SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing
Abstract
:1. Introduction
2. Results
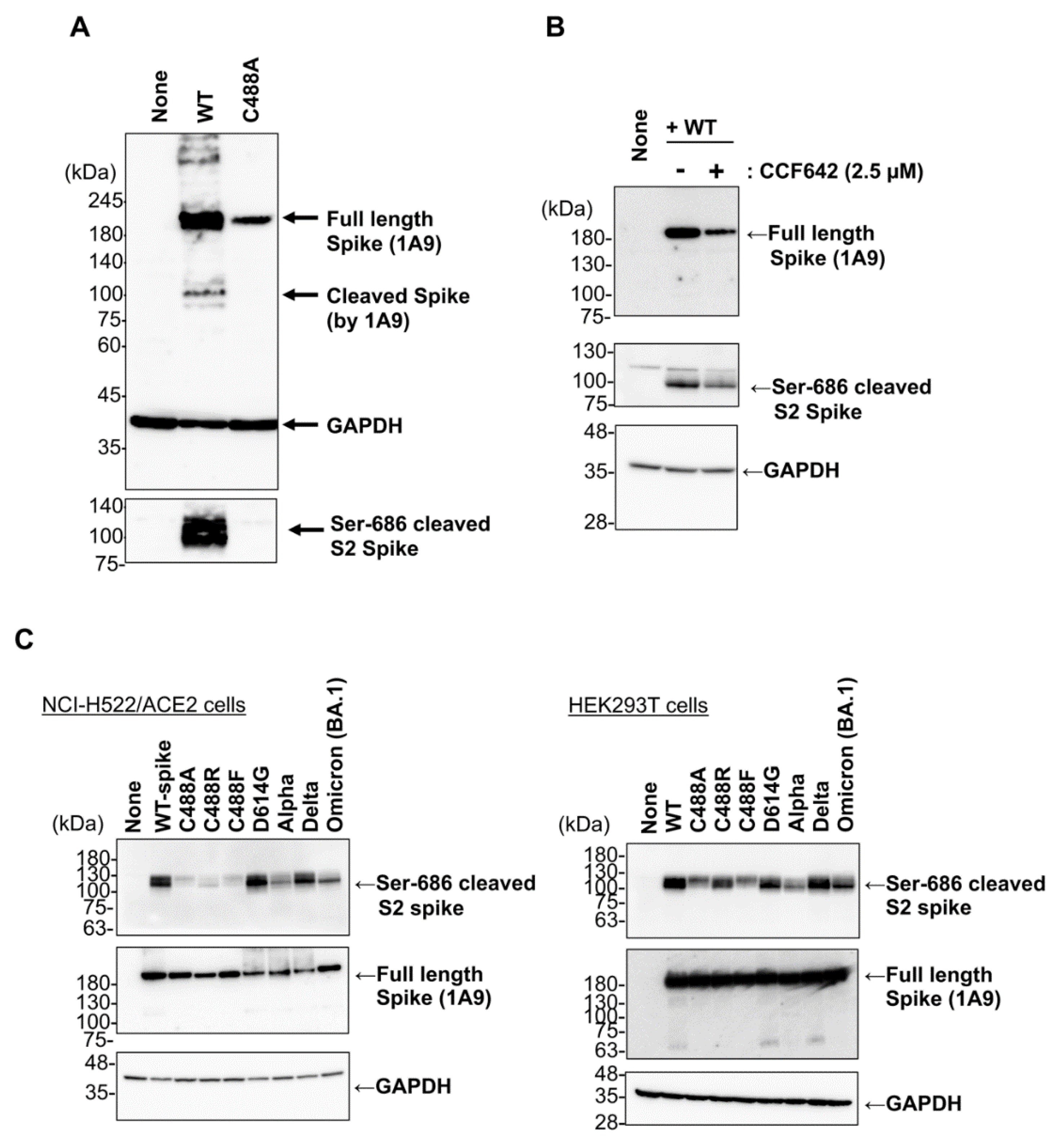
2.1. C488 Is Important for the Fusogenic Activity and Cell-Surface Expression of the Spike Protein
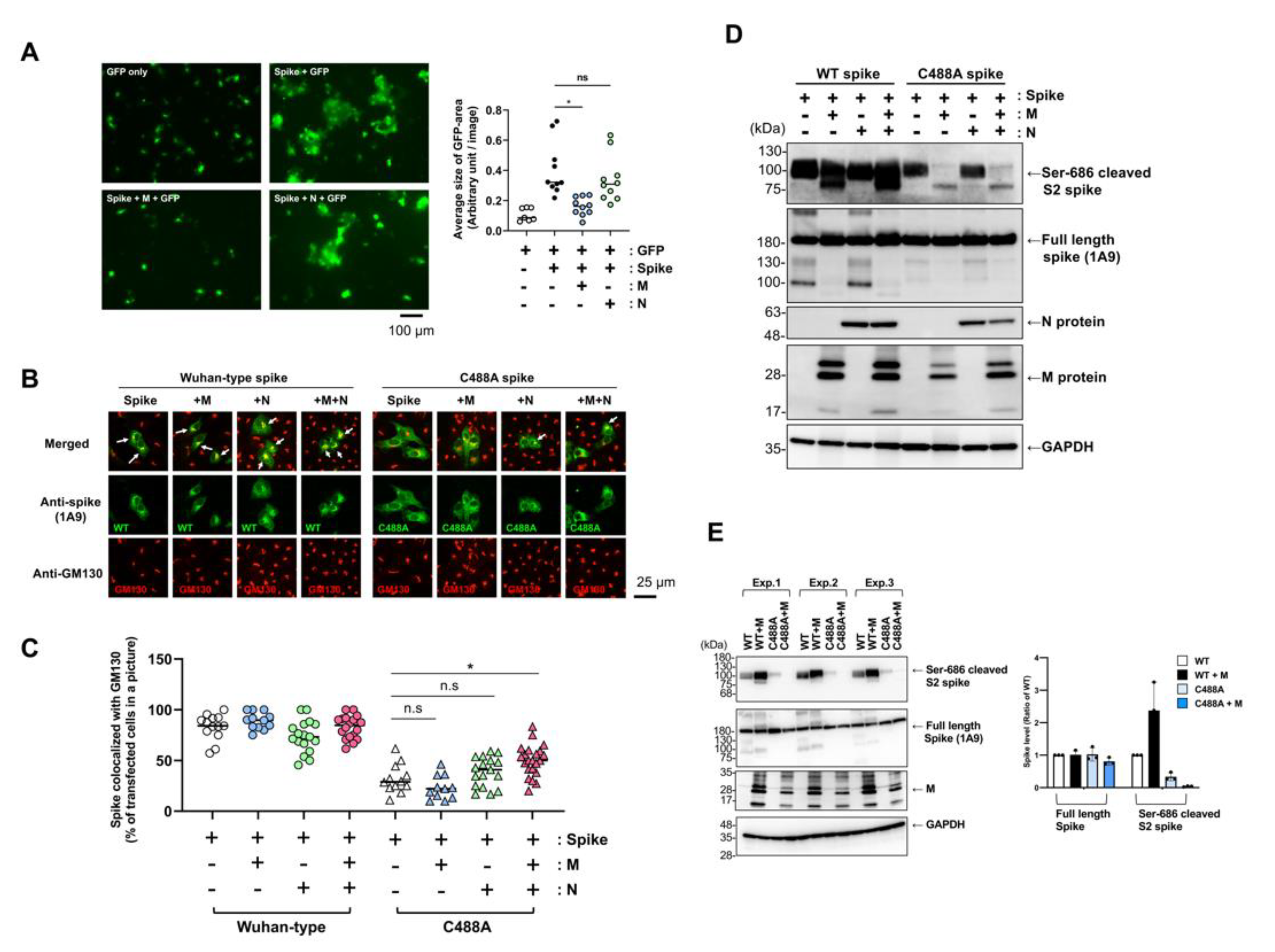
2.2. C488 Is Involved in Golgi-Targeting by the Spike Protein
2.3. C488 Mutation Suppresses S1/S2 Processing of the Spike Protein
2.4. SARS-CoV-2 Membrane Protein Does Not Compensate C488 Mutation of Spike Protein
2.5. Brefeldin A Inhibits SARS-CoV-2 Spike Protein Processing
3. Discussion
4. Materials and Methods
4.1. Cells, Plasmids, and Reagents
4.2. Transfection
4.3. Immunofluorescence Microscopic Analysis and Antibodies
4.4. Hybridoma Production of mAbs against SARS-CoV-2
4.5. Immunoblotting
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cattin-Ortolá, J.; Welch, L.G.; Maslen, S.L.; Papa, G.; James, L.C.; Munro, S. Sequences in the Cytoplasmic Tail of SARS-CoV-2 Spike Facilitate Expression at the Cell Surface and Syncytia Formation. Nat. Commun. 2021, 12, 5333. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Zhu, N.; Wang, W.; Liu, Z.; Liang, C.; Wang, W.; Ye, F.; Huang, B.; Zhao, L.; Wang, H.; Zhou, W.; et al. Morphogenesis and Cytopathic Effect of SARS-CoV-2 Infection in Human Airway Epithelial Cells. Nat. Commun. 2020, 11, 3910. [Google Scholar] [CrossRef]
- Buchrieser, J.; Dufloo, J.; Hubert, M.; Monel, B.; Planas, D.; Rajah, M.M.; Planchais, C.; Porrot, F.; Guivel-Benhassine, F.; van der Werf, S.; et al. Syncytia Formation by SARS-CoV-2-infected Cells. EMBO J. 2020, 39, e106267. [Google Scholar] [CrossRef]
- Saraste, J.; Prydz, K. Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells 2021, 10, 503. [Google Scholar] [CrossRef]
- McBride, C.E.; Li, J.; Machamer, C.E. The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein. J. Virol. 2007, 81, 2418–2428. [Google Scholar] [CrossRef] [Green Version]
- Jennings, B.C.; Kornfeld, S.; Doray, B. A Weak COPI Binding Motif in the Cytoplasmic Tail of SARS-CoV-2 Spike Glycoprotein Is Necessary for Its Cleavage, Glycosylation, and Localization. FEBS Lett. 2021, 595, 1758–1767. [Google Scholar] [CrossRef]
- Murae, M.; Shimizu, Y.; Yamamoto, Y.; Kobayashi, A.; Houri, M.; Inoue, T.; Irie, T.; Gemba, R.; Kondo, Y.; Nakano, Y.; et al. The Function of SARS-CoV-2 Spike Protein Is Impaired by Disulfide-Bond Disruption with Mutation at Cysteine-488 and by Thiol-Reactive N-Acetyl-Cysteine and Glutathione. Biochem. Biophys. Res. Commun. 2022, 597, 30–36. [Google Scholar] [CrossRef]
- Hackstadt, T.; Chiramel, A.I.; Hoyt, F.H.; Williamson, B.N.; Dooley, C.A.; Beare, P.A.; de Wit, E.; Best, S.M.; Fischer, E.R. Disruption of the Golgi Apparatus and Contribution of the Endoplasmic Reticulum to the SARS-CoV-2 Replication Complex. Viruses 2021, 13, 1798. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G. Furin at the Cutting Edge: From Protein Traffic to Embryogenesis and Disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, W.; Stroh, A.; Berghöfer, S.; Seiler, J.; Vey, M.; Kruse, M.L.; Kern, H.F.; Klenk, H.D.; Garten, W. Two Independent Targeting Signals in the Cytoplasmic Domain Determine Trans-Golgi Network Localization and Endosomal Trafficking of the Proprotein Convertase Furin. EMBO J. 1995, 14, 2424–2435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Go, E.P.; Ding, H.; Anang, S.; Kappes, J.C.; Desaire, H.; Sodroski, J.G. Analysis of Glycosylation and Disulfide Bonding of Wild-Type SARS-CoV-2 Spike Glycoprotein. J. Virol. 2022, 96, e01626-2. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.-L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 Envelope and Membrane Proteins Modulate Maturation and Retention of the Spike Protein, Allowing Assembly of Virus-like Particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef]
- Scherer, K.M.; Mascheroni, L.; Carnell, G.W.; Wunderlich, L.C.S.; Makarchuk, S.; Brockhoff, M.; Mela, I.; Fernandez-Villegas, A.; Barysevich, M.; Stewart, H.; et al. SARS-CoV-2 Nucleocapsid Protein Adheres to Replication Organelles before Viral Assembly at the Golgi/ERGIC and Lysosome-Mediated Egress. Sci. Adv. 2022, 8, eabl4895. [Google Scholar] [CrossRef]
- Misumi, A.; Misumi, Y.; Miki, K.; Takatsuki, A.; Tamura, G.; Ikehara, Y. Novel Blockade by Brefeldin A of Intracellular Transport of Secretory Proteins in Cultured Rat Hepatocytes. J. Biol. Chem. 1986, 261, 11398–11403. [Google Scholar] [CrossRef]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Manček-Keber, M.; Hafner-Bratkovič, I.; Lainšček, D.; Benčina, M.; Govednik, T.; Orehek, S.; Plaper, T.; Jazbec, V.; Bergant, V.; Grass, V.; et al. Disruption of Disulfides within RBD of SARS-CoV-2 Spike Protein Prevents Fusion and Represents a Target for Viral Entry Inhibition by Registered Drugs. FASEB J. 2021, 35, e21651. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.-M.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. A Study on the Nature of SARS-CoV-2 Using the Shell Disorder Models: Reproducibility, Evolution, Spread, and Attenuation. Biomolecules 2022, 12, 1353. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; van Zyl, M.; Fielding, B. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tien, C.-F.; Tsai, W.-T.; Chen, C.H.; Chou, H.-J.; Zhang, M.M.; Lin, J.-J.; Lin, E.-J.; Dai, S.-S.; Ping, Y.-H.; Yu, C.-Y.; et al. Glycosylation and S-Palmitoylation Regulate SARS-CoV-2 Spike Protein Intracellular Trafficking. iScience 2022, 25, 104709. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Fernandes, M.H.V.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional Evaluation of the P681H Mutation on the Proteolytic Activation of the SARS-CoV-2 Variant B.1.1.7 (Alpha) Spike. iScience 2022, 25, 103589. [Google Scholar] [CrossRef] [PubMed]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; de Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S.; et al. SARS-CoV-2 Omicron Is an Immune Escape Variant with an Altered Cell Entry Pathway. Nat. Microbiol. 2022, 7, 1161–1179. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Datir, R.; Choi, J.; Bradley, J.R.; Smith, K.G.C.; Lee, J.H.; Gupta, R.K.; Baker, S.; Dougan, G.; Hess, C.; et al. SARS-CoV-2 Spike N-Terminal Domain Modulates TMPRSS2-Dependent Viral Entry and Fusogenicity. Cell Rep. 2022, 40, 111220. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Jeremiah, S.S.; Funabashi, R.; Miyakawa, K.; Morita, T.; Mihana, Y.; Kato, H.; Ryo, A. Characterization and Utilization of Disulfide-Bonded SARS-CoV-2 Receptor Binding Domain of Spike Protein Synthesized by Wheat Germ Cell-Free Production System. Viruses 2022, 14, 1461. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]



| Plasmid Names | Remarks, Protein Sequence and Mutation |
|---|---|
| pEGFP-C1 | BD Biotech Clontech |
| pcDNA3.1–spike/WT | Wuhan Hu-1 spike, YP_009724390.1 |
| pcDNA3.1–M | Wuhan Hu M, YP_009724393 |
| pcDNA3.1 SARS-CoV-2 N | pcDNA3.1 SARS-CoV-2 N was a gift from Jeremy Luban (Addgene plasmid # 158079; http://n2t.net/addgene:158079, accessed on 15 November 2022; RRID:Addgene_158079) [29] |
| pcDNA3.1–spike/D614G | D614G |
| pcDNA3.1–spike/C488A | C488A |
| pcDNA3.1–spike/C488R | C488R |
| pcDNA3.1-spike/C488F | C488F |
| pcDNA3.1–spike/Alpha | H69/V70-del, Y144-del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| pcDNA3.1–spike/Delta | L5F, T19R, E156G, F157/R158-del, L452R, T478K, D614G, P681R, D950N |
| pcDNA3.1–spike/Omicron (BA.1) | A67V, H69/V70-del, T95I, G142D, del143–145, ins214EPE, NL211–212I, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| pcDNA3.1-hACE2-DYK | GenScript, Cat No:OHu20260D |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, Y.; Inoue, T.; Inoue, M.; Murae, M.; Fukasawa, M.; Kaneko, M.K.; Kato, Y.; Noguchi, K. SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing. Int. J. Mol. Sci. 2022, 23, 15834. https://doi.org/10.3390/ijms232415834
Yamamoto Y, Inoue T, Inoue M, Murae M, Fukasawa M, Kaneko MK, Kato Y, Noguchi K. SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing. International Journal of Molecular Sciences. 2022; 23(24):15834. https://doi.org/10.3390/ijms232415834
Chicago/Turabian StyleYamamoto, Yuichiro, Tetsuya Inoue, Miyu Inoue, Mana Murae, Masayoshi Fukasawa, Mika K. Kaneko, Yukinari Kato, and Kohji Noguchi. 2022. "SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing" International Journal of Molecular Sciences 23, no. 24: 15834. https://doi.org/10.3390/ijms232415834
APA StyleYamamoto, Y., Inoue, T., Inoue, M., Murae, M., Fukasawa, M., Kaneko, M. K., Kato, Y., & Noguchi, K. (2022). SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing. International Journal of Molecular Sciences, 23(24), 15834. https://doi.org/10.3390/ijms232415834







