclease activity | 0.00484046 | 2 | PLD6/ANG |
| GO:0016298 | MF | lipase activity | 0.00484046 | 2 | PLD6/CES1 |
| GO:0016614 | MF | oxidoreductase activity, acting on CH-OH group of donors | 0.00536449 | 2 | DHRS3/RDH10 |
| GO:0015271 | MF | outward rectifier potassium channel activity | 0.01056786 | 1 | KCNK2 |
| GO:0004518 | MF | nuclease activity | 0.01238517 | 2 | PLD6/ANG |
| GO:0008191 | MF | metalloendopeptidase inhibitor activity | 0.01299175 | 1 | RARRES1 |
| GO:0022841 | MF | potassium ion leak channel activity | 0.01299175 | 1 | KCNK2 |
| GO:0008392 | MF | arachidonic acid epoxygenase activity | 0.01379848 | 1 | CYP2J2 |
| GO:0042625 | MF | ATPase-coupled ion transmembrane transporter activity | 0.01379848 | 1 | ATP6V0E2 |
| GO:0044769 | MF | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism | 0.01379848 | 1 | ATP6V0E2 |
| GO:0046961 | MF | proton-transporting ATPase activity, rotational mechanism | 0.01379848 | 1 | ATP6V0E2 |
| GO:0022840 | MF | leak channel activity | 0.0154101 | 1 | KCNK2 |
| GO:0022842 | MF | narrow pore channel activity | 0.0154101 | 1 | KCNK2 |
| GO:0009678 | MF | pyrophosphate hydrolysis-driven proton transmembrane transporter activity | 0.01621498 | 1 | ATP6V0E2 |
| GO:0008391 | MF | arachidonic acid monooxygenase activity | 0.01701926 | 1 | CYP2J2 |
| GO:0016866 | MF | intramolecular transferase activity | 0.02183201 | 1 | CYP2J2 |
| Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) | |
|---|---|---|
| YTHDF1 | ACCTGTCCAGCTATTACCCG | TGGTGAGGTATGGAATCGGAG |
| METTL3 | CATTGCCCACTGATGCTGTG | AGGCTTTCTACCCCATCTTGA |
| ALKBH5 | AGTTCCAGTTCAAGCCTATTCG | TGAGCACAGTCACGCTTCC |
| METTL14 | GAACACAGAGCTTAAATCCCCA | TGTCAGCTAAACCTACATCCCTG |
| YTHDF3 | GCTATCCACCTAGTTCTCTTGGG | ATGCCAGGCACCTTACTCAAA |
| YTHDF2 | CCTTAGGTGGAGCCATGATTG | TCTGTGCTACCCAACTTCAGT |
| RBMX | CTTCAGGACCAGTTCGCAGTA | TCACGACCACTTGAGTAGAGAT |
| LRPPRC | GCTCATAGGATATGGGACACACT | CCAGGAAATCAGTTGGTGAGAAT |
| RBM15B | TGATTGGTCGCAACCCCATTA | CAGTGACGTGTTAGGTCCCAG |
| FMR1 | TATGCAGCATGTGATGCAACT | TTGTGGCAGGTTTGTTGGGAT |
| HNRNPC | TCCTCCTCCTATTGCTCGGG | GTGTTTCCTGATACACGCTGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Sun, J.; Jin, L. The N6-Methyladenosine Regulator ALKBH5 Mediated Stromal Cell–Macrophage Interaction via VEGF Signaling to Promote Recurrent Spontaneous Abortion: A Bioinformatic and In Vitro Study. Int. J. Mol. Sci. 2022, 23, 15819. https://doi.org/10.3390/ijms232415819
Zhao Y, Sun J, Jin L. The N6-Methyladenosine Regulator ALKBH5 Mediated Stromal Cell–Macrophage Interaction via VEGF Signaling to Promote Recurrent Spontaneous Abortion: A Bioinformatic and In Vitro Study. International Journal of Molecular Sciences. 2022; 23(24):15819. https://doi.org/10.3390/ijms232415819
Chicago/Turabian StyleZhao, Yongbo, Jiani Sun, and Liping Jin. 2022. "The N6-Methyladenosine Regulator ALKBH5 Mediated Stromal Cell–Macrophage Interaction via VEGF Signaling to Promote Recurrent Spontaneous Abortion: A Bioinformatic and In Vitro Study" International Journal of Molecular Sciences 23, no. 24: 15819. https://doi.org/10.3390/ijms232415819
APA StyleZhao, Y., Sun, J., & Jin, L. (2022). The N6-Methyladenosine Regulator ALKBH5 Mediated Stromal Cell–Macrophage Interaction via VEGF Signaling to Promote Recurrent Spontaneous Abortion: A Bioinformatic and In Vitro Study. International Journal of Molecular Sciences, 23(24), 15819. https://doi.org/10.3390/ijms232415819





