TRH Regulates the Synthesis and Secretion of Prolactin in Rats with Adenohypophysis through the Differential Expression of miR-126a-5p
Abstract
1. Introduction
2. Results
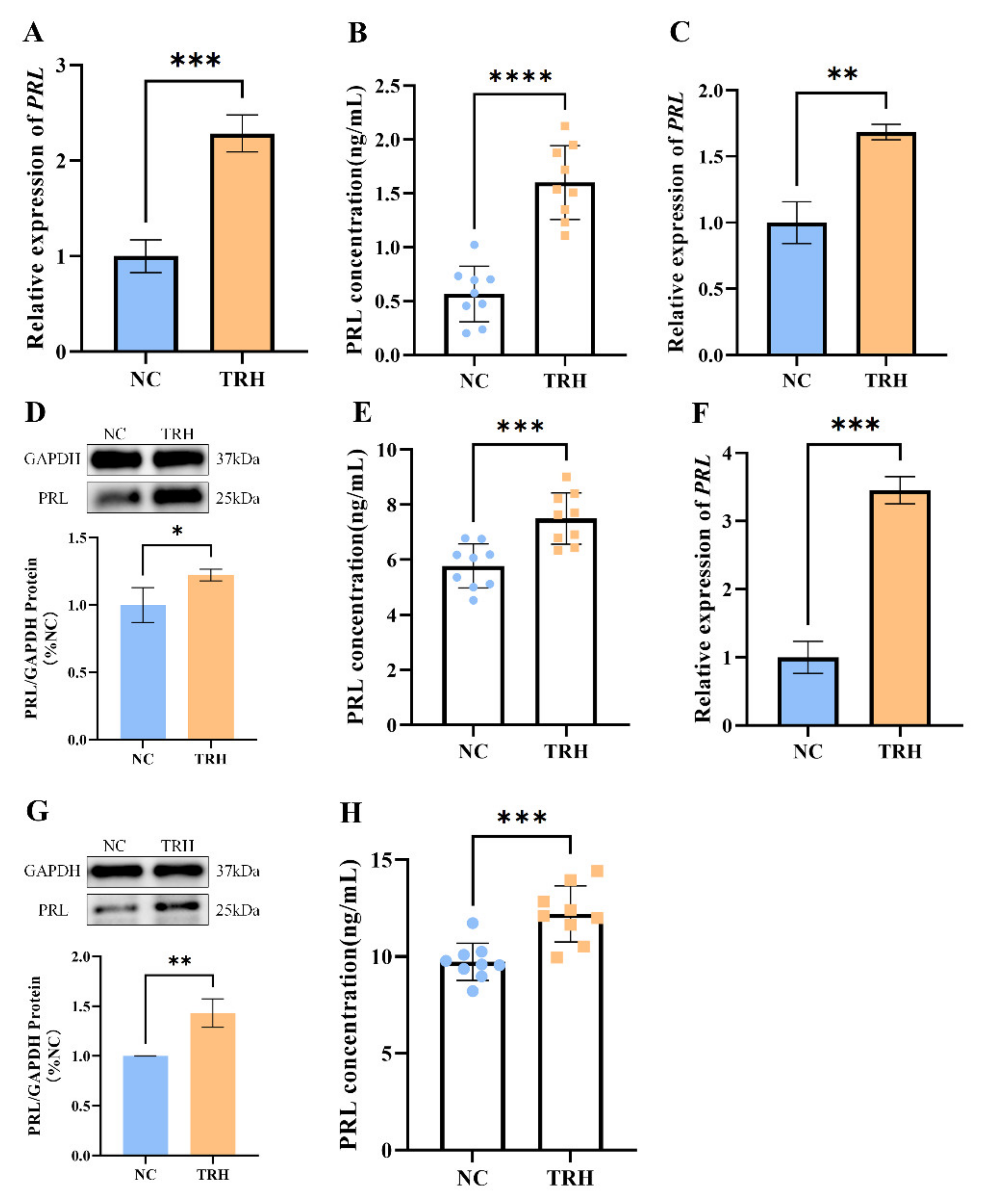
2.1. TRH Treatment Can Promote the Synthesis and Secretion of PRL in Rats
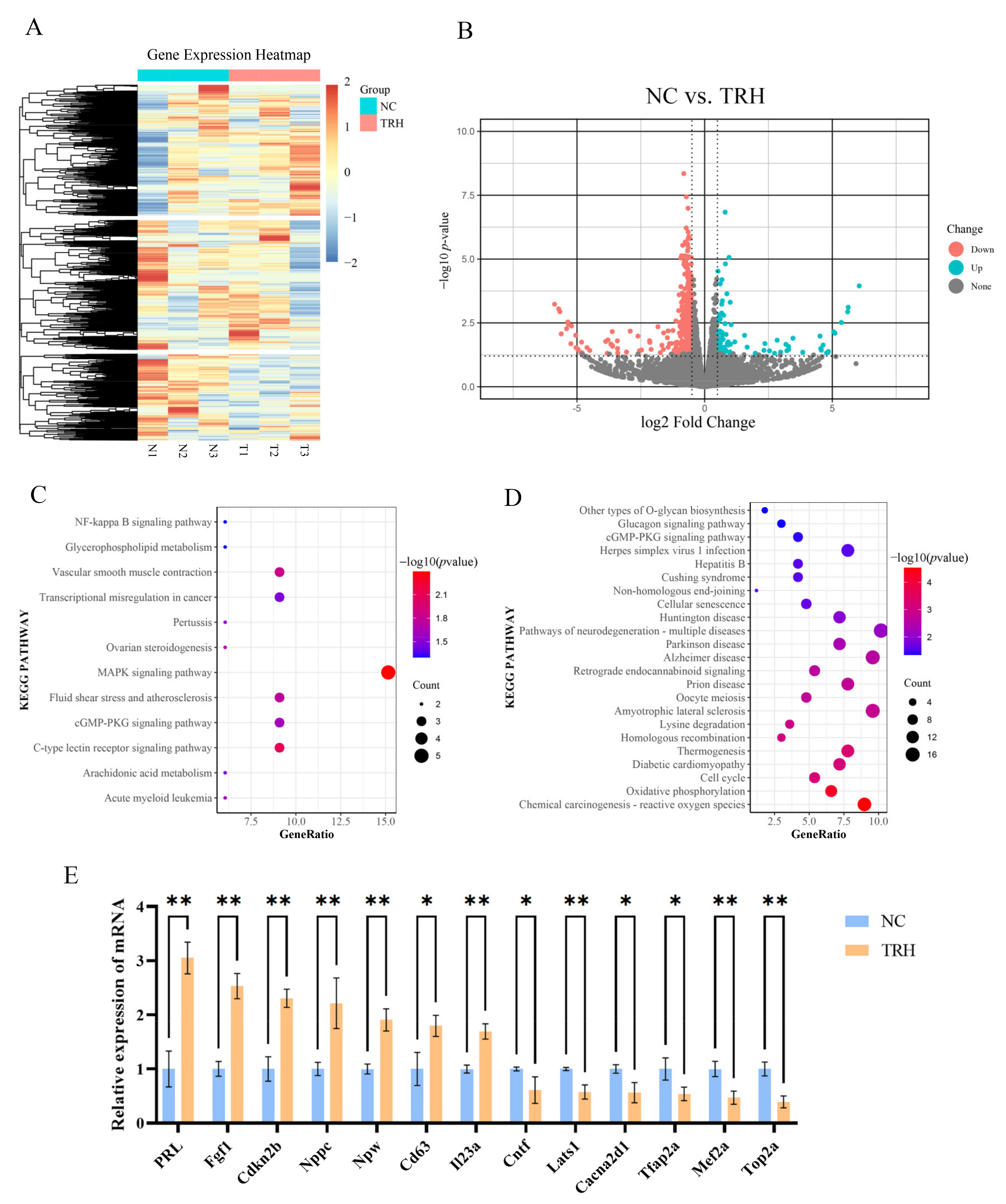
2.2. Analysis of Differentially Expressed Genes before and after TRH Treatment
2.3. miR-126a-5p Targets the 3′UTR of PRL mRNA
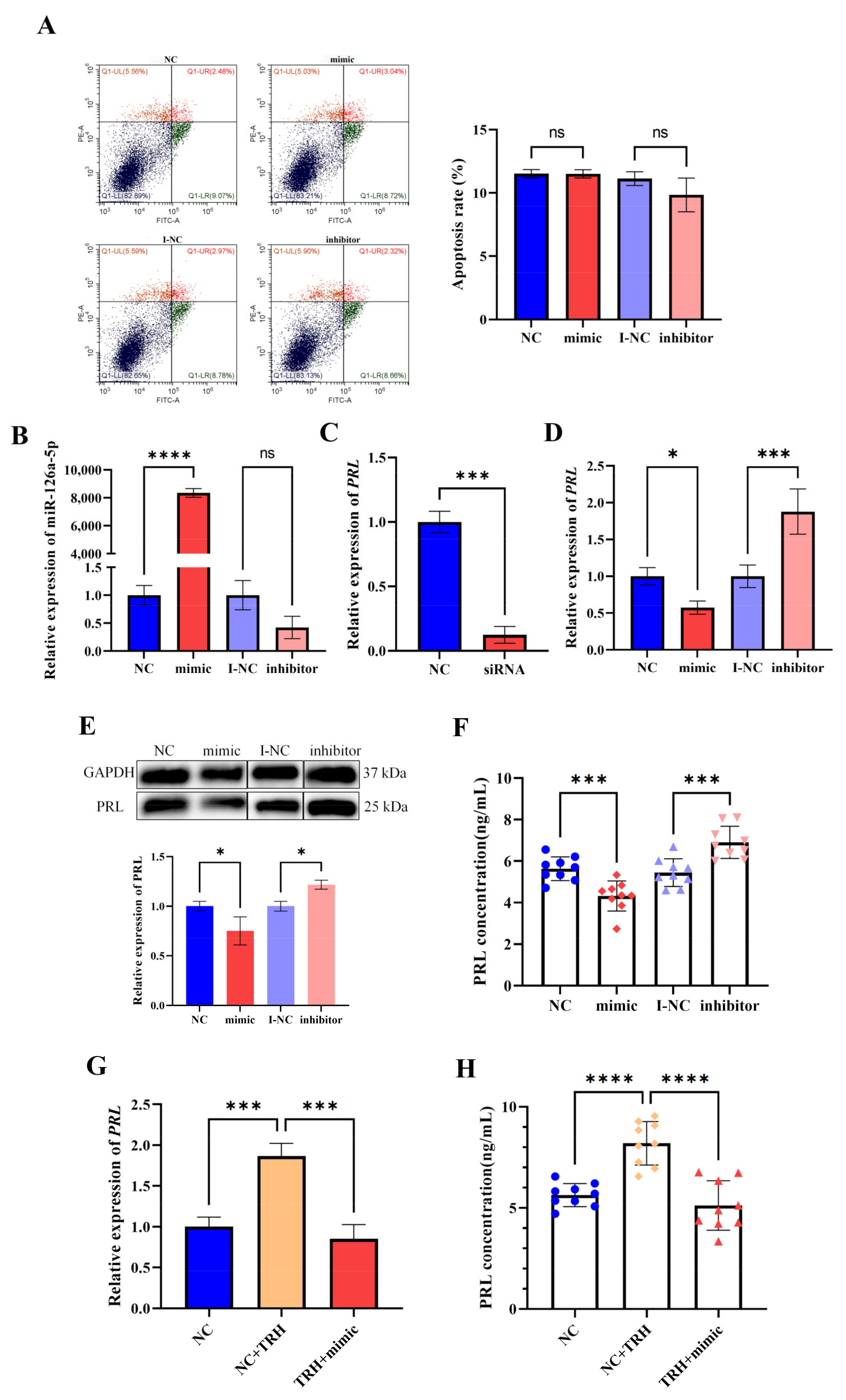
2.4. miR-126a-5p Regulates the Level of PRL mRNA Expression and PRL Secretion in GH3 Cells
2.5. miR-126a-5p Regulates the Level of PRL mRNA Expression and PRL Secretion in Rat Pituitary Cells
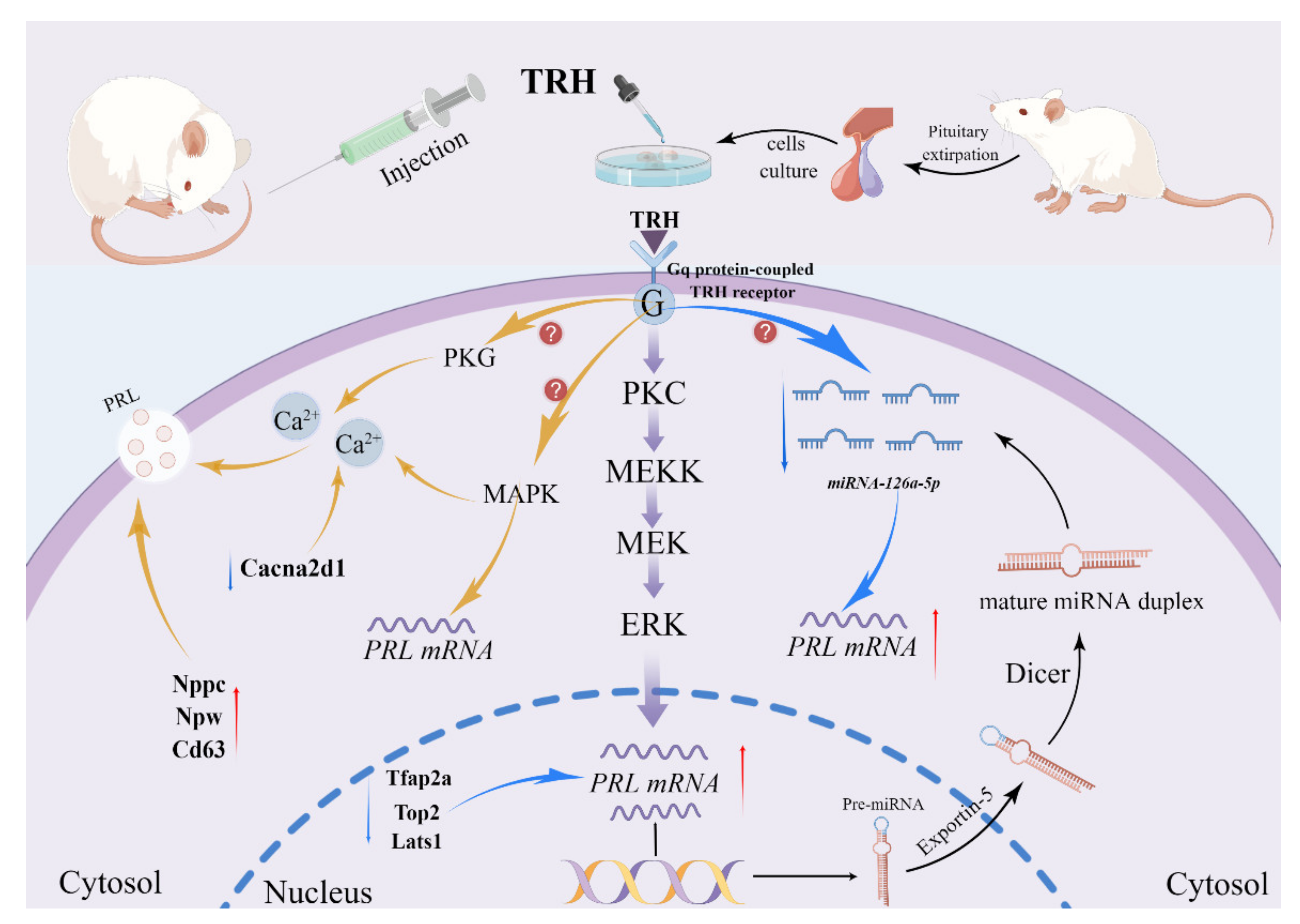
3. Discussion
4. Materials and Methods
4.1. Animals and Ethics
4.2. Cell Culture
4.3. Rat TRH Treatment
4.4. TRH Treatment of Cells
4.5. RNA-Seq
4.6. Data Evaluation and Quality Control
4.7. RNA-Seq Evaluation
4.8. Gene Structure Analysis
4.9. Expression Level Analysis
4.10. Analysis of Differential Expression
4.11. Gene Enrichment Analysis
4.12. Prediction of Potential PRL Gene Targets
4.13. Cell Transfection
4.14. RNA Extraction and Real-Time Quantitative PCR
4.15. Dual Luciferase Reporter Gene Assay
4.16. Protein Blotting Analysis
4.17. Enzyme-Linked Immunosorbent Assay
4.18. Apoptosis Analysis
4.19. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Freeman, M.E.; Kanyicska, B.; Lerant, A.; Nagy, G. Prolactin: Structure, Function, and Regulation of Secretion. Physiol. Rev. 2000, 80, 1523–1631. [Google Scholar] [CrossRef] [PubMed]
- Bole-Feysot, C.; Goffin, V.; Edery, M.; Binart, N.; Kelly, P.A. Prolactin (PRL) and Its Receptor: Actions, Signal Transduction Pathways and Phenotypes Observed in PRL Receptor Knockout Mice. Endocr. Rev. 1998, 19, 225–268. [Google Scholar] [CrossRef] [PubMed]
- Morishige, W.K.; Rothchild, I. Temporal Aspects of the Regulation of Corpus Luteum Function by Luteinizing Hormone, Prolactin and Placental Luteotrophin During the First Half of Pregnancy in the Rat. Endocrinology 1974, 95, 260–274. [Google Scholar] [CrossRef] [PubMed]
- Borba, V.V.; Zandman-Goddard, G.; Shoenfeld, Y. Prolactin and Autoimmunity. Front. Immunol. 2018, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jonathan, N.; Hugo, E.R.; Brandebourg, T.D.; LaPensee, C.R. Focus on prolactin as a metabolic hormone. Trends Endocrinol. Metab. 2006, 17, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Lechan, R.M. Neuroendocrinology of Pituitary Hormone Regulation. Endocrinol. Metab. Clin. North Am. 1987, 16, 475–501. [Google Scholar] [CrossRef]
- Lamberts, S.W.; MacLeod, R.M. Regulation of prolactin secretion at the level of the lactotroph. Physiol. Rev. 1990, 70, 279–318. [Google Scholar] [CrossRef]
- Ben-Jonathan, N.; Hnasko, R. Dopamine as a Prolactin (PRL) Inhibitor. Endocr. Rev. 2001, 22, 724–763. [Google Scholar] [CrossRef]
- Burgus, R.; Dunn, T.F.; Desiderio, D.; Ward, D.N.; Vale, W.; Guillemin, R. Characterization of Ovine Hypothalamic Hypophysiotropic TSH-releasing Factor. Nature 1970, 226, 321–325. [Google Scholar] [CrossRef]
- Galas, L.; Raoult, E.; Tonon, M.-C.; Okada, R.; Jenks, B.G.; Castaño, J.P.; Kikuyama, S.; Malagon, M.; Roubos, E.W.; Vaudry, H. TRH acts as a multifunctional hypophysiotropic factor in vertebrates. Gen. Comp. Endocrinol. 2009, 164, 40–50. [Google Scholar] [CrossRef]
- Kanasaki, H.; Oride, A.; Mijiddorj, T.; Kyo, S. Role of thyrotropin-releasing hormone in prolactin-producing cell models. Neuropeptides 2015, 54, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Winitz, S.; Russell, M.; Qian, N.X.; Gardner, A.; Dwyer, L.; Johnson, G.L. Involvement of Ras and Raf in the Gi-coupled acetylcholine muscarinic m2 receptor activation of mitogen-activated protein (MAP) kinase kinase and MAP kinase. J. Biol. Chem. 1993, 268, 19196–19199. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.J.; Gorelick, F.S.; Dannies, P.S. Calcium/calmodulin-dependent protein kinase-II activation in rat pituitary cells in the presence of thyrotropin-releasing hormone and dopamine. Endocrinology 1994, 134, 2245–2250. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, J.; Ju, B.-G.; Rosenfeld, M.G. Signaling and epigenetic regulation of pituitary development. Curr. Opin. Cell Biol. 2007, 19, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jonathan, N.; Chen, S.; Dunckley, J.A.; LaPensee, C.; Kansra, S. Estrogen Receptor-α Mediates the Epidermal Growth Factor-Stimulated Prolactin Expression and Release in Lactotrophs. Endocrinology 2009, 150, 795–802. [Google Scholar] [CrossRef]
- Stallings, C.E.; Kapali, J.; Evans, B.W.; McGee, S.R.; Ellsworth, B.S. FOXO Transcription Factors Are Required for Normal Somatotrope Function and Growth. Endocrinology 2022, 2, 163. [Google Scholar] [CrossRef]
- Sosa, L.D.V.; Gutiérrez, S.; Petiti, J.P.; Palmeri, C.M.; Mascanfroni, I.D.; Soaje, M.; De Paul, A.L.; Torres, A.I. 17β-Estradiol modulates the prolactin secretion induced by TRH through membrane estrogen receptors via PI3K/Akt in female rat anterior pituitary cell culture. Am. J. Physiol. Metab. 2012, 302, E1189–E1197. [Google Scholar] [CrossRef]
- Kang, C.W.; Han, Y.E.; Lee, M.K.; Cho, Y.H.; Kang, N.; Koo, J.; Ku, C.R.; Lee, E.J. Olfactory marker protein regulates prolactin secretion and production by modulating Ca2+ and TRH signaling in lactotrophs. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Tong, Y.; Zhou, J.; Mizutani, J.; Fukuoka, H.; Ren, S.-G.; Gutierrez-Hartmann, A.; Koeffler, H.P.; Melmed, S. CEBPD Suppresses Prolactin Expression and Prolactinoma Cell Proliferation. Mol. Endocrinol. 2011, 25, 1880–1891. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Liu, H.; Sun, H.; Xiang, J.; Wang, X.; Jiang, C.; Ma, L.; Cao, Z. MiR-361-3p/Nfat5 Signaling Axis Controls Cementoblast Differentiation. J. Dent. Res. 2019, 98, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Q.; Hu, Y.; Xu, L.; Jiang, Y.; Zhang, C.; Ding, L.; Jiang, R.; Sun, J.; Sun, H.; et al. miR-181a increases FoxO1 acetylation and promotes granulosa cell apoptosis via SIRT1 downregulation. Cell Death Dis. 2017, 8, e3088. [Google Scholar] [CrossRef] [PubMed]
- Komoll, R.-M.; Hu, Q.; Olarewaju, O.; von Döhlen, L.; Yuan, Q.; Xie, Y.; Tsay, H.-C.; Daon, J.; Qin, R.; Manns, M.P.; et al. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J. Hepatol. 2021, 74, 122–134. [Google Scholar] [CrossRef]
- Ye, X.; Lu, Q.; Yang, A.; Rao, J.; Xie, W.; He, C.; Wang, W.; Li, H.; Zhang, Z. MiR-206 regulates the Th17/Treg ratio during osteoarthritis. Mol. Med. 2021, 27, 64. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Zhang, D.; Xu, G.; Zhang, J.; Cui, S. miR-7 mediates the signaling pathway of NE affecting FSH and LH synthesis in pig pituitary. J. Endocrinol. 2020, 244, 459–471. [Google Scholar] [CrossRef]
- Ye, R.-S.; Li, M.; Li, C.-Y.; Qi, Q.-E.; Chen, T.; Cheng, X.; Wang, S.-B.; Shu, G.; Wang, L.-N.; Zhu, X.-T.; et al. miR-361-3p regulates FSH by targeting FSHB in a porcine anterior pituitary cell model. Reproduction 2017, 153, 341–349. [Google Scholar] [CrossRef]
- Yu, Z.-W.; Gao, W.; Feng, X.-Y.; Zhang, J.-Y.; Guo, H.-X.; Wang, C.-J.; Chen, J.; Hu, J.-P.; Ren, W.-Z.; Yuan, B. Roles of differential expression of miR-543-5p in GH regulation in rat anterior pituitary cells and GH3 cells. PLoS ONE 2019, 14, e0222340. [Google Scholar] [CrossRef]
- Xiong, S.; Tian, J.; Ge, S.; Li, Z.; Long, Z.; Guo, W.; Huang, P.; He, Y.; Xiao, T.; Gui, J.-F.; et al. The miRNA-200 cluster on chromosome 23 is required for oocyte maturation and ovulation in zebrafish. Biol. Reprod. 2020, 103, 769–778. [Google Scholar] [CrossRef]
- Gangisetty, O.; Jabbar, S.; Wynne, O.; Sarkar, D.K. MicroRNA-9 regulates fetal alcohol-induced changes in D2 receptor to promote prolactin production. J. Endocrinol. 2017, 235, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, T.; Xiong, J.; Hu, B.; Luo, J.; Xi, Q.; Jiang, Q.; Sun, J.; Zhang, Y. MiR-130a-3p Inhibits PRL Expression and Is Associated with Heat Stress-Induced PRL Reduction. Front. Endocrinol. 2020, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, J.; Zhang, D.; Liu, H.; Gou, K.; Cui, S. miR-375 acts as a novel factor modulating pituitary prolactin synthesis through Rasd1 and Esr1. J. Endocrinol. 2021, 250, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Boockfor, F.R.; Schwarz, L.K. Cultures of Gh3 Cells Contain Both Single and Dual Hormone Secretors. Endocrinology 1988, 122, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.M.G.; Barrett, J.F.; Bowers, C.Y.; Schally, A.V. Structure of porcine thyrotropin-releasing hormone. Biochemistry 1970, 9, 1103–1106. [Google Scholar] [CrossRef]
- Vale, W.; Blackwell, R.; Grant, G.; Guillemin, R. TRF and Thyroid Hormones on Prolactin Secretion by Rat Anterior Pituitary Cells In Vitro. Endocrinology 1973, 93, 26–33. [Google Scholar] [CrossRef]
- Brand, A.V.D.; Rubinstein, E.; Berg, M.V.D.; van Duursen, M. GH3 and RC-4BC cell lines are not suitable as in vitro models to study prolactin modulation and AHR responsiveness in rat pituitary. Mol. Cell. Endocrinol. 2019, 496, 110520. [Google Scholar] [CrossRef]
- Mirczuk, S.; Scudder, C.; Read, J.; Crossley, V.; Regan, J.; Richardson, K.; Simbi, B.; McArdle, C.; Church, D.; Fenn, J.; et al. Natriuretic Peptide Expression and Function in GH3 Somatolactotropes and Feline Somatotrope Pituitary Tumours. Int. J. Mol. Sci. 2021, 22, 1076. [Google Scholar] [CrossRef]
- Mallo, F.; Wilson, E.; Whorwood, C.B.; Singh, S.; Sheppard, M.C. Basic and acidic fibroblast growth factor increase prolactin mRNA in a dose-dependent and specific manner in GH3 cells. Mol. Cell. Endocrinol. 1995, 114, 117–125. [Google Scholar] [CrossRef]
- Shimomura, Y.; Harada, M.; Goto, M.; Sugo, T.; Matsumoto, Y.; Abe, M.; Watanabe, T.; Asami, T.; Kitada, C.; Mori, M.; et al. Identification of Neuropeptide W as the Endogenous Ligand for Orphan G-protein-coupled Receptors GPR7 and GPR8. J. Biol. Chem. 2002, 277, 35826–35832. [Google Scholar] [CrossRef]
- Pandey, S.; Tuma, Z.; Peroni, E.; Monasson, O.; Papini, A.; Dvorakova, M.C. Identification of NPB, NPW and Their Receptor in the Rat Heart. Int. J. Mol. Sci. 2020, 21, 7827. [Google Scholar] [CrossRef]
- Xekouki, P.; Lodge, E.; Matschke, J.; Santambrogio, A.; Apps, J.R.; Sharif, A.; Jacques, T.S.; Aylwin, S.; Prevot, V.; Li, R.; et al. Non-secreting pituitary tumours characterised by enhanced expression of YAP/TAZ. Endocr. Relat. Cancer 2019, 26, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Duran, P.; Sandoval, A.; González-Ramírez, R.; Zarco, N.; Felix, R. Regulation of the Ca2+channel α2δ-1 subunit expression by epidermal growth factor via the ERK/ELK-1 signaling pathway. Am. J. Physiol. Metab. 2020, 319, E232–E244. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Voltage-Gated Calcium Channels. Cold Spring Harb. Perspect. Biol. 2011, 3, a003947. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.-J.; Hong, S.-H.; Lee, J.-E.; Kim, S.C.; Yang, H.-S.; Yi, P.I.; Lee, S.-M.; An, B.-S. Pregnenolone sulfate regulates prolactin production in the rat pituitary. J. Endocrinol. 2016, 230, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Iglesias, A.E.; Jiang, Y.; Tomic, M.; Kretschmannova, K.; Andric, S.; Zemkova, H.; Stojilkovic, S.S. Dependence of Electrical Activity and Calcium Influx-Controlled Prolactin Release on Adenylyl Cyclase Signaling Pathway in Pituitary Lactotrophs. Mol. Endocrinol. 2006, 20, 2231–2246. [Google Scholar] [CrossRef]
- Cataldi, M.; Secondo, A.; D’Alessio, A.; Sarnacchiaro, F.; Colao, A.; Amoroso, S.; Di Renzo, G.; Annunziato, L. Involvement of phosphodiesterase-cGMP-PKG pathway in intracellular Ca2+ oscillations in pituitary GH3 cells. Biochim. Biophys. Acta 1999, 1449, 186–193. [Google Scholar] [CrossRef][Green Version]
- Camoratto, A.M.; Grandison, L. Evidence Supporting a Correlation between Arachidonic Acid Release and Prolactin Secretion from GH 3 Cells*. Endocrinology 1985, 116, 1506–1513. [Google Scholar] [CrossRef]
- Canonico, P.L.; Cronin, M.J.; Sortino, M.A.; Speciale, C.; Scapagnini, U.; MacLeod, R.M. Phospholipid Metabolism and Prolactin Secretion in vitro. Horm. Res. 1985, 22, 164–171. [Google Scholar] [CrossRef]
- Geymayer, S.; Doppler, W. Activation of NF-κB p50/p65 is regulated in the developing mammary gland and inhibits STAT5-mediated β-casein gene expression. FASEB J. 2000, 14, 1159–1170. [Google Scholar] [CrossRef]
- Dudas, B.; Merchenthaler, I. Thyrotropin-releasing hormone axonal varicosities appear to innervate dopaminergic neurons in the human hypothalamus. Anat. Embryol. 2020, 225, 2193–2201. [Google Scholar] [CrossRef]
- Myers, L.S.; Steele, M.K. The Brain Renin-Angiotensin System and Prolactin Secretion in the Male Rat*. Endocrinology 1991, 129, 1744–1748. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Iglesias, A.E.; Murano, T.; Li, S.; Tomic, M.; Stojilkovic, S.S. Dopamine Inhibits Basal Prolactin Release in Pituitary Lactotrophs through Pertussis Toxin-Sensitive and -Insensitive Signaling Pathways. Endocrinology 2008, 149, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Q.; Wang, W.-H.; Chen, C.-Z.; Guo, H.-X.; Jiang, H.; Yuan, B.; Zhang, J.-B. Regulation of FSH Synthesis by Differentially Expressed miR-488 in Anterior Adenohypophyseal Cells. Animals 2021, 11, 3262. [Google Scholar] [CrossRef] [PubMed]
- Han, D.-X.; Xiao, Y.; Wang, C.-J.; Jiang, H.; Gao, Y.; Yuan, B.; Zhang, J.-B. Regulation of FSH expression by differentially expressed miR-186-5p in rat anterior adenohypophyseal cells. PLoS ONE 2018, 13, e0194300. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, W.; Pan, S.; Li, Y.; Wang, H.; Wang, B.; Khalil, R.A. MiR-126a-5p limits the formation of abdominal aortic aneurysm in mice and decreases ADAMTS-4 expression. J. Cell. Mol. Med. 2020, 24, 7896–7906. [Google Scholar] [CrossRef]
- Xu, Y.-P.; He, Q.; Shen, Z.; Shu, X.-L.; Wang, C.-H.; Zhu, J.-J.; Shi, L.-P.; Du, L.-Z. MiR-126a-5p is involved in the hypoxia-induced endothelial-to-mesenchymal transition of neonatal pulmonary hypertension. Hypertens. Res. 2017, 40, 552–561. [Google Scholar] [CrossRef]
- Yu, Z.; Fang, X.; Liu, W.; Sun, R.; Zhou, J.; Pu, Y.; Zhao, M.; Sun, D.; Xiang, Z.; Liu, P.; et al. Microglia Regulate Blood–Brain Barrier Integrity via MiR-126a-5p/MMP9 Axis during Inflammatory Demyelination. Adv. Sci. 2022, 9, e2105442. [Google Scholar] [CrossRef]
- Zhao, Q.; Sun, H.; Yin, L.; Wang, L. miR-126a-5p-Dbp and miR-31a-Crot/Mrpl4 interaction pairs crucial for the development of hypertension and stroke. Mol. Med. Rep. 2019, 20, 4151–4167. [Google Scholar] [CrossRef]
- Du, X.; Zhu, M.; Zhang, T.; Wang, C.; Tao, J.; Yang, S.; Zhu, Y.; Zhao, W. The Recombinant Eg.P29-Mediated miR-126a-5p Promotes the Differentiation of Mouse Naive CD4+ T Cells via DLK1-Mediated Notch1 Signal Pathway. Front. Immunol. 2022, 13, 773276. [Google Scholar] [CrossRef]
- Kuśnierz-Cabala, B.; Nowak, E.; Sporek, M.; Kowalik, A.; Kuźniewski, M.; Enguita, F.J.; Stępień, E. Serum levels of unique miR-551-5p and endothelial-specific miR-126a-5p allow discrimination of patients in the early phase of acute pancreatitis. Pancreatology 2015, 15, 344–351. [Google Scholar] [CrossRef]
- Xin, G.; Chen, R.; Zhang, X. Identification of key microRNAs, transcription factors and genes associated with congenital obstructive nephropathy in a mouse model of megabladder. Gene 2018, 650, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhou, F.; Yang, D.; Zhang, X.; Zeng, M.; Wan, L. MicroRNA-126a-5p Exerts Neuroprotective Effects on Ischemic Stroke via Targeting NADPH Oxidase 2. Neuropsychiatr. Dis. Treat. 2021, 17, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. Rna 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Han, D.-X.; Sun, X.-L.; Xu, M.-Q.; Chen, C.-Z.; Jiang, H.; Gao, Y.; Yuan, B.; Zhang, J.-B. Roles of differential expression of microRNA-21-3p and microRNA-433 in FSH regulation in rat anterior pituitary cells. Oncotarget 2017, 8, 36553–36565. [Google Scholar] [CrossRef] [PubMed]
- Helena, C.V.; Cristancho-Gordo, R.; Gonzalez-Iglesias, A.E.; Tabak, J.; Bertram, R.; Freeman, M.E. Systemic oxytocin induces a prolactin secretory rhythm via the pelvic nerve in ovariectomized rats. Am. J. Physiol. Integr. Comp. Physiol. 2011, 301, R676–R681. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, G.-K.; Zheng, Y.; Guo, H.-X.; Wang, H.-Q.; Ji, Z.-H.; Wang, T.; Yu, S.; Zhang, J.-B.; Yuan, B.; Ren, W.-Z. TRH Regulates the Synthesis and Secretion of Prolactin in Rats with Adenohypophysis through the Differential Expression of miR-126a-5p. Int. J. Mol. Sci. 2022, 23, 15914. https://doi.org/10.3390/ijms232415914
Zhao G-K, Zheng Y, Guo H-X, Wang H-Q, Ji Z-H, Wang T, Yu S, Zhang J-B, Yuan B, Ren W-Z. TRH Regulates the Synthesis and Secretion of Prolactin in Rats with Adenohypophysis through the Differential Expression of miR-126a-5p. International Journal of Molecular Sciences. 2022; 23(24):15914. https://doi.org/10.3390/ijms232415914
Chicago/Turabian StyleZhao, Guo-Kun, Yi Zheng, Hai-Xiang Guo, Hao-Qi Wang, Zhong-Hao Ji, Tian Wang, Song Yu, Jia-Bao Zhang, Bao Yuan, and Wen-Zhi Ren. 2022. "TRH Regulates the Synthesis and Secretion of Prolactin in Rats with Adenohypophysis through the Differential Expression of miR-126a-5p" International Journal of Molecular Sciences 23, no. 24: 15914. https://doi.org/10.3390/ijms232415914
APA StyleZhao, G.-K., Zheng, Y., Guo, H.-X., Wang, H.-Q., Ji, Z.-H., Wang, T., Yu, S., Zhang, J.-B., Yuan, B., & Ren, W.-Z. (2022). TRH Regulates the Synthesis and Secretion of Prolactin in Rats with Adenohypophysis through the Differential Expression of miR-126a-5p. International Journal of Molecular Sciences, 23(24), 15914. https://doi.org/10.3390/ijms232415914







