Hepatoprotective Activity of Lignin-Derived Polyphenols Dereplicated Using High-Resolution Mass Spectrometry, In Vivo Experiments, and Deep Learning
Abstract
:1. Introduction
2. Results
2.1. Overview of the Study Pipeline
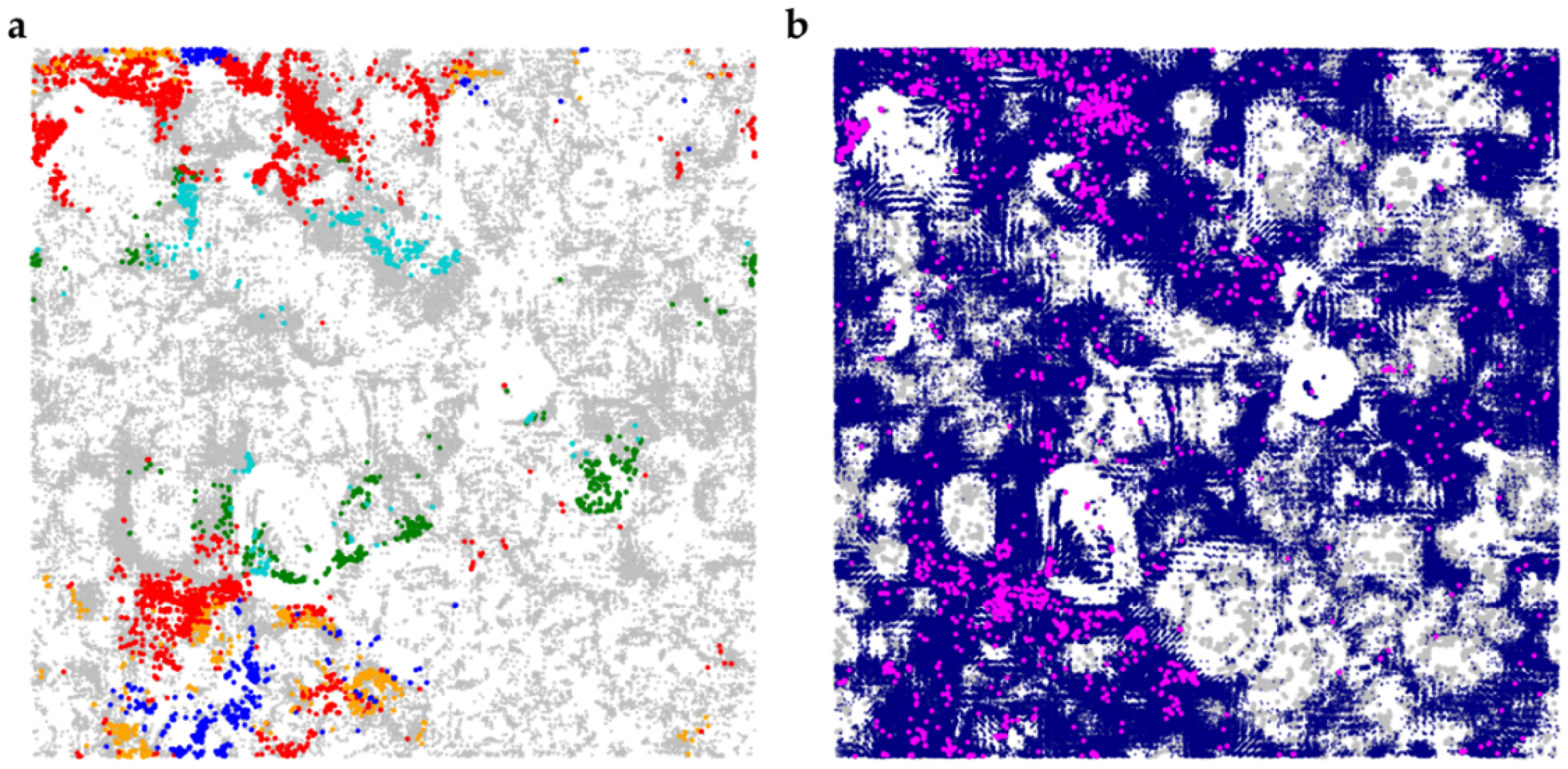
2.2. Determination of BP-Cx-1 Components in Mice Liver
2.3. Fractionation of BP-Cx-1 for Enrichment of Targeted Component
2.4. Assessment of Biological Activity of Parent and Fractionated BP-Cx-1
2.5. Examination of BP-Cx-1 Interaction with KEAP1
2.6. Generation of Structural Candidates for BP-Cx-1 Active Components Using Deep Learning
2.7. Molecular Docking of Structural Candidates to KEAP-1 Structure

3. Discussion
4. Materials and Methods
4.1. Enumeration of Functional Groups and H/D Exchanges
4.2. Fractionation of BP-Cx-1
4.3. Animal Welfare
4.4. Induction of NAFLD Accompanied by Type 2 Diabetes
4.5. Extraction of BP-Cx-1 Components from Mice Liver Tissue
4.6. BP-Cx-1-KEAP Interaction
4.7. ChEMBL Bioactivity Data Mining
4.8. Generation of Structural Candidates Using Deep Learning
4.8.1. Generation of Compounds
4.8.2. Filtering of Generated Compounds Using Labeling Data
4.8.3. Chemical Space Analysis
4.9. Molecular Docking of Structural Candidates to KEAP-1 Structure
4.9.1. Preparation of Receptor
4.9.2. Binding Site Detection
4.9.3. Preparation of Ligands
4.9.4. Molecular Docking
4.9.5. Visualization and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, X.; Zheng, L.; Wang, M.; Du, Y.; Jiang, J. Prevalence Trends in Non-Alcoholic Fatty Liver Disease at the Global, Regional and National Levels, 1990–2017: A Population-Based Observational Study. BMJ Open 2020, 10, e036663. [Google Scholar] [CrossRef] [PubMed]
- Vento, S.; Cainelli, F. Chronic Liver Diseases Must Be Reduced Worldwide: It Is Time to Act. Lancet Glob. Health 2022, 10, e471–e472. [Google Scholar] [CrossRef] [PubMed]
- Riazi, K.; Azhari, H.; Charette, J.H.; Underwood, F.E.; King, J.A.; Afshar, E.E.; Swain, M.G.; Congly, S.E.; Kaplan, G.G.; Shaheen, A.-A. The Prevalence and Incidence of NAFLD Worldwide: A Systematic Review and Meta-Analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, M. Pathophysiology of Non-Alcoholic Steatohepatitis and Basis for Treatment. Dig. Dis. 2011, 29, 243–248. [Google Scholar] [CrossRef]
- Umemura, A.; Kataoka, S.; Okuda, K.; Seko, Y.; Yamaguchi, K.; Moriguchi, M.; Okanoue, T.; Itoh, Y. Potential Therapeutic Targets and Promising Agents for Combating NAFLD. Biomedicines 2022, 10, 901. [Google Scholar] [CrossRef]
- Pydyn, N.; Miękus, K.; Jura, J.; Kotlinowski, J. New Therapeutic Strategies in Nonalcoholic Fatty Liver Disease: A Focus on Promising Drugs for Nonalcoholic Steatohepatitis. Pharmacol. Rep. 2020, 72, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Barapatre, A.; Meena, A.S.; Mekala, S.; Das, A.; Jha, H. In Vitro Evaluation of Antioxidant and Cytotoxic Activities of Lignin Fractions Extracted from Acacia Nilotica. Int. J. Biol. Macromol. 2016, 86, 443–453. [Google Scholar] [CrossRef]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The Role of Polyphenols in Human Health and Food Systems: A Mini-Review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef] [Green Version]
- Balasaheb Nimse, S.; Pal, D. Free Radicals, Natural Antioxidants, and Their Reaction Mechanisms. RSC Adv. 2015, 5, 27986–28006. [Google Scholar] [CrossRef] [Green Version]
- Forman, H.J.; Zhang, H. Targeting Oxidative Stress in Disease: Promise and Limitations of Antioxidant Therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Fedoros, E.I.; Baldueva, I.A.; Perminova, I.V.; Badun, G.A.; Chernysheva, M.G.; Grozdova, I.D.; Melik-Nubarov, N.S.; Danilova, A.B.; Nekhaeva, T.L.; Kuznetsova, A.I.; et al. Exploring Bioactivity Potential of Polyphenolic Water-Soluble Lignin Derivative. Environ. Res. 2020, 191, 110049. [Google Scholar] [CrossRef] [PubMed]
- Madrigal-Santillán, E.; Madrigal-Bujaidar, E.; Álvarez-González, I.; Sumaya-Martínez, M.T.; Gutiérrez-Salinas, J.; Bautista, M.; Morales-González, Á.; García-Luna y González-Rubio, M.; Aguilar-Faisal, J.L.; Morales-González, J.A. Review of Natural Products with Hepatoprotective Effects. World J. Gastroenterol. 2014, 20, 14787–14804. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Chen, J.; Zhang, T.; Yuan, X.; Ge, A.; Wang, S.; Xu, H.; Zeng, L.; Ge, J. Efficacy and Safety of Dietary Polyphenol Supplementation in the Treatment of Non-Alcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Front. Immunol. 2022, 13, 949746. [Google Scholar] [CrossRef]
- Abenavoli, L.; Larussa, T.; Corea, A.; Procopio, A.C.; Boccuto, L.; Dallio, M.; Federico, A.; Luzza, F. Dietary Polyphenols and Non-Alcoholic Fatty Liver Disease. Nutrients 2021, 13, 494. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication Strategies in Natural Product Research: How Many Tools and Methodologies behind the Same Concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Allard, P.-M.; Kubo, M.; Queiroz, E.F. Metabolomics Strategies for the Dereplication of Polyphenols and Other Metabolites in Complex Natural Extracts. In Recent Advances in Polyphenol Research; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 183–205. ISBN 978-1-119-42789-6. [Google Scholar]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT Online: Collection of Open Natural Products Database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef]
- Hertkorn, N.; Frommberger, M.; Witt, M.; Koch, B.P.; Schmitt-Kopplin, P.H.; Perdue, E.M. Natural Organic Matter and the Event Horizon of Mass Spectrometry. Anal. Chem. 2008, 80, 8908–8919. [Google Scholar] [CrossRef]
- Leyva, D.; Tariq, M.U.; Jaffé, R.; Saeed, F.; Lima, F.F. Unsupervised Structural Classification of Dissolved Organic Matter Based on Fragmentation Pathways. Environ. Sci. Technol. 2022, 56, 1458–1468. [Google Scholar] [CrossRef]
- Verrillo, M.; Parisi, M.; Savy, D.; Caiazzo, G.; Di Caprio, R.; Luciano, M.A.; Cacciapuoti, S.; Fabbrocini, G.; Piccolo, A. Antiflammatory Activity and Potential Dermatological Applications of Characterized Humic Acids from a Lignite and a Green Compost. Sci. Rep. 2022, 12, 2152. [Google Scholar] [CrossRef]
- Zykova, M.V.; Brazovsky, K.S.; Veretennikova, E.E.; Danilets, M.G.; Logvinova, L.A.; Romanenko, S.V.; Trofimova, E.S.; Ligacheva, A.A.; Bratishko, K.A.; Yusubov, M.S.; et al. New Artificial Network Model to Estimate Biological Activity of Peat Humic Acids. Environ. Res. 2020, 191, 109999. [Google Scholar] [CrossRef] [PubMed]
- Orlov, A.A.; Zherebker, A.; Eletskaya, A.A.; Chernikov, V.S.; Kozlovskaya, L.I.; Zhernov, Y.V.; Kostyukevich, Y.; Palyulin, V.A.; Nikolaev, E.N.; Osolodkin, D.I.; et al. Examination of Molecular Space and Feasible Structures of Bioactive Components of Humic Substances by FTICR MS Data Mining in ChEMBL Database. Sci. Rep. 2019, 9, 12066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikhnevich, T.A.; Vyatkina (Turkova), A.V.; Grigorenko, V.G.; Rubtsova, M.Y.; Rukhovich, G.D.; Letarova, M.A.; Kravtsova, D.S.; Vladimirov, S.A.; Orlov, A.A.; Nikolaev, E.N.; et al. Inhibition of Class a β-Lactamase (TEM-1) by Narrow Fractions of Humic Substances. ACS Omega 2021, 6, 23873–23883. [Google Scholar] [CrossRef]
- Badun, G.A.; Chernysheva, M.G.; Zhernov, Y.V.; Poroshina, A.S.; Smirnov, V.V.; Pigarev, S.E.; Mikhnevich, T.A.; Volkov, D.S.; Perminova, I.V.; Fedoros, E.I. A Use of Tritium-Labeled Peat Fulvic Acids and Polyphenolic Derivatives for Designing Pharmacokinetic Experiments on Mice. Biomedicines 2021, 9, 1787. [Google Scholar] [CrossRef] [PubMed]
- Fedoros, E.I.; Orlov, A.A.; Zherebker, A.; Gubareva, E.A.; Maydin, M.A.; Konstantinov, A.I.; Krasnov, K.A.; Karapetian, R.N.; Izotova, E.I.; Pigarev, S.E.; et al. Novel Water-Soluble Lignin Derivative BP-Cx-1: Identification of Components and Screening of Potential Targets in Silico and in Vitro. Oncotarget 2018, 9, 18578–18593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlak, T.W.; Saleh, M.; Higginson, D.S.; Paul, B.D.; Juluri, K.R.; Snyder, S.H. Bilirubin and Glutathione Have Complementary Antioxidant and Cytoprotective Roles. Proc. Natl. Acad. Sci. USA 2009, 106, 5171–5176. [Google Scholar] [CrossRef] [Green Version]
- Segler, M.H.S.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.-L.; Kensler, T.W.; et al. Therapeutic Targeting of the NRF2 and KEAP1 Partnership in Chronic Diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [Green Version]
- Unni, S.; Deshmukh, P.; Krishnappa, G.; Kommu, P.; Padmanabhan, B. Structural Insights into the Multiple Binding Modes of Dimethyl Fumarate (DMF) and Its Analogs to the Kelch Domain of Keap1. FEBS J. 2021, 288, 1599–1613. [Google Scholar] [CrossRef]
- Cleasby, A.; Yon, J.; Day, P.J.; Richardson, C.; Tickle, I.J.; Williams, P.A.; Callahan, J.F.; Carr, R.; Concha, N.; Kerns, J.K.; et al. Structure of the BTB Domain of Keap1 and Its Interaction with the Triterpenoid Antagonist CDDO. PLoS ONE 2014, 9, e98896. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- AlphaFold Protein Structure Database. Available online: https://alphafold.ebi.ac.uk/ (accessed on 25 October 2021).
- Ooi, B.K.; Chan, K.-G.; Goh, B.H.; Yap, W.H. The Role of Natural Products in Targeting Cardiovascular Diseases via Nrf2 Pathway: Novel Molecular Mechanisms and Therapeutic Approaches. Front Pharm. 2018, 9, 1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baird, L.; Swift, S.; Llères, D.; Dinkova-Kostova, A.T. Monitoring Keap1–Nrf2 Interactions in Single Live Cells. Biotechnol. Adv. 2014, 32, 1133–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Musso, G.; Cassader, M.; Gambino, R. Non-Alcoholic Steatohepatitis: Emerging Molecular Targets and Therapeutic Strategies. Nat. Rev. Drug Discov. 2016, 15, 249–274. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Emerging Molecular Targets for the Treatment of Nonalcoholic Fatty Liver Disease. Annu. Rev. Med. 2010, 61, 375–392. [Google Scholar] [CrossRef]
- Meyers, J.; Fabian, B.; Brown, N. De Novo Molecular Design and Generative Models. Drug Discov. Today 2021, 26, 2707–2715. [Google Scholar] [CrossRef]
- Zherebker, A.Y.A.; Rukhovich, G.D.; Kharybin, O.N.; Fedoros, E.I.; Perminova, I.V.; Nikolaev, E.N. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry for the Analysis of Molecular Composition and Batch-to-Batch Consistency of Plant-Derived Polyphenolic Ligands Developed for Biomedical Application. Rapid Commun. Mass Spectrom. 2020, 34, e8850. [Google Scholar] [CrossRef]
- Vinardell, M.P.; Mitjans, M. Lignins and Their Derivatives with Beneficial Effects on Human Health. Int. J. Mol. Sci. 2017, 18, 1219. [Google Scholar] [CrossRef] [Green Version]
- George, N.; Andersson, A.A.M.; Andersson, R.; Kamal-Eldin, A. Lignin is the Main Determinant of Total Dietary Fiber Differences between Date Fruit (Phoenix Dactylifera L.) Varieties. NFS J. 2020, 21, 16–21. [Google Scholar] [CrossRef]
- Zherebker, A.; Lechtenfeld, O.J.; Sarycheva, A.; Kostyukevich, Y.; Kharybin, O.; Fedoros, E.I.; Nikolaev, E.N. Refinement of Compound Aromaticity in Complex Organic Mixtures by Stable Isotope Label Assisted Ultrahigh-Resolution Mass Spectrometry. Anal. Chem. 2020, 92, 9032–9038. [Google Scholar] [CrossRef] [PubMed]
- Zherebker, A.; Perminova, I.V.; Kostyukevich, Y.; Kononikhin, A.S.; Kharybin, O.; Nikolaev, E. Structural Investigation of Coal Humic Substances by Selective Isotopic Exchange and High-Resolution Mass Spectrometry. Faraday Discuss. 2019, 218, 172–190. [Google Scholar] [CrossRef]
- Zherebker, A.; Shirshin, E.; Rubekina, A.; Kharybin, O.; Kononikhin, A.; Kulikova, N.A.; Zaitsev, K.V.; Roznyatovsky, V.A.; Grishin, Y.K.; Perminova, I.V.; et al. Optical Properties of Soil Dissolved Organic Matter Are Related to Acidic Functions of Its Components as Revealed by Fractionation, Selective Deuteromethylation, and Ultrahigh Resolution Mass Spectrometry. Environ. Sci. Technol. 2020, 54, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Zherebker, A.; Rukhovich, G.D.; Sarycheva, A.; Lechtenfeld, O.J.; Nikolaev, E.N. Aromaticity Index with Improved Estimation of Carboxyl Group Contribution for Biogeochemical Studies. Environ. Sci. Technol. 2022, 56, 2729–2737. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Wilson, R.D. Experimentally Induced Rodent Models of Type 2 Diabetes. In Animal Models in Diabetes Research; Joost, H.-G., Al-Hasani, H., Schürmann, A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; pp. 161–174. ISBN 978-1-62703-068-7. [Google Scholar]
- Furman, B.L. Streptozotocin-Induced Diabetic Models in Mice and Rats. Curr. Protoc. 2021, 1, e78. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [Green Version]
- ChEMBL Ftp Site. Available online: https://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_29/ (accessed on 1 September 2021).
- Mysql-Connector-Python Library. Available online: https://pypi.org/project/mysql-connector-python/ (accessed on 25 October 2021).
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Team, T.P.D. Pandas Python Library. Available online: https://pandas.pydata.org/ (accessed on 25 October 2021).
- RDKit: Open-Source Cheminformatics. Available online: http://Www.Rdkit.Org (accessed on 25 October 2021).
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.-P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for Checking the Quality of Protein Structures Solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sippl, M.J. Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An Open Source Platform for Ligand Pocket Detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular Graphics for All Devices—From Smartphones to Workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]



| Group | Total Protein, g/L | Albumin, g/L | Globulins, g/L | Amylase, U/L | AST, U/L | ALT, U/L |
|---|---|---|---|---|---|---|
| 1 | 44.4 ± 0.8 | 28.0 ± 0.47 | 16.4 ± 0.90 | 664 ± 34.7 | 196 ± 16.3 | 70 ± 5.4 |
| 2 | 60.9 ± 3.33 | 27.2 ± 1.14 | 33.7 ± 2.74 | 1250 ± 64.5 | 341 ± 10.7 | 139 ± 37.0 |
| 3 | 52.4 ± 2.37 | 25.5 ± 0.42 | 33.3 ± 5.03 | 1725 ± 343.1 | 287 ± 31.6 | 137 ± 12.1 |
| 4 | 46.8 ± 1.52 | 22.9 ± 0.53 | 24.2 ± 1.55 | 1462 ± 133.6 | 286 ± 21.7 | 164 ± 12.5 |
| Group | Total bilirubin, µmol/L | Conjugated bilirubin, µmol/L | Conjugated bilirubin, % | Cholesterol, mmol/L | Glucose, mmol/L | LDH, U/L |
| 1 | 10.9 ± 0.34 | 7.7 ± 0.37 | 71 ± 1.3 | 1.67 ± 0.016 | 3.38 ± 0.066 | 1974 ± 156 |
| 2 | 18.4 ± 0.68 | 17.2 ± 1.13 | 93 ± 2.7 | 3.54 ± 0.257 | 5.83 ± 0.561 | 2985 ± 345 |
| 3 | 19.5 ± 1.74 | 17.8 ± 0.42 | 93 ± 7.2 | 3.32 ± 0.332 | 6.30 ± 0.600 | 2835 ± 319 |
| 4 | 15.4 ± 0.67 | 13.5 ± 1.07 | 87 ± 3.3 | 2.83 ± 0.084 | 5.12 ± 0.425 | 2839 ± 207 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlov, A.; Semenov, S.; Rukhovich, G.; Sarycheva, A.; Kovaleva, O.; Semenov, A.; Ermakova, E.; Gubareva, E.; Bugrova, A.E.; Kononikhin, A.; et al. Hepatoprotective Activity of Lignin-Derived Polyphenols Dereplicated Using High-Resolution Mass Spectrometry, In Vivo Experiments, and Deep Learning. Int. J. Mol. Sci. 2022, 23, 16025. https://doi.org/10.3390/ijms232416025
Orlov A, Semenov S, Rukhovich G, Sarycheva A, Kovaleva O, Semenov A, Ermakova E, Gubareva E, Bugrova AE, Kononikhin A, et al. Hepatoprotective Activity of Lignin-Derived Polyphenols Dereplicated Using High-Resolution Mass Spectrometry, In Vivo Experiments, and Deep Learning. International Journal of Molecular Sciences. 2022; 23(24):16025. https://doi.org/10.3390/ijms232416025
Chicago/Turabian StyleOrlov, Alexey, Savva Semenov, Gleb Rukhovich, Anastasia Sarycheva, Oxana Kovaleva, Alexander Semenov, Elena Ermakova, Ekaterina Gubareva, Anna E. Bugrova, Alexey Kononikhin, and et al. 2022. "Hepatoprotective Activity of Lignin-Derived Polyphenols Dereplicated Using High-Resolution Mass Spectrometry, In Vivo Experiments, and Deep Learning" International Journal of Molecular Sciences 23, no. 24: 16025. https://doi.org/10.3390/ijms232416025
APA StyleOrlov, A., Semenov, S., Rukhovich, G., Sarycheva, A., Kovaleva, O., Semenov, A., Ermakova, E., Gubareva, E., Bugrova, A. E., Kononikhin, A., Fedoros, E. I., Nikolaev, E., & Zherebker, A. (2022). Hepatoprotective Activity of Lignin-Derived Polyphenols Dereplicated Using High-Resolution Mass Spectrometry, In Vivo Experiments, and Deep Learning. International Journal of Molecular Sciences, 23(24), 16025. https://doi.org/10.3390/ijms232416025








